Abstract
In this study, we investigated the role of a transcription factor, PU.1, in the regulation of CD80 and CD86 expression in dendritic cells (DCs). A chromatin immunoprecipitation assay revealed that PU.1 is constitutively bound to the CD80 and CD86 promoters in bone marrow–derived DCs. In addition, co-expression of PU.1 resulted in the transactivation of the CD80 and CD86 promoters in a reporter assay. The binding of PU.1 to cis-enhancing regions was confirmed by electromobility gel-shift assay. As expected, inhibition of PU.1 expression by short interfering RNA (siRNA) in bone marrow–derived DCs resulted in marked down-regulation of CD80 and CD86 expression. Moreover, overexpression of PU.1 in murine bone marrow–derived lineage-negative cells induced the expression of CD80 and CD86 in the absence of monocyte/DC-related growth factors and/or cytokines. Based on these results, we conclude that PU.1 is a critical factor for the expression of CD80 and CD86. We also found that subcutaneous injection of PU.1 siRNA or topical application of a cream-emulsified PU.1 siRNA efficiently inhibited murine contact hypersensitivity. Our results suggest that PU.1 is a potential target for the treatment of immune-related diseases.
Introduction
T-cell initiation requires 2 signals from antigen-presenting cells (APCs). The first signal comes from ligation of the T-cell receptor and the major histocompatibility complex (MHC)/antigen presented on the surface of APCs, and the second signal is via additional costimulatory molecules, including the interaction between the CD28 family on T cells and B7, eg, CD80 (B7-1) and CD86 (B7-2), expressed on the APC.1-3 CD80 and CD86 are members of the immunoglobulin supergene family encoded by separate genes, and are expressed on dendritic cells (DCs) or up-regulated on activated macrophages/monocytes, B cells, and activated T cells.1-6 CD80 or CD86 can provide costimulatory signals by engaging CD28 or CTLA4 (cytotoxic T-lymphocyte antigen 4; CD152) on T cells. The engagement of CD28 with CD80 or CD86 leads to multiple effects on the immune response, including T-cell activation and differentiation and tissue migration.7-9
In contrast to CD28, the outcome of CTLA4 engagement on T cells is to suppress proliferation by transmitting an inhibitory signal.10 In addition, a subset of T cells with potent immunoregulatory properties, CD4+CD25+ regulatory T cells, constitutively expresses CTLA4.11,12 Thus, interactions between CTLA4 and CD80 or CD86 provides an immunosuppressive function in modulating T-cell proliferation and plays a role in immune tolerance. Based on these observations, the regulated expression of costimulatory molecules is critical for immune function. Therefore, revealing the molecular mechanisms of gene expression of costimulatory molecules is essential for an understanding of the regulation of T cell–mediated immune responses.
Despite the importance of CD80 and CD86, the critical transcription factor for gene expression of CD80 or CD86 is still unknown. Transcription factors involved in stimulus-induced expression of CD80 or CD86 have been observed in some promoter structure analyses; for example, interferon regulatory factor 7 (IRF7) regulates lipopolysaccharide (LPS)–induced human CD80 transcription through the activation of Jun N-terminal kinase in monocytes,13 nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) plays a role in stimulation-induced transactivation of murine CD80 in B-cell lines,14 human CD86 in B cells,15 and CD86 in DCs.16 However, the specific transcription factors regulating the basic and constitutive expression of CD80 and CD86 have not been identified to date.
PU.1 is a transcription factor belonging to the Ets family, which is involved in hematopoietic cell development. Although PU.1 is required for the development of thymic and myeloid DCs, including some specific gene expression by DCs in the analysis of PU.1−/− animals,17,18 the effects of PU.1 on the expression of CD80 and CD86 have not been verified in those reports. In our previous studies, mast cells and their progenitors overexpressing PU.1 acquired some DC-specific gene expression, suggesting that PU.1 is a master regulator for gene expression of DCs.19-21 These findings prompted us to analyze the role of PU.1 in the expression of CD80 and CD86 in DCs. In the present study, we show that PU.1 plays a key role in the gene expression of CD80 and CD86 in granulocyte-macrophage colony-stimulating factor (GM-CSF)–cultured, bone marrow–derived DCs (BMDCs).
Methods
Cells
BMDCs were generated from the femoral and tibial bone marrow cells of female BALB/c mice (Japan SLC) as described previously.22 All animal experiments were performed according to the approved manual of the institutional review board of Juntendo University School of Medicine, Japan. Cells were incubated in RPMI 1640 (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal calf serum (JRH Biosciences), 100 U/mL of penicillin, 100 μg/mL of streptomycin, 100μM 2-mercaptoethanol, 10μM Minimum Essential Medium nonessential amino acid solution (Invitrogen), and 20 ng/mL of murine GM-CSF (Wako Pure Chemical Industries) at 37°C in a humidified atmosphere in the presence of 5% CO2. The purity of CD11c+ BMDCs isolated by the MACS separation system with mouse CD11c MicroBeads and an autoMACS (all Miltenyi Biotech) was confirmed to be over 98% by flow cytometry. Lineage-negative (Lin−) cells were prepared from the femoral and tibial bone marrow cells of female BALB/c mice by the MACS separation system according to the manufacturer's instructions. Briefly, bone-marrow cells were treated with biotinylated monoclonal antibodies (mAbs) against mouse CD5, CD45R (B220), CD11b, Ly-6G (Gr-1), 7-4, and Ter-119. The cell suspension treated with biotinylated mAbs was then reacted with MicroBeads conjugated with anti–biotin mAbs (clone: Bio3-18E7.2; mouse IgG) and was applied to an autoMACS. The purity of the isolated Lin− cells, which were collected as the negative fraction using this MACS separation system, was over than 95% in FACS analysis. The simian kidney cell line CV-1 was purchased from the RIKEN Cell Bank. CV-1 and the mouse mono-macrophage cell lines RAW264.7 and J774 were maintained in Dulbecco modified Eagle medium (Sigma-Aldrich) with 10% heat-inactivated fetal calf serum, 100 U/mL of penicillin, and 100 μg/mL of streptomycin.
LPS and interferonγ stimulation
Cells were stimulated in the presence or absence of 1 μg/mL of LPS (Sigma) and/or 100 ng/mL of interferonγ (IFNγ; PeproTech).
Flow cytometric analysis
Phycoerythrin (PE)–conjugated anti–mouse antibodies against CD80, CD86, I-Ad, CD11b, and CD11c and fluorescein isothiocyanate (FITC)–conjugated anti–mouse antibodies against CD86, I-Ad, CD11b, CD11c (all of which were purchased from BD Pharmingen); PE-conjugated anti–CD205 (from Miltenyi Biotech); PE-conjugated anti–CD207 and anti–CD209 (eBioscience); FITC-conjugated anti–Toll-like receptor 2 (anti–TLR2) and PE-conjugated anti–TLR4 (BioLegend) were used to stain each cell-surface molecule by incubation for 0.5-1 hour at 4°C after blocking Fc receptors with 2.4G2 (BD Pharmingen). After washing with phosphate-buffered saline, cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences).
5′-rapid amplification of cDNA ends procedure
Total RNA was prepared from BALB/c BMDCs and was used for 5′-rapid amplification of cDNA ends (5′-RACE) with a GeneRacer kit (Invitrogen) according to the manufacturer's instructions. Briefly, total RNA was treated with calf intestinal phosphatase to remove the 5′ phosphates, and the product was then treated with tobacco acid pyrophosphatase to remove the 5′ cap structure. The decapped mRNA was ligated with the GeneRacer RNA oligo and reverse-transcribed using random primers. To obtain 5′ ends, the first-strand cDNA was amplified using the following primers: forward primer, GeneRacer 5′ primer (Invitrogen); reverse primers, CD80-R1 (5′-TCCACCCGGCAGATGCTA-3′), CD80-R3 (5′-TCCACCCGGCAGATGCTA-3′), and CD86-R1 (5′-TCCACCCGGCAGATGCTA-3′). To confirm that these polymerase chain reaction (PCR) products were indeed the expected products, nested PCR was performed with the following primers: forward primer, GeneRacer 5′ Nested primer (Invitrogen); reverse primers, CD80-R1-nest (5′-TCCACCCGGCAGATGCTA-3′), CD80-R2 (5′-TCCACCCGGCAGATGCTA-3′), and CD86-R1-nest (5′-TCCACCCGGCAGATGCTA-3′). TOPO (Invitrogen) cloning was the performed with PCR products, and the products were sequenced.
Quantitative reverse-transcriptase PCR
Total RNA was isolated from each cell or ear tissue with the RNeasy Mini Kit or the RNeasy Lipid Tissue Mini Kit (QIAGEN) per the supplier's instructions, and was used as a template for reverse transcription. Reverse transcription to synthesize cDNA for CD80-1, CD80-2, CD80-common, CD86-1, CD86-2, CD86-common, and β-actin was performed using high-capacity cDNA reverse-transcription kits (Applied Biosystems) or the ReverTra Ace qPCR RT kit (TOYOBO). Quantitative PCR was performed using TaqMan Universal PCR Master Mix (Applied Biosystems) and a 7500 real-time PCR system with TaqMan gene expression assays of mouse target genes,23 including CD80 (Mm00711660-m1), CD86 (Mm00444543-m1), and an endogenous control (β-actin), and the primers and TaqMan probes listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The expression levels of CD80-1, CD80-2, CD80-common, CD86-1, CD86-2, and CD86-common are given relative to those of β-actin by calculation of the cycle threshold values in amplification plots with 7500 SDS software Version 1.2.2 (Applied Biosystems), as recommended by the supplier.
Chromatin immunoprecipitation assay
A chromatin immunoprecipitation (ChIP) assay was performed using a kit (Upstate Biotechnology) as described previously.24 Anti–PU.1 goat IgG antibody (D19; Santa Cruz Biotechnologies), anti–NF-kB rabbit IgG (C-20; Santa Cruz Biotechnologies), anti–IRF4 goat IgG antibody (M17; Santa Cruz Biotechnologies), rabbit IgG (Sigma), and goat IgG (Sigma) were used. The amount of chromosomal DNA immunoprecipitated by each antibody was determined by quantitative PCR. Primers and TaqMan probe sequences used for this analysis are as listed in supplemental Table 2. The amount of target DNA bound with PU.1, NF-κB p65, or IRF4 was quantified from the cycle threshold value, which was determined with 7500 SDS software (Applied Biosystems). Briefly, the ratio of a specific DNA fragment in each immunoprecipitate to that fragment in the DNA before immunoprecipitation (input DNA) was calculated from each cycle threshold value.
Plasmid construction
A series of murine CD80 or CD86 promoter fragments were amplified from murine genomic DNA purified from BALB/c mice by PCR. Primers with restriction endonuclease recognition sequences are synthesized as listed in supplemental Table 3. Amplified PCR products were subcloned into the pCR4-TOPO vector, and the nucleotide sequences were confirmed. The correct insertions were subcloned into the XhoI/HindIII- or XhoI/BglII-digested pGL4.10 (Promega), the basic luciferase reporter plasmid, to generate 4 reporter plasmids, pGL4.10-CD80-1, CD80-2, CD86-1, and CD86-2. A series of reporter plasmids with various lengths of each promoter were generated by introduction of XhoI recognition sequences by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene). The 5′-region of XhoI-sequence-introduced each promoter was removed by XhoI digestion and subsequent self-ligation, resulting in the generation of 5′-deleted promoter plasmid.
To generate the IRF4 expression plasmid pCR-IRF4, mouse IRF4 cDNA (NM_013674) amplified by PCR using 5′-GCACGCGTCATGAACTTGGAGAC-3′ and 5′-CGGCTGCCCTGTCAGAGTATTTC-3′ as primers and cDNA synthesized from total RNA of RAW264.7 cells as a template, respectively, was cloned into pCR3.1 (Invitrogen). Other expression plasmids, pCR-PU.125 and pCR3.1-IRF8,26 have been generated in our previous studies. The cDNA encoding the Ets domain (DNA-binding domain) of PU.1 was amplified from pCR-PU.1 by PCR using 5′-GGGGAGGAATTCAGCAAGAAGAAGATTCGT-3′, and 5′-GTCTCGAGCTAAGAACTGAGTGGAGACAC-3′ (EcoRI and XhoI recognition sequences are italic) as primers, and PU.1 cDNA of pMX-puro-PU.119 was replaced with the PU.1 Ets domain by EcoRI/XhoI digestion and ligation, resulting in pMX-puro-Ets for retrovirus vector expressing PU.1 Ets-domain fused with the Flag-tag at the N-terminus (Figure 5B). Mouse interleukin 12 (IL12) p40 promoter, −315/+14, was amplified from Balb/c genomic DNA by PCR using 5′-GGAGATCTTAGCCATTGCCGCCTCTATTCACC-3′ and 5′-GCAAGCTTCCTGGTCTGATGTGCCTGGACACC-3′ (BglII and HindIII recognition sequences are italic) as primers, and inserted into BglII/HindIII site of pGL3-Basic (Promega).
Luciferase assay
CV-1 or J774 cells (1 × 106) were transfected with 2 μg of reporter plasmid (pGL4.10-CD80-1, CD80-2, CD86-1, CD86-2, and their deletion variants), 2 μg of expression plasmid (pCR-PU.1, pCR-IRF4, pCR-IRF8,23 pCR-3.1) and 2 ng of pRL-CMV (Promega) or 125 ng of pRL-null (Promega) as an internal control of transfection efficiency by FuGene6 (Roche Diagnostics) were used in accordance with the manufacturer's instructions. Measurement of luciferase activity in cells harvested after 24 hours of incubation and calculation of relative luciferase activity were performed as described previously.24
Electromobility gel-shift assay
The electromobility gel-shift assay (EMSA) was performed based on a method described previously.27 Synthesized oligonucleotides of the target sequence and its complementary sequence, both of which were labeled with FITC at the 5′-end, were annealed to prepare the probe of double-stranded DNA. PU.1, IRF4, and IRF8 proteins were prepared using a TnT T7 quick-coupled transcription/translation system (Promega) using pCR-PU.1, pCR-IRF4, and pCR-IRF8 as templated, respectively. Nuclear proteins were extracted from RAW264.7 transfected with pCR-PU.1 using Nucleofector II (Amaxa) as described previously.24,25 The band shift on polyacrylamide gel was analyzed with a fluorescence detector (Fluor Imager 595; GE Healthcare).
Introduction of PU.1 cDNA in Lin− cells by retrovirus vector
Retroviral infection of Lin− cells was performed according to a previously reported method.19,20 Briefly, pMX-IG (mock vector)28 and pMX-IG-PU.121 were transiently introduced into Plat-E29 with Fugene6 (Roche Diagnostics). Lin− cells were incubated with the harvested culture medium of transfected Plat-E containing infectious viruses for 3 days in the presence of: 10 ng/mL of recombinant murine IL3, 10 ng/mL of recombinant murine IL6, 10 ng/mL of recombinant murine SCF, 10 ng/mL of recombinant murine thrombopoietin, 10 ng/mL of recombinant murine GM-CSF, 10 ng/mL of recombinant murine G-CSF, 10 ng/mL of recombinant murine M-CSF, and 10 ng/mL of recombinant murine Flt3L (all from Wako Pure Chemical Industries); 10 ng/mL of recombinant murine erythropoietin (R&D Systems); and 10 μg/mL of polybrene (Sigma-Aldrich). Transfectants were selected as green fluorescent protein (GFP)–positive cells. When pMX-puro-series were used for retrovirus transfection, transfectants were concentrated by puromycin selection.19,21 Western blotting was performed as described previously.21,30
Short interfering RNA–mediated inhibition of PU.1 expression
Five microliters of 20μM double-stranded stealth short interfering RNA (siRNA) of PU.1, CD80, or CD86 (Invitrogen: PU.1, Sfpi1-Mss247676; CD80, Cd80-Mss202820; CD86, Cd86-Mss202829) was introduced into 1.0 × 106 BMDCs with Nucleofector II set at program Y-001 using a mouse macrophage nucleofector kit (Amaxa) according to the manufacturer's instructions. Stealth RNAi-negative control medium GC Duplex (Invitrogen, 12 935-300) was used as a nonsilencing control. Messenger RNA levels for PU.1, CD80, and CD86 in transfected BMDCs at 24 or 48 hours of incubation after siRNA introduction were analyzed by quantitative real-time PCR, and cell-surface CD80 and CD86 were measured by FACS. Other DC characters, including cell-surface expression of DC-related molecules, T cell–stimulation activity,21 and cytokine production following LPS stimulation,21,23 were also compared between control DCs and PU.1–knocked-down DCs.
Induction of contact hypersensitivity and treatment with siRNA
For induction of contact hypersensitivity (CHS) to trinitrochlorobenzene (TNCB, picryl chloride; Nacalai Tesque), 100 μL of 1% TNCB dissolved in acetone was painted onto the shaved abdominal skin of mice on day 0. On day 7, 20 μL of 0.2% TNCB dissolved in acetone:olive oil = 4:1 was applied to the right ear for challenge. Ear thickness was measured before and at 24 and 48 hours after challenge. For application of siRNA, 5.6 μL of a cream-based ointment (baby lotion, no fragrance; Johnson & Johnson) mixed with 9.4 μL of 20μM siRNA (approximately 0.2 nmol) was painted onto the right ear skin 24 hours before challenge. For injection of siRNA, mice were subcutaneously injected in the right ear with 10 μL of 20μM siRNA (0.2 nmol) at 24 hours before challenge.
Statistical analysis
Unpaired Student t test was used. P <.05 was regarded as statistically significant.
Results
Transcription start sites of the murine CD80 and CD86 genes in BMDCs
Although there have been various reports regarding the structure of the murine CD80 and CD86 genes and/or promoters in B-cell lymphoma and spleen cells, the transcription start sites in DCs have not yet been verified.31-35 Therefore, we determined the transcription start sites of murine CD80 and CD86 genes in BMDCs because studies have reported that the BMDCs generated with GM-CSF constitutively express CD80 and CD86.4,22
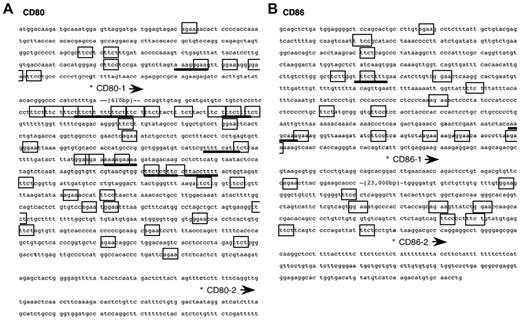
We initially confirmed that CD80 and CD86 are constitutively expressed by murine BMDCs using FACS (supplemental Figure 1). We then performed 5′-RACE with mRNA from murine BMDCs using the gene-specific primers for the nucleotide sequences of CD80 and CD86 cDNAs to determine the transcription start sites in DCs (supplemental Figure 2). Clones of the 2 different CD80 transcripts and of the 2 different CD86 transcripts were thus obtained. This suggests that both the CD80 and CD86 genes possess at least 2 major promoters in BMDCs. The nucleotide sequences around the promoter regions are shown in Figure 1. As a matter of convenience, we use the terms “CD80-1,” “CD80-2,” “CD86-1,” and “CD86-2” for each transcript, as shown in Figure 1. Use of the identified promoters for expression of CD80 and CD86 was confirmed by quantitative real-time PCR using primers and probes designed by referring to the determined sequences of CD80-1, CD80-2, CD86-1, and CD86-2 mRNAs (supplemental Figures 2-3).
Nucleotide sequence of the mouse CD80 (A) and CD86 (B) promoters. The position of the transcription start site is marked by (*). In addition to the consensus Ets-family protein binding sequence (GGAA; boxed), AGAA25 (boxed) is also bound with PU.1. The IRF recognition sequence (AANNGAAA)–like is underlined. PU.1 binds genomic DNA via GGAA and AGAA as a monomer and GGAANNGAAA as a heterodimer with IRF4 or IRF8.54
Nucleotide sequence of the mouse CD80 (A) and CD86 (B) promoters. The position of the transcription start site is marked by (*). In addition to the consensus Ets-family protein binding sequence (GGAA; boxed), AGAA25 (boxed) is also bound with PU.1. The IRF recognition sequence (AANNGAAA)–like is underlined. PU.1 binds genomic DNA via GGAA and AGAA as a monomer and GGAANNGAAA as a heterodimer with IRF4 or IRF8.54
PU.1 binds to CD80 and CD86 promoters
To analyze the binding of transcription factors on the CD80 and CD86 promoters, we performed a quantitative ChIP assay using BMDCs in the presence or absence of LPS stimulation, which induces DC maturation and up-regulation of CD80 and CD86 and/or IFNγ stimulation, which up-regulates CD86 expression on DCs (supplemental Figures 1-3).4 For immunoprecipitation, we used antibodies for the transcription factors PU.1, NF-κB p65 (Rel A), IRF4, and IRF8 for the following reasons.
It has been reported that NF-κB is involved in the stimulation-induced transactivation of CD80 in B cells.14 In studies of IRF4−/− mice, GM-CSF–cultured BMDCs from IRF4−/− mice lacked CD80 expression36 or exhibited a reduced capacity for LPS-induced up-regulation of CD80 and CD86.37 Although IRF8 is reported to be involved in the development of plasmacytoid DCs and CD8α+ DCs,38-40 the requirement of IRF8 for CD80 and CD86 expression by GM-CSF–mediated DCs is not as striking as that of IRF4.36,41,42 Moreover, PU.1, which functions alone or after forming a heterodimer with IRF4 or IRF8, is necessary for the development of DCs,17,18,43 although the involvement of PU.1 in the expression of CD80 and CD86 is unknown.
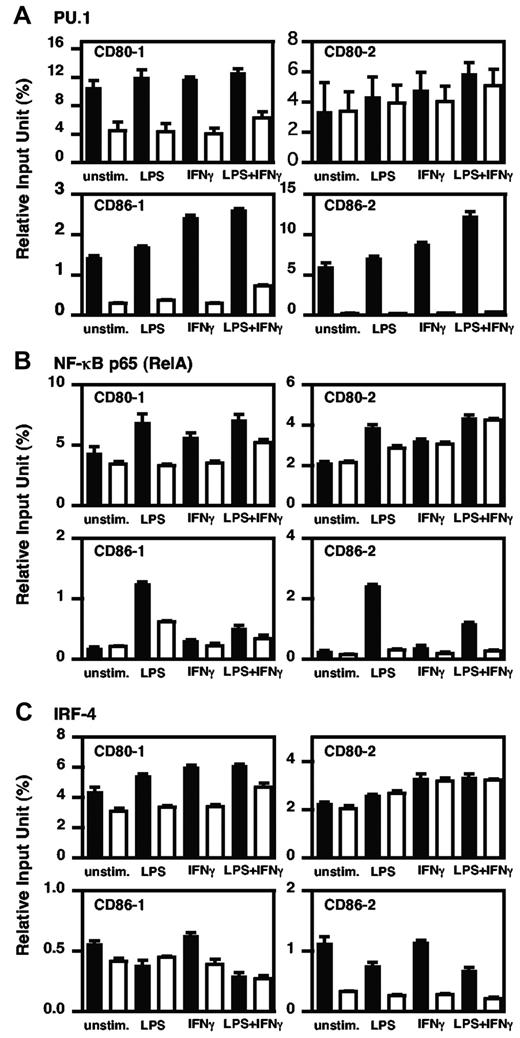
In the ChIP assay, a larger amount of chromatin containing the promoter regions of CD80-1, CD86-1, and CD86-2 was immunoprecipitated by anti–PU.1 antibody compared with each control antibody in BMDCs with or without stimulations (Figure 2A). In contrast, when performed with the anti–NF-κB antibody, the amount of immunoprecipitated chromatin containing each promoter region was increased by LPS stimulation, whereas there was little or no difference between specific antibody and control antibody in unstimulated or IFNγ-stimulated BMDCs (Figure 2B). Using the anti–IRF4 antibody, the apparent amount of CD80-1 was immunoprecipitated under each condition (Figure 2C top left), and the amounts of both CD86 promoters (CD86-1 and CD86-2) bound with IRF4 decreased with LPS stimulation (Figure 2C bottom). ChIP assay with anti–IRF8 antibody showed a similar profile as IRF4 (data not shown). These results indicate that PU.1 is involved in the basic expression of CD80 and CD86 through binding via the upper promoter and both promoters, respectively.
In vivo binding of PU.1, NF-κB p65, and IRF4 to the CD80 or CD86 promoter in BMDCs. Quantitative analysis of PU.1 (A), NF-κB p65 (B), and IRF4 (C) binding to the CD80 or CD86 promoter region was performed by ChIP assay using real-time PCR. The results are expressed as means + SD for 3 PCRs with duplicates using murine BMDCs treated with IFNγ (100 ng/mL) for 24 hours and/or following stimulation by LPS (1 μg/mL) for 1 hour or without treatment. Closed bars, specific antibody; open bars, control antibody. Similar results were obtained in 2 additional experiments.
In vivo binding of PU.1, NF-κB p65, and IRF4 to the CD80 or CD86 promoter in BMDCs. Quantitative analysis of PU.1 (A), NF-κB p65 (B), and IRF4 (C) binding to the CD80 or CD86 promoter region was performed by ChIP assay using real-time PCR. The results are expressed as means + SD for 3 PCRs with duplicates using murine BMDCs treated with IFNγ (100 ng/mL) for 24 hours and/or following stimulation by LPS (1 μg/mL) for 1 hour or without treatment. Closed bars, specific antibody; open bars, control antibody. Similar results were obtained in 2 additional experiments.
PU.1 transactivates the CD80-1, CD86-1, and CD86-2 promoters
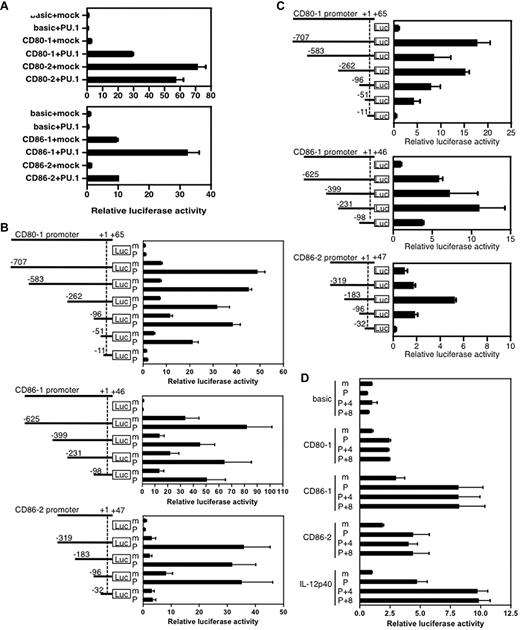
To examine the effects of PU.1 on the promoter activity of CD80 or CD86, we used a reporter assay with the CV-1 cells, which were previously shown to be useful in co-expression studies of hematopoietic cell–specific transcription factors.24 The luciferase activity derived from CV-1 cells transfected with pGL4.10-CD80-1, pGL4.10-CD86-1, or pGL4.10-CD86-2 was significantly increased by the expression of exogenous PU.1 (Figure 3A). In contrast, PU.1 did not activate the CD80-2 promoter. These results were consistent with those of ChIP assay, thus demonstrating that PU.1 binds to the CD80-1, CD86-1, and CD86-2 promoters, but not to the CD80-2 promoter.
Transcriptional activity of the CD80 or CD86 promoter. (A) Effects of PU.1 on each promoter. (B) Reporter assay with deletion constructs to identify PU.1-responsive elements. CV-1 cells were transfected with 2 μg of reporter plasmid, 2 μg of expression plasmid, and 2 ng of pRL-CMV (Promega) as an internal control of transfection efficiency. After 24 hours of culture, cells were harvested, luciferase activity was measured, and relative luciferase activity was calculated as described previously.24 m, co-expression of mock (pCR-3.1); P, co-expression of PU.1 (pCR-PU.1). (C) Transcriptional activity of the CD80 or CD86 promoter in hematopoietic cells. J774 cells were transfected with 2 μg of reporter plasmid and 125 ng of pRL-null. (D) Effects of IRF4/8 on transactivation activity of PU.1. CV-1 cells were transfected with 2 μg of reporter plasmid, a total of 2μg of expression plasmid (adjusted with mock vector as in our previous study25 ), and 125 ng of pRL-null. CD80-1 (−51/+65), CD86-1 (−98/+46), CD86-2 (−96/+47), and IL12 p40 (−315/+14) were used. m, pCR-3.1; P, pCR-PU.1; 4, pCR-IRF4; 8, pCR-IRF8. Relative luciferase activity is represented as the ratio of activity to that of pGL4-Basic (C) with pCR3.1-mock (A, B, D). Data represent the mean + SD of triplicate samples. A representative result of 3 independent experiments is shown.
Transcriptional activity of the CD80 or CD86 promoter. (A) Effects of PU.1 on each promoter. (B) Reporter assay with deletion constructs to identify PU.1-responsive elements. CV-1 cells were transfected with 2 μg of reporter plasmid, 2 μg of expression plasmid, and 2 ng of pRL-CMV (Promega) as an internal control of transfection efficiency. After 24 hours of culture, cells were harvested, luciferase activity was measured, and relative luciferase activity was calculated as described previously.24 m, co-expression of mock (pCR-3.1); P, co-expression of PU.1 (pCR-PU.1). (C) Transcriptional activity of the CD80 or CD86 promoter in hematopoietic cells. J774 cells were transfected with 2 μg of reporter plasmid and 125 ng of pRL-null. (D) Effects of IRF4/8 on transactivation activity of PU.1. CV-1 cells were transfected with 2 μg of reporter plasmid, a total of 2μg of expression plasmid (adjusted with mock vector as in our previous study25 ), and 125 ng of pRL-null. CD80-1 (−51/+65), CD86-1 (−98/+46), CD86-2 (−96/+47), and IL12 p40 (−315/+14) were used. m, pCR-3.1; P, pCR-PU.1; 4, pCR-IRF4; 8, pCR-IRF8. Relative luciferase activity is represented as the ratio of activity to that of pGL4-Basic (C) with pCR3.1-mock (A, B, D). Data represent the mean + SD of triplicate samples. A representative result of 3 independent experiments is shown.
To further identify the critical regions for transactivation by PU.1, a series of reporter plasmids with various 5′-deleted promoters were used for co-expression reporter assay (Figure 3B). When the CD80-1 promoter region was deleted to −11/+65, the exogenous PU.1 did not affect promoter activity, whereas other longer promoters were significantly transactivated by PU.1 (Figure 3B top), indicating the presence of a region critical for response to PU.1 at between −51 and −12. Similarly, the PU.1-responsive region was identified as between −96 and −33 on the CD86-2 promoter (Figure 3B bottom) and between −98 and +1 on the CD86-1 promoter (Figure 3B middle). When J774, a mouse monocytic cell line, was used for the reporter assay as a more relevant hematopoietic host cell, the regions of CD80-1 (−51/+65), CD86-1 (−98/+46), and CD86-2 (−96/+47) exhibited promoter activity (Figure 3C), suggesting that these regions possess the cis elements for minimum promoter, which are essential for transactivation of the minimum promoters by endogenous transcription factors. To evaluate the effect of IRF4 and IRF8 on PU.1-mediated transactivation of these promoters, reporter analysis was performed with expression plasmid for IRF4 or IRF8 in addition to the PU.1 expression plasmid. As shown in Figure 3D, additional expression of IRF4/8 did not affect the PU.1-mediated activation of the CD80 and CD86 promoters, whereas the IL12 p40 promoter44 carrying the IRF motif was further activated by IRF4/8. Therefore, the IRF-like motifs between −38 and −31 of CD80-1 and between −90 and −83 of CD86-1 (Figure 1) may not be functional.
Direct binding of PU.1 to cis-enhancing elements in the CD80-1, CD86-1, and CD86-2 promoters
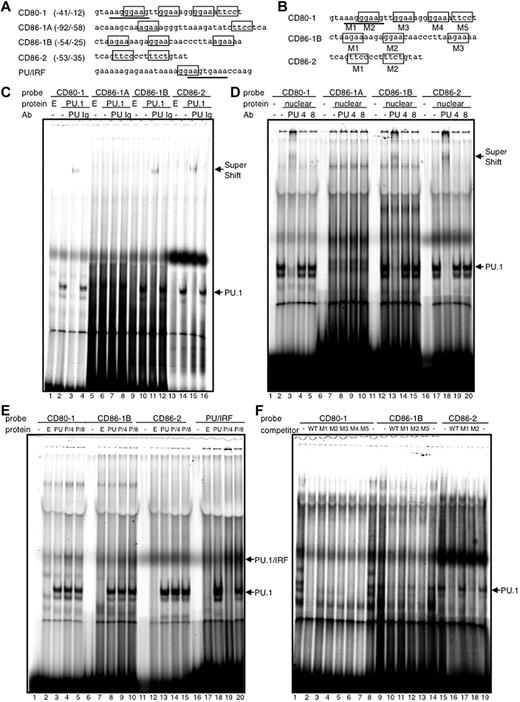
EMSA was performed to examine the direct binding of PU.1 to these identified regions in each promoter. Several PU.1-recognizable sequences were found in the regions limited by the reporter assay (Figure 4A). When PU.1 protein or nuclear extract of a mouse mono-macrophage cell line was added to the probe of CD80-1 (−41/−12), a specific shifted band appeared (Figure 4C-D lane 2), which was further super-shifted in the presence of anti–PU.1 antibody (lane 3), indicating that PU.1 directly binds to one or more of the possible sequences. EMSA also showed that the PU.1-binding sequences are located between −54 and −25 of the CD86-1 promoter and between −53 and −35 of the CD86-2 promoter (Figure 4C-D). Binding of the PU.1/IRF-complex to probes of CD80-1, CD86-1B, and CD86-2 was not observed, whereas the PU/IRF probe26 formed a ternary complex with PU.1/IRF4 or IRF8, accompanied by the disappearance or decrease of the probe/PU.1 complex, when IRF4 or IRF8 was added to the EMSA mixture (Figure 4E). In the competition assay with mutant oligonucleotides, M2 of CD86-1B and M1 of CD86-2 did not completely inhibit the binding between each probe and PU.1 (Figure 4F lanes 12 and 17), suggesting that PU.1 binds CD86-1B and CD86-2 via mainly GGAA at −43/−40 and TTCC at −49/−46, respectively. Complete inhibition of binding between CD80-1 and PU.1 by all mutant competitors (Figure 4F lanes 3-7) suggests that PU.1 binds CD80-1 via multiple Ets sites in −41/−12.
Direct binding of PU.1 to cis-enhancing sequences in the CD80 and CD86 promoters. EMSA with each probe and in vitro–translated PU.1 protein. Common Ets-motif (GGAA) and PU.1-recognizable sequence (AGAA) in the probes are boxed (top). PU/IRF probe is bound with PU.1/IRF4 or IRF8 heterodimer via overlapping Ets (GGAA; boxed) and IRF (AANNGAAA; underlined) sequence.26 Protein, in vitro transcription/translation reacted mixture using empty vector pCR3.1 (E), pCR-PU.1 (PU.1, PU, or P), pCR-IRF (4), pCR-IRF8 (8), or nuclear extract prepared from PU.1-overexpressing RAW264.7 (nuclear). antibody, anti–PU.1 goat IgG antibody (PU), anti–IRF4 goat IgG antibody (4), anti–IRF8 goat IgG antibody (8), or control goat IgG (Ig). CD80-1 M1-5, CD86-1B M1-3, and CD86-2 M1 and M2 (F) are competitive oligonucleotides with mutant sequences at each indicated site (B). WT is each self competitor with a wild-type sequence (F).
Direct binding of PU.1 to cis-enhancing sequences in the CD80 and CD86 promoters. EMSA with each probe and in vitro–translated PU.1 protein. Common Ets-motif (GGAA) and PU.1-recognizable sequence (AGAA) in the probes are boxed (top). PU/IRF probe is bound with PU.1/IRF4 or IRF8 heterodimer via overlapping Ets (GGAA; boxed) and IRF (AANNGAAA; underlined) sequence.26 Protein, in vitro transcription/translation reacted mixture using empty vector pCR3.1 (E), pCR-PU.1 (PU.1, PU, or P), pCR-IRF (4), pCR-IRF8 (8), or nuclear extract prepared from PU.1-overexpressing RAW264.7 (nuclear). antibody, anti–PU.1 goat IgG antibody (PU), anti–IRF4 goat IgG antibody (4), anti–IRF8 goat IgG antibody (8), or control goat IgG (Ig). CD80-1 M1-5, CD86-1B M1-3, and CD86-2 M1 and M2 (F) are competitive oligonucleotides with mutant sequences at each indicated site (B). WT is each self competitor with a wild-type sequence (F).
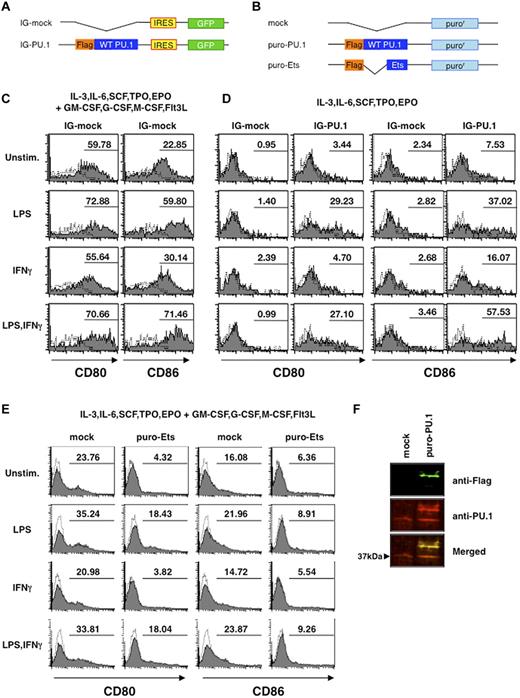
Overexpression of PU.1 induced the expression of CD80 and CD86
We then evaluated the effects of PU.1 on the expression of the CD80 and CD86 molecules by transfecting Lin− cells with a retrovirus vector directing production of GFP with wild-type PU.1 or with an empty vector (mock) in medium supplemented with a cytokine cocktail (IL3, IL6, SCF, TPO, EPO, GM-CSF, G-CSF, M-CSF, and Flt3L) that can result in multi-lineage development. The cytokine cocktail induced high levels of CD80 and CD86 expression, even in mock transfectants (Figure 5C), and no significant differences were seen between PU.1 and mock transfectants (supplemental Figure 4). We then repeated this experiment using a cytokine cocktail without GM-CSF, G-CSF, M-CSF, and Flt3L, which potentially induce the development of the DC/macrophage lineage. As expected, overexpression of PU.1 in Lin− cells in the presence of cytokine cocktail without GM-CSF, G-CSF, M-CSF, and Flt3L induced the expression of CD80 and CD86 on the cell surface, whereas mock transfectants did not exhibit such expression (Figure 5D). Moreover, when stimulated with LPS, both CD80 and CD86 were expressed at much higher levels in PU.1 transfectants than in mock transfectants. The expression levels of CD86 with stimulation by LPS and IFNγ were comparable to those in the presence of the cytokine cocktail containing GM-CSF, G-CSF, M-CSF, and Flt3L. To further clarify the effect of PU.1, the Ets domain of PU.1 was overexpressed in Lin− as a dominant-negative form of PU.1 (Figure 5B). After 10 days of culture for puromycin selection, the population of CD80- or CD86-positive cells was markedly reduced by overexpression of the Ets domain (Figure 5E). This result suggests that the Ets domain suppresses the CD80 and CD86 expression induced by endogenous PU.1. Western blotting was performed to compare endogenous and exogenous PU.1 protein levels (Figure 5F). Although endogenous PU.1 expression was down-regulated by maintenance in cytokine cocktail without GM/G/M-CSF and Flt3L for 10 days, when PU.1 was exogenously produced, the exogenous protein (which was found in a much higher amount than that of the endogenous PU.1 in the mock transfectants) induced higher endogenous expression. These data indicate that PU.1 plays a crucial role in the regulation and expression of CD80 and CD86.
Effects of enforced expression of PU.1 on cell-surface expression of CD80 and CD86. (A) Schematic drawing of plasmids pMX-IG (IG-mock) and pMX-IG-PU.1 (IG-PU.1) used in Figures 5C-D and 7D). (B) Schematic drawing of plasmids pMX-puro (mock), pMX-puro-PU.1 (puro-PU.1), and pMX-puro-Ets (puro-Ets) used in Figure 5E and F. Lin− cells were transfected with the pMX-IRES/GFP series and transfectants were monitored as GFP-positive cells. Cells were incubated with infectious viruses for 3 days in the presence of IL3, IL6, SCF, TPO, and EPO with (C) or without (D) GM-CSF, G-CSF, M-CSF, and Flt3L. Three days after infection, cells were stimulated with or without 1 μg/mL of LPS and/or 100 ng/mL of IFNγ for 24 hours, followed by staining with PE-labeled mAbs, and transfectants were monitored as GFP-positive cells (C-D). Lin− cells transfected with the pMX-puro series were incubated with the cytokine mixture as in panel C (for E) or panel D (for F). To concentrate transfectants, cells were maintained for 10 days in the presence of puromycin (2 μg/mL). Solid line histogram represents cells with each antibody. Dotted line histogram represents negative control with 2.4G2 alone. Representative results are shown. (F) Western blotting to compare endogenous and exogenous PU.1 protein levels in retrovirus transfectants. Green, anti–Flag antibody; red, anti–PU.1 antibody.
Effects of enforced expression of PU.1 on cell-surface expression of CD80 and CD86. (A) Schematic drawing of plasmids pMX-IG (IG-mock) and pMX-IG-PU.1 (IG-PU.1) used in Figures 5C-D and 7D). (B) Schematic drawing of plasmids pMX-puro (mock), pMX-puro-PU.1 (puro-PU.1), and pMX-puro-Ets (puro-Ets) used in Figure 5E and F. Lin− cells were transfected with the pMX-IRES/GFP series and transfectants were monitored as GFP-positive cells. Cells were incubated with infectious viruses for 3 days in the presence of IL3, IL6, SCF, TPO, and EPO with (C) or without (D) GM-CSF, G-CSF, M-CSF, and Flt3L. Three days after infection, cells were stimulated with or without 1 μg/mL of LPS and/or 100 ng/mL of IFNγ for 24 hours, followed by staining with PE-labeled mAbs, and transfectants were monitored as GFP-positive cells (C-D). Lin− cells transfected with the pMX-puro series were incubated with the cytokine mixture as in panel C (for E) or panel D (for F). To concentrate transfectants, cells were maintained for 10 days in the presence of puromycin (2 μg/mL). Solid line histogram represents cells with each antibody. Dotted line histogram represents negative control with 2.4G2 alone. Representative results are shown. (F) Western blotting to compare endogenous and exogenous PU.1 protein levels in retrovirus transfectants. Green, anti–Flag antibody; red, anti–PU.1 antibody.
Inhibition of PU.1 expression down-regulates CD80 and CD86 expression
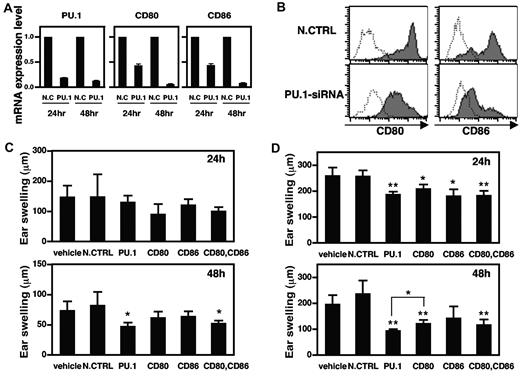
To confirm the involvement of PU.1 in CD80 and CD86 expression, PU.1 siRNA was introduced into murine BMDCs by electroporation. PU.1 mRNA levels were reduced by more than 80% at 24 hours after PU.1 siRNA electroporation, and this effect was maintained for at least 48 hours. At 48 hours after electroporation, PU.1 siRNA reduced both CD80 and CD86 mRNA levels by more than 90% compared with control siRNA (Figure 6A). These results suggest that PU.1 siRNA successfully down-regulates PU.1 expression in BMDCs, resulting in the subsequent suppression of CD80 and CD86 transcription. FACS analysis demonstrated that the cell-surface expression levels of CD80 and CD86 were markedly decreased by PU.1 knock down (Figure 6B). From these results, we conclude that PU.1 is essential for the expression of CD80 and CD86 genes and proteins.
Suppression of CD80 and CD86 expression and CHS by PU.1 siRNA. Murine BMDCs were transfected with 2 μg of PU.1 siRNA or negative control siRNA (N.CTRL; A-B). (A) After a 24- or 48-hour culture, total RNA was extracted from each transfectant, and the amounts of PU.1, CD80, CD86, and β-actin mRNAs were analyzed by ABI7500. PU.1, CD80, and CD86 mRNA levels are represented as a ratio relative to those in negative controls. Results are expressed as means + SD for 3 PCRs performed in duplicate. (B) After a 48-hour culture, cells were harvested and subjected to analysis for CD80 or CD86 by flow cytometry. Solid line histogram represents cells with each antibody. Dotted line histogram represents negative control with 2.4G2 alone. Representative results of 3 independent experiments are shown. CHS against TNCB was induced as described in “Introduction of contact hypersensitivity and treatment with siRNA” (C-D). Nonsilencing control (N.CTRL), PU.1 siRNA, CD80 siRNA, CD86 siRNA, or a CD80/CD86 siRNA combination mixed with a cream-based ointment was applied (C), or each siRNA was subcutaneously injected (D) and the ear was challenged with TNCB. Ear swelling was measured at 24 and 48 hours after challenge. Sterile water mixed with the cream-based ointment was used as a control (vehicle, C). Diethylpyrocarbonate-treated water was subcutaneously injected as a control (vehicle, D). Data are representative of 2 or 3 independent experiments (n = 4∼6) and significant differences from the nonsilencing control siRNA group (*P < .1; **P < .01) were observed. Forty-eight hours after the challenge, ear swelling in the PU.1 siRNA group was significantly reduced compared with the CD80 siRNA group (*P < .05).
Suppression of CD80 and CD86 expression and CHS by PU.1 siRNA. Murine BMDCs were transfected with 2 μg of PU.1 siRNA or negative control siRNA (N.CTRL; A-B). (A) After a 24- or 48-hour culture, total RNA was extracted from each transfectant, and the amounts of PU.1, CD80, CD86, and β-actin mRNAs were analyzed by ABI7500. PU.1, CD80, and CD86 mRNA levels are represented as a ratio relative to those in negative controls. Results are expressed as means + SD for 3 PCRs performed in duplicate. (B) After a 48-hour culture, cells were harvested and subjected to analysis for CD80 or CD86 by flow cytometry. Solid line histogram represents cells with each antibody. Dotted line histogram represents negative control with 2.4G2 alone. Representative results of 3 independent experiments are shown. CHS against TNCB was induced as described in “Introduction of contact hypersensitivity and treatment with siRNA” (C-D). Nonsilencing control (N.CTRL), PU.1 siRNA, CD80 siRNA, CD86 siRNA, or a CD80/CD86 siRNA combination mixed with a cream-based ointment was applied (C), or each siRNA was subcutaneously injected (D) and the ear was challenged with TNCB. Ear swelling was measured at 24 and 48 hours after challenge. Sterile water mixed with the cream-based ointment was used as a control (vehicle, C). Diethylpyrocarbonate-treated water was subcutaneously injected as a control (vehicle, D). Data are representative of 2 or 3 independent experiments (n = 4∼6) and significant differences from the nonsilencing control siRNA group (*P < .1; **P < .01) were observed. Forty-eight hours after the challenge, ear swelling in the PU.1 siRNA group was significantly reduced compared with the CD80 siRNA group (*P < .05).
Topical application of PU.1 siRNA inhibits CHS
Treatment with an anti–CD80 or an anti–CD86 mAb at the time of challenge partially inhibited CHS in a mouse model.45 In addition, Ritprajak et al reported that treatment with CD86 siRNA inhibited CHS.46 Based on these reports and the results of the present study, we expected that PU.1 siRNA would inhibit CHS. Before verifying the effect of siRNA in vivo, we used BMDCs to confirm that CD80 and CD86 siRNA reduced the expression of CD80 and CD86, respectively, and that neither siRNA had any effect on the expression of PU.1 mRNA (supplemental Figure 5). PU.1 siRNA, CD80 siRNA, CD86 siRNA, a combination of CD80 siRNA and CD86 siRNA, nonsilencing control siRNA formulated as a cream-based ointment, or cream-based ointment alone was applied to the right ears of sensitized BALB/c mice 24 hours before challenge. Although no significant differences were observed in ear swelling at 48 hours after challenge between the group treated with cream-based ointment alone and the group treated with cream-emulsified nonsilencing control siRNA, ear swelling in the group treated with cream-emulsified PU.1 siRNA or a combination of CD80 and CD86 siRNA was significantly reduced (Figure 6C). However, a significant suppression of ear swelling was not seen in any group at 24 hours after challenge. We believe that this was due to the time necessary for the siRNA to infiltrate into the dermis; therefore, we injected siRNA subcutaneously into the right ears of sensitized mice at 24 hours before the challenge. As a result, PU.1 siRNA and the combination of CD80 and CD86 siRNA suppressed ear swelling significantly at 24 hours after challenge, and the effects of PU.1 siRNA were sustained at 48 hours (Figure 6D). These results suggest that PU.1 siRNA down-regulates PU.1, CD80, and CD86 transcription and expression in vivo, thereby reducing CHS.
Roles of PU.1 in other phenotypes and functions of DCs
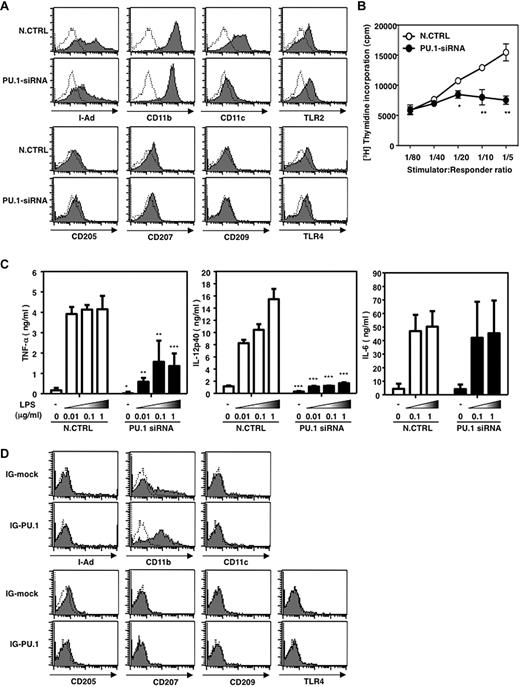
To evaluate the effect of PU.1 on overall DC phenotype and function, cell-surface expression of DC-related molecules, T cell–stimulation activity, and TLR responses of PU.1 siRNA-introduced BMDCs were analyzed. Expression of MHC class II, CD11b, and CD11c were reduced in PU.1 siRNA-introduced BMDCs, whereas marked reduction was not observed in the expression of other molecules, including TLR2 and TLR4 (Figure 7A). T cell–stimulation activity was significantly suppressed by PU.1 knock-down (Figure 7B). Although LPS-induced production of tumor necrosis factor-α (TNF-α) and IL12 p40 was down-regulated by PU.1 siRNA, the IL6 production level was comparable between PU.1–knocked-down BMDCs and the control (Figure 7C). Furthermore, overexpression of PU.1 in Lin− induced the expression of CD11b but not the expression of the others (Figure 7D); profiles of unstimulated cells are shown and similar results were observed in cells stimulated by LPS and/or IFN-γ [data not shown]). The results shown in Figure 7A and D indicate that PU.1 is involved in the expression of MHC class II, CD11b, and CD11c, and that PU.1 is enough for CD11b induction but needs the cooperation of additional factor(s) for the expression of MHC class II and CD11c.
Effects of PU.1 on phenotype and function of DCs. (A) Cell-surface expression of DC-related molecules in PU.1-siRNA–introduced BMDCs. Solid line histogram represents cells with each antibody. Dotted line histogram represents negative control with 2.4G2 alone (A and D). Representative results of 3 independent experiments are shown. (B) T cell–stimulation activity of PU.1–knocked-down BMDCs (*P < .05; **P < .005). LPS-stimulated BMDCs (Balb/c) transfected with control siRNA (○) or PU.1 siRNA (●) were used as a stimulator for T cells from B6 mice. Representative results of 2 independent experiments are shown. (C) Cytokine production of PU.1–knocked-down BMDCs in response to LPS stimulation (*P < .05; **P < .005; ***P < .001). Cytokine concentration in the supernatant at 6 hours (for TNF-α and IL6) and at 24 hours (for IL12 p40) after the addition of various concentrations of LPS was measured by ELISA. Results are expressed as means + SD of 2 independent experiments performed in duplicate. (D) Cell-surface expression of DC-related molecules in PU.1-overexpressing hematopoietic stem cells. Lin− cells were transfected by retrovirus vector carrying IG-mock or IG-PU.1 (Figure 5A) and maintained in the same condition as that of Figure 5D with supplementation of IL3, IL6, SCF, TPO, and EPO. Similar results were obtained in 2 additional independent experiments.
Effects of PU.1 on phenotype and function of DCs. (A) Cell-surface expression of DC-related molecules in PU.1-siRNA–introduced BMDCs. Solid line histogram represents cells with each antibody. Dotted line histogram represents negative control with 2.4G2 alone (A and D). Representative results of 3 independent experiments are shown. (B) T cell–stimulation activity of PU.1–knocked-down BMDCs (*P < .05; **P < .005). LPS-stimulated BMDCs (Balb/c) transfected with control siRNA (○) or PU.1 siRNA (●) were used as a stimulator for T cells from B6 mice. Representative results of 2 independent experiments are shown. (C) Cytokine production of PU.1–knocked-down BMDCs in response to LPS stimulation (*P < .05; **P < .005; ***P < .001). Cytokine concentration in the supernatant at 6 hours (for TNF-α and IL6) and at 24 hours (for IL12 p40) after the addition of various concentrations of LPS was measured by ELISA. Results are expressed as means + SD of 2 independent experiments performed in duplicate. (D) Cell-surface expression of DC-related molecules in PU.1-overexpressing hematopoietic stem cells. Lin− cells were transfected by retrovirus vector carrying IG-mock or IG-PU.1 (Figure 5A) and maintained in the same condition as that of Figure 5D with supplementation of IL3, IL6, SCF, TPO, and EPO. Similar results were obtained in 2 additional independent experiments.
Discussion
Because of their role in immune function, CD80 and CD86 are potential targets for clinical treatments in autoimmunity or transplantation.9,47 However, the critical factor for the regulation and expression of CD80 and CD86 remains unknown. In the present study, we describe the essential transcription factor in the regulation of CD80 and CD86.
Previous reports have suggested the involvement of transcription factors such as NF-κB or IRF-family proteins in the regulation and expression of CD80 and CD86. However, we predicted the involvement of another transcription factor, PU.1, which is necessary for the development of DCs. As expected, the ChIP assay showed that the binding of PU.1 to the promoter region of CD80-1, CD86-1, and CD86-2 was constitutive. In contrast, PU.1 binding to the promoter region of CD80-2 was not observed, and this was supported by the reporter assay showing that co-expressed PU.1 did not affect CD80-2 promoter activity. On the other hand, promoter activity of CD80-1, CD86-1, and CD86-2 were significantly increased by co-expression of PU.1. These results indicate that PU.1 transactivates the CD80-1, CD86-1, and CD86-2 promoters via binding to these promoter regions. A reporter assay with deleted constructs and EMSA suggested that the PU.1-recognizing, cis-enhancing elements are any of 4 motifs in −41/−12 of CD80-1, 3 motifs in −54/−25 (probably −43/−40) of CD86-1, and 2 motifs in −53/−35 (probably −49/−46) of CD86-2. Further detailed analyses to identify the elements critical to PU.1-mediated promoter activity, including reporter assay with mutant plasmids lacking each of candidate motifs, will be useful in confirming this hypothesis.
Although significant binding and transactivation of PU.1 on the CD80-2 promoter were not detected, a possible role for PU.1 in the CD80-2 promoter cannot be excluded. Considering the adjacent locations of CD80-1 and CD80-2 (approximately 1.5 kb apart), PU.1 binding to cis-enhancing element(s) in the CD80-1 promoter region may participate in the activation of RNA polymerase machinery regulating the CD80-2 promoter. PU.1 siRNA reduced CD80 mRNA and CD86 mRNA to a similar degree (Figure 6), suggesting the possibility that PU.1 knock-down successfully suppresses not only the CD80-1 promoter but also the CD80-2 promoter. Regardless, these results demonstrate that PU.1 siRNA is a useful tool for the suppression of CD80 and CD86 expression.
Based on the results of the ChIP assay, we expected that PU.1 was the key player in the expression of CD80 and CD86, so we examined the role of PU.1 in their expression. The co-expression reporter assay, the overproduction of PU.1, and the inhibition of PU.1 by siRNA confirmed that PU.1 is a critical factor in the expression and regulation of CD80 and CD86. The results of overproduction and inhibition of PU.1 were particularly surprising, because simply regulating a single transcriptional factor caused marked changes in the expression levels of cell-surface molecules without any extracellular stimulation. This is the first report describing the induction of the costimulatory molecules CD80 and CD86 in hematopoietic stem cells without growth factors and cytokines such as GM-CSF, TNF-α, and IL4, but with the targeting of a single transcription factor.
Considering that IRF4 functions by forming a heterodimer with PU.1, PU.1 siRNA may suppress the gene expression regulated by PU.1 alone and by PU.1/IRF4. The critical requirement for IRF4 in CD80 expression36 or LPS-induced up-regulation of CD80 and CD8637 on GM-CSF-induced BMDCs has been shown in previous studies, although the role of the PU.1/IRF4 heterodimer with regard to the CD80 and CD86 promoters has not been demonstrated to date. In the present study, the involvement of PU.1/IRF4 in the function of the CD80 and CD86 promoters was not observed. IRF4 may regulate the CD80 and CD86 promoters through other regions. A further detailed analysis targeting other regions (including far upstream of the minimum promoters) may be useful in clarifying this possibility. Alternatively, a study using PU.1 mutants lacking the proline, glutamate, serine, and threonine (PEST) domain, which is necessary for interaction with IRF4, is required to address this issue.
Most reports have focused on CD80 and CD86 as direct targets of clinical treatment with antibody, siRNA, or forced expression.48-50 Another report recently showed that NF-kB pathway suppression by RelB siRNA succeeded in inhibiting DC maturation with CD80 and CD86 repression, resulting in the prevention of allograft rejection.51 In the present study, we demonstrated the significant effects of PU.1 siRNA treatment in a murine CHS model. The use of PU.1 siRNA for the treatment and/or prevention of other autoimmune diseases and for transplantation should be explored, because while PU.1 siRNA regulates the basic expression of CD80 and CD86, its effects on such diseases are unknown.
The effects of PU.1 siRNA were comparable to treatment by the CD80/CD86 siRNA combination in this study. Thus, one of the main effects of PU.1 siRNA is to suppress the expression of CD80 and CD86, and it may also cause the suppression of antigen-specific T-cell responses in the regional lymph nodes. Although the mechanisms of hypersensitivity suppression by PU.1 siRNA are uncertain, we have shown that transcription factor PU.1 may be a target for clinical treatment. PU.1 is involved in the development of a certain subsets of DCs, including myeloid DCs, by regulating cell-type–specific gene expression, suggesting the possibility that the effects of PU.1 siRNA are not limited to CD80 and CD86. In fact, in addition to its role in CD80 and CD86 expression, PU.1 siRNA suppressed some of the characters of DCs, including MHC class II expression and its subsequent T cell–stimulation activity and TNF-α and IL12 production. A slightly stronger reduction of CHS by PU.1 siRNA than by the combination of siRNAs for CD80 and CD86 was possibly due to the suppression of other functions of DCs. Although PU.1 overexpression induced the expression of CD80 and CD86 in Lin− with LPS/IFNγ stimulation, MHC class II and CD11c were not expressed in the same conditions, whereas CD11b expression was simply induced by PU.1 itself. Furthermore, PU.1 siRNA reduced the LPS-induced production of TNF-α and IL12 but not IL6. These observations indicate that the role of PU.1 in the expression of DC-related molecules is different. Regardless, further studies analyzing the gene-expression profile of PU.1 siRNA-introduced DCs and revealing the mechanism of PU.1-mediated expression of each gene should be performed to clarify the effects of PU.1 siRNA.
In the present study, we succeeded in both the suppression and induction of CD80 and CD86 expression by targeting a single transcription factor. One crucial role of CD80 and CD86 is the costimulation of the T-cell response and the induction of T cell–mediated immunity, such as the rejection of tumors.52,53 Conversely, their other crucial role is to block the development of several autoimmune diseases or to induce donor-specific tolerance to allografts by abrogation of CD28/B7 signals.47-50 Through these apparently opposing roles of CD80 and CD86, PU.1 can be targeted for clinical treatment due to its inducible and inhibitory effects.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to members of Atopy Research Center and Department of Immunology for helpful discussions and for critical advice in completing the manuscript. We also thank Ms M. Matsumoto for secretarial assistance.
This work was supported by a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to C.N.).
Authorship
Contribution: S.K. performed experiments, analyzed data, and wrote the paper; C.N. designed the research, performed experiments, and wrote the paper; N.N., R.S, K.M., M.H., and N.K. performed experiments and analyzed the data; and H.O. and K.O. designed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Chiharu Nishiyama, Atopy (Allergy) Research Center, Juntendo University School of Medicine, 2-1-1 Hongo, Bunkyo-ku, Tokyo 113-8421, Japan; e-mail: chinishi@juntendo.ac.jp.
References
Author notes
S.K. and C.N. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal