Abstract
Of the genetic changes in primary central nervous system lymphoma (PCNSL), little is known. To detect copy number alterations and differentially expressed genes in PCNSL, we analyzed a total of 12 PCNSL samples with high-resolution array-based comparative genomic hybridization and performed expression profiling in 7 of the 12 samples. The most frequent deletion found in 8 patients (66.7%) occurred in 9p21.3 containing CDKN2A. We compiled the top 96 genes (family-wise error rate, P < .05) showing the greatest differential expression between PCNSL and normal lymph node tissues. From these, we selected 8 candidate genes (NPFFR2, C4orf7, OSMR, EMCN, TPO, FNDC1, COL12A1, and MSC) in which expression changes were associated with copy number aberrations. All 8 genes showed both down-regulation in expression microarray and deletion in array-based comparative genomic hybridization analyses. These genes participate in cell signaling or cell adhesion. In addition, low mRNA expression of C4orf7 was significantly associated with poor survival (P = .0425). Using gene set enrichment analysis, we identified several signal transduction pathways, such as Janus kinase-signal transducers and activators of transcription pathway and adhesion-related pathways, which may be involved in pathogenesis of PCNSL. In conclusion, this study identified novel tumor suppressor genes that may serve as therapeutic targets of PCNSL.
Introduction
Primary diffuse large B-cell lymphoma (DLBCL) of the central nervous system (CNS) is a distinct form of extranodal lymphoma, which is recognized as a DLBCL localized in the eye or brain parenchyma. This diagnosis excludes meningeal and intravascular DLBCL. The tumor cells are mainly large, with numerous apoptotic cells or widespread necrosis, which commonly hinders diagnosis in small biopsies.1,2 Because the brain is segregated from systemic circulation by the blood-brain barrier, the phenotype and cell of origin of primary central nervous system lymphoma (PCNSL) have been topics of investigation.
PCNSL has been reported to show an activated B-cell-like or germinal center B-cell-like subtype of DLBCL3-5 ; however, recent studies of genome-wide gene expression in PCNSL compared with non-CNS DLBCL suggest that PCNSL has specific signatures to be distinguished from non-CNS DLBCL and greater molecular heterogeneity.6,7 Tumor cells of PCNSL carry a heavy load of ongoing somatic hypermutation with biased use of VH genes in immunoglobulin gene analysis, suggesting an antigen-dependent proliferation.8 A recent study of genome-wide gene expression in PCNSL compared with non-CNS DLBCL showed significant differential expression of multiple extracellular matrix- and adhesion-related pathways.6 These indicate that primary CNS DLBCL is derived from peripheral circulating B cells activated by antigenic stimulation and localized in the brain through interactions between various extracellular matrix and adhesion molecules.
Of the genetic changes that lead to PCNSL, very little is known and no characteristic genetic alterations have been defined thus far. Genomic alterations commonly seen in DLBCLs, regardless of the site of origin, include abnormalities of the 3q27 region involving the BCL6 gene in up to 30% of cases and translocation of the BCL2 gene t(14;18) in 20% to 30% of cases.9,10 A MYC rearrangement is observed in up to 10% of DLBCLs and in 3% of PCNSLs11,12 ; however, higher expression of MYC in PCNSL than nodal DLBCL suggests that a mechanism other than a MYC translocation may be responsible for the MYC overexpression in PCNSL.7,12 In PCNSL, inactivation of CDKN2A by either homozygous deletion or DNA methylation is commonly observed,13,14 but this is also common in non-CNS DLBCLs. A single study using comparative genomic hybridization reported frequent losses of 6q21–22, 12q, 18q, and chromosome 22q.15
In this study, we applied high resolution array-based comparative genomic hybridization (aCGH) to a panel of 12 PCNSL samples from patients and 13 peripheral blood samples from healthy men to obtain a genomic profile of PCNSL. In addition, gene expression analysis was performed with 7 tumor samples and 7 normal lymph node samples to identify genes differentially expressed in PCNSL, and to identify pathways specifically affected in PCNSL, a pathway analysis of the PCNSL gene expression dataset was performed. The identified genes were validated using quantitative real-time reverse-transcribed polymerase chain reaction (RT-PCR). The copy number alteration data were combined with the dataset of genes differentially expressed to confirm whether those genetic alterations had an impact on the expression level of affected genomic regions. Finally, we investigated the prognostic significance of those candidate genes using 23 additional formalin-fixed paraffin-embedded (FFPE) tissue samples of primary CNS DLBCL.
Methods
Tissues
Fresh-frozen samples were obtained from 13 patients with primary CNS DLBCL using a protocol that the Samsung Medical Center Institutional Review Board approved, in accordance with the Declaration of Helsinki. Of the samples, 8 were obtained from resection and 5 were collected from stereotactic needle biopsies. All samples were confirmed by routine histologic evaluation. The quality of frozen samples was assessed by hematoxylin and eosin and CD20 immunohistochemical stains. All samples contained more than 80% of malignant B cells except for one, which was excluded from the study. A total of 12 patients were enrolled in the study, including 6 men and 6 women. Age at diagnosis ranged from 32 to 68 years (median, 54.5 years). All 12 samples were included in aCGH analysis. Of the 12 samples, only 8 for RNA study were used for further expression array analysis because of the quantity of tissue available after aCGH. For normal control samples, we used peripheral blood from 13 healthy men for aCGH and 8 fresh-frozen normal lymph nodes for expression array.
Extraction of genomic DNA and total RNA
For aCGH, DNA was extracted from 12 fresh-frozen tumor samples using standard proteinase K/RNAse treatment and phenol-chloroform extraction. Normal DNA was obtained from the peripheral blood of 13 healthy male donors. For expression array, total RNA was prepared from 8 matched fresh tumor tissues and 8 fresh-frozen normal lymph nodes using trizol-chloroform extraction.
aCGH
aCGH was performed using the Agilent Human Genome CGH Microarray Kit 4 × 44K containing approximately 44 000 probes composed of 60-mer oligonucleotides designed by Agilent. Labeling and hybridization were performed by protocols provided by the manufacturer. Briefly, 1.5 μg of test DNA and reference DNAs were digested with AluI and Rsa I (Promega) and purified with the QIAprep Spin Miniprep kit (QIAGEN). Test sample DNA and reference DNA were labeled by random priming with either Cy3-dUTP or Cy5-dUTP using the Agilent Genomic DNA Labeling Kit PLUS. After the labeling reaction, the individually labeled samples and reference samples were combined and concentrated using Microcon YM-30 filters (Millipore). After probe denaturation and preannealing with Cot-1 DNA, hybridization was performed at 65°C with rotation for 40 hours. Four washing steps were completed with Agilent Oligo CGH washes: wash buffer 1 at room temperature for 5 minutes, wash buffer 2 at 37°C for 1 minute, an acetonitrile rinse at room temperature for 1 minute, and a 30-second wash at room temperature in Agilent's Stabilization and Drying Solution. All slides were scanned on an Agilent DNA microarray scanner. Data were obtained using Agilent Feature Extraction Software Version 9, and analyzed with Agilent CGH Analytics Version 4.0 Software, using the statistical algorithms z-score and common aberration detection method 2 with sensitivity thresholds at 2.5 and 6.0, respectively, and a moving average window of 0.2 Mb.
Gene expression microarray
Expression profiling analysis was performed using Agilent Oligo Microarray Kit 4 × 44K according to Agilent One-Color Microarray-based Gene Expression Analysis Protocol. Briefly, 0.5 μg of total RNA was amplified and labeled using Agilent Quick Amp Labeling Kit. After labeling, cRNA was purified with RNeasy Mini-spin columns (QIAGEN) and quantified using NanoDrop ND-1000 UV-VIS Spectrophotometer for cyanine 3 dye concentration, RNA absorbance ratio (260 nm/280 nm), and cRNA concentration. Cyanine 3–labeled linearly amplified cRNA was mixed with 10× blocking agent, nuclease-free water, and 25× fragmentation buffer provided in the Agilent Gene Expression Hybridization Kit (Agilent Technologies), and incubated for 30 minutes at 60°C to fragment RNA. After adding 2× GEx hybridization buffer Hi-RPM, the amplified fragmented RNA was hybridized on a 4 × 44K Whole Human Genome Oligo Microarray Chip (Agilent Technologies) containing 44 000 cDNA clones using a SureHyb DNA Microarray Hybridization Chamber in a DNA Microarray Hybridization Oven (Agilent Technologies) at 10 rpm, 65°C, for 17 hours. After hybridization, slides were washed in Gene Expression wash buffers 1 and 2 for 1 minute each and then scanned with an Agilent DNA microarray scanner (Agilent Microarray Scanner-G2565BA; Agilent Technologies). Feature Extraction Software Version 9 (Agilent Technologies), was used to extract and analyze the signals. To validate the amplification process, amplified and unamplified RNAs were labeled, hybridized, and compared.
Gene expression analysis
To identify genes differentially expressed between the tumors and normal lymph nodes, the family-wise error rate (FWER) for control of multiple-testing error was calculated using software R (Version 2.9.1; http://www.r-project.org) after normalization by the variance-stabilizing transformation method (VSN) proposed by Huber et al.16 Significantly different genes (FWER, P < .05) identified by expression array analysis were then imported into DAVID (the database for annotation, visualization and integrated discovery) bioinformatics resources17,18 to extract biologic features and meaning associated with large gene lists. To identify genomic alterations in the different genes expressed, the identified genes were combined with aCGH data obtained by Agilent CGH Analytics Version 4.0 Software, to correlate common aberrant regions from aCGH. In addition, to identify pathways specifically affected in PCNSL, a pathway analysis of the PCNSL gene expression dataset was performed using gene set enrichment analysis19,20 with KEGG/Biocarta pathway gene sets.
Quantitative real-time RT PCR
To validate expression data, cDNA was synthesized from 2 μg of total RNA from 6 fresh tumor samples and 5 normal reactive lymph node samples using Superscript III RNA Reverse Transcriptase (Invitrogen) according to the manufacturer's protocol. Real-time RT-PCR was performed using the ABI 7900 HT Fast Real-Time PCR System (Applied Biosystems). The NPFFR2, C4orf7, OSMR, EMCN, TPO, FNDC1, COL12A1, and MSC genes and a reference gene (GAPDH) were amplified and detected using dsDNA-specific CYBR Green dye (Applied Biosystems). PCR reactions were prepared in a final volume of 10 μL, with 5 μL of 2 × Power SYBR Green PCR Master Mix (Applied Biosystems), 0.5 μL of cDNA, and 0.2 μL each of the 10 pmol/μL forward and reverse primers. Thermal cycling commenced with DNA polymerase activation at 95°C for 10 minutes and continued with 45 cycles of denaturation at 95°C for 15 seconds, followed by annealing and extension at 60°C for 1 minute. Each measurement was performed in duplicate. PCR product quality was monitored using post-PCR melt-curve analysis. Threshold cycle (Ct), the fractional cycle number at which the amount of amplified target reached a fixed threshold, was determined. The concentration ratios of each of the 8 genes to GAPDH were calculated using 2−ΔCt (ΔCt = ΔCtested genes − ΔCreference gene).
Primers were as follows: COL12A1: forward (5′-AGTGCATCAACGGGATACAG), reverse (5′-ACATGCAATTCCCTCTCTGC); C4orf7: forward (5′-CATATCCATTTCGCCCACTT), reverse (5′-TTTCGCTAGG AAGGGGAGTT); FNDC1: forward (5′-TCCCCAATGACCTGAAGAAG), reverse (5′-GCTTTGTCCCAGTCCACAAT); NPFFR2: forward (5′-GCCCCTGTGG ACTCTAATGA), reverse (5′-TGGGATTGAC ACTGCTGTTG); MSC: forward (5′-CGCTTAAATCGGACTGGAAC), reverse (5′-GGGAAGGGTCAAGGAGAATC); EMCN: forward (5′-TCACACCAACAACTGGAACAA), reverse (5′-TCAGTGGTTGTGGCTTTCAA); OSMR: forward (5′-TCTGAGCTCCCTTTGGAATG), reverse (5′-GGAACTCCAGTTGCTCCAGA); TPO: forward (5′-GAGGGATTACATCCCCAGGA), reverse (5′-GAACACGTTGGACACAGTGG); and GAPDH: forward (5′-GCACCGTCAAGGCTGAGAA), reverse (5′-AGCATCGCCCCACTTGATT).
Statistical analysis
Differences in mRNA expression levels between tumors and normal samples were calculated using the Mann-Whitney U test. Overall survival was determined using the Kaplan-Meier method, and survival curves were compared using the log-rank test. Survival was measured from the date of surgery, and all patients were followed through April 2010. All tests were 2-sided, and statistical significance was set at a threshold of P less than .05. Statistical analyses were performed using SPSS Version 18 (SPSS Inc).
Results
Copy number aberrations in PCNSL
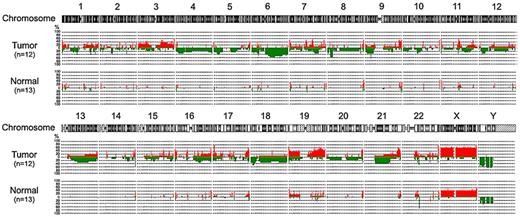
Data have been deposited in Gene Expression Omnibus (accession number GSE25298). All of the 12 PCNSLs showed frequent genomic aberrations, whereas all normal samples from healthy men showed few genomic aberrations (Figure 1). Regions of recurrent gain (≥ 50%) were located on chromosomes 1p36.33, 3q21.3, 3q22.1, 3q29, 7p22.3, 7q11.23, 7q22.1, 7q36.1, 8q24.3, 9q34.11, 9q34.3, 10q26.3, 11p15.5, 11q13.1, 11q23.3, 14q32.33, 15q24.1, 16p13.3, 16p11.2, 16q24.3, 17p13.3, 17q25.3, 19p13.3, 19q13.2, 19q13.31–33, 19q13.41–43, 20q13.33, 21q22.3, and 22q13.33. Regions of most frequent gain were 17p13.3 containing TMEM93 and P2RX5, and 22q13.33 containing IL17REL and MLC1 in 9 patients (75%). Regions of recurrent loss (≥ 50%) were located on chromosomes 6p21.32, 6q14.1–3, 6q15, 6q16.1–3, 6q21, 6q22.1–2, 6q22.31–33, 6q23.1–3, 6q24.1–2, 6q26, 8p22, 8p21.3, 9p21.3, 14q32.33, p11.31, and 22q11.23. Deletions were most frequently observed in 9p21.3 (8 patients; 66.7%), which contains CDKN2A.
Genome-wide frequency plots of copy number alterations identified in 12 PCNSLs and 13 normal peripheral blood samples.
Genome-wide frequency plots of copy number alterations identified in 12 PCNSLs and 13 normal peripheral blood samples.
Expression signatures of PCNSL compared with normal lymph node
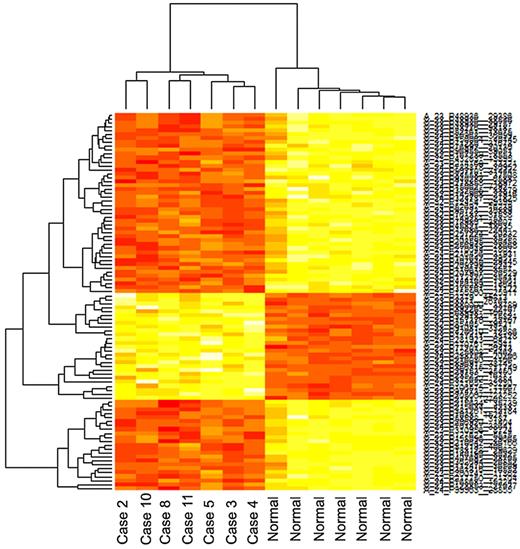
Data have been deposited in Gene Expression Omnibus (accession number GSE25297). From the raw data from 8 tumors and 8 normal lymph nodes, we excluded one tumor and one normal sample that showed inadequate quality. After normalization using the VSN method, we detected 96 significant genes (FWER, P < .05) that were differentially expressed in the tumor group compared with the normal group. Clustering of these top 96 genes showed a distinct separation between the 2 groups (Figure 2). The genes included 27 up-regulated genes (Table 1) and 69 down-regulated genes (Table 2). Functional annotation analysis using DAVID bioinformatics resources produced 111 enriched biologic terms. Many of these terms are related to signaling, extracellular matrix (ECM) properties, and cellular adhesion. A functional annotation group, including OSMR, CCND3, TPO, and IL3RA, is associated with Janus kinase (JAK)-signal transducers and activators of transcription (STAT) signaling. Biologic terms that met the significance threshold of P less than .01 are summarized in Table 3.
Clustering of the top 96 genes (P < .05, FWER) differentially expressed between the tumor group and normal group shows a distinct separation between the groups.
Clustering of the top 96 genes (P < .05, FWER) differentially expressed between the tumor group and normal group shows a distinct separation between the groups.
Up-regulated genes in PCNSL
| PCNSL versus normal LN (present study) . | PCNSL versus non-CNS DLBCL . | |||||
|---|---|---|---|---|---|---|
| Probe ID . | Gene . | P (FWER) . | Rank . | Gene by Tun et al6 . | Gene by Tun et al6 . | Gene by Rubenstein et al7 . |
| A_23_P2181 | CYB5R2 | .001 | 1 | SPP1 | RGS13 | MYC |
| A_24_P157370 | IL 17RB | .002 | 4 | TF | FCGR3A | MINA |
| A_24_P366315 | KIF21A | .003 | 7 | DDR1 | CNP | EIF5 |
| A_32_P85405 | Not found | .006 | 8 | NM_178012 | FBP1 | EIF4A |
| A_23_P3371 | RASGRF1 | .006 | 10 | SERPINA3 | BC000525 | CUL5 |
| A_32_P125808 | Not found | .008 | 14 | S-100B | BMP7 | E2F5 |
| A_23_P97473 | MRPL20 | .009 | 17 | C9orf58 | NM_013332 | BUB3 |
| A_24_P880000 | Not found | .011 | 20 | BC020630 | C1QB | PIM1 |
| A_32_P23525 | Not found | .012 | 22 | CRYAB | IRF4 | JAK1 |
| A_23_P325642 | PDIA2 | .015 | 23 | NM_018584 | DECR2 | XBP1 |
| A_23_P77201 | C15orf41 | .015 | 26 | AF034208 | ATF6 | |
| A_23_P3379 | RASGRF1 | .016 | 29 | CXCL13 | PDCD4 | |
| A_23_P420610 | FCHO2 | .017 | 31 | NM_152680 | FAF1 | |
| A_24_P392265 | EXOC5 | .023 | 38 | TCL1A | CFLAR0 | |
| A_23_P205007 | IPO5 | .025 | 42 | DKK3 | ITGAE | |
| A_24_P31165 | GFAP | .026 | 45 | FOXG1B | CTSD | |
| A_23_P170761 | PDLIM5 | .026 | 46 | UGT2B17 | ||
| A_24_P281923 | DLAT | .026 | 48 | TACSTD1 | ||
| A_23_P329133 | GOT2 | .026 | 50 | FEZ1 | ||
| A_24_P288754 | PIGA | .027 | 52 | CHI3L1 | ||
| A_23_P68998 | MIOX | .030 | 61 | CA2 | ||
| A_23_P216080 | HOOK3 | .031 | 67 | MGST1 | ||
| A_32_P89691 | SORD | .033 | 73 | DNAJC12 | ||
| A_23_P117411 | FOXG1 | .039 | 78 | AK097976 | ||
| A_23_P106575 | GOT2 | .040 | 79 | NM_018357 | ||
| A_23_P29153 | RTDR1 | .042 | 82 | HBA2 | ||
| A_23_P91081 | TACSTD1 | .045 | 89 | NM_024897 | ||
| PCNSL versus normal LN (present study) . | PCNSL versus non-CNS DLBCL . | |||||
|---|---|---|---|---|---|---|
| Probe ID . | Gene . | P (FWER) . | Rank . | Gene by Tun et al6 . | Gene by Tun et al6 . | Gene by Rubenstein et al7 . |
| A_23_P2181 | CYB5R2 | .001 | 1 | SPP1 | RGS13 | MYC |
| A_24_P157370 | IL 17RB | .002 | 4 | TF | FCGR3A | MINA |
| A_24_P366315 | KIF21A | .003 | 7 | DDR1 | CNP | EIF5 |
| A_32_P85405 | Not found | .006 | 8 | NM_178012 | FBP1 | EIF4A |
| A_23_P3371 | RASGRF1 | .006 | 10 | SERPINA3 | BC000525 | CUL5 |
| A_32_P125808 | Not found | .008 | 14 | S-100B | BMP7 | E2F5 |
| A_23_P97473 | MRPL20 | .009 | 17 | C9orf58 | NM_013332 | BUB3 |
| A_24_P880000 | Not found | .011 | 20 | BC020630 | C1QB | PIM1 |
| A_32_P23525 | Not found | .012 | 22 | CRYAB | IRF4 | JAK1 |
| A_23_P325642 | PDIA2 | .015 | 23 | NM_018584 | DECR2 | XBP1 |
| A_23_P77201 | C15orf41 | .015 | 26 | AF034208 | ATF6 | |
| A_23_P3379 | RASGRF1 | .016 | 29 | CXCL13 | PDCD4 | |
| A_23_P420610 | FCHO2 | .017 | 31 | NM_152680 | FAF1 | |
| A_24_P392265 | EXOC5 | .023 | 38 | TCL1A | CFLAR0 | |
| A_23_P205007 | IPO5 | .025 | 42 | DKK3 | ITGAE | |
| A_24_P31165 | GFAP | .026 | 45 | FOXG1B | CTSD | |
| A_23_P170761 | PDLIM5 | .026 | 46 | UGT2B17 | ||
| A_24_P281923 | DLAT | .026 | 48 | TACSTD1 | ||
| A_23_P329133 | GOT2 | .026 | 50 | FEZ1 | ||
| A_24_P288754 | PIGA | .027 | 52 | CHI3L1 | ||
| A_23_P68998 | MIOX | .030 | 61 | CA2 | ||
| A_23_P216080 | HOOK3 | .031 | 67 | MGST1 | ||
| A_32_P89691 | SORD | .033 | 73 | DNAJC12 | ||
| A_23_P117411 | FOXG1 | .039 | 78 | AK097976 | ||
| A_23_P106575 | GOT2 | .040 | 79 | NM_018357 | ||
| A_23_P29153 | RTDR1 | .042 | 82 | HBA2 | ||
| A_23_P91081 | TACSTD1 | .045 | 89 | NM_024897 | ||
LN indicates lymph node.
Down-regulated genes in PCNSL
| PCNSL versus normal LN (present study) . | PCNSL versus non-CNS DLBCL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe ID . | Gene . | P (FWER) . | Rank . | Probe ID . | Gene . | P (FWER) . | Rank . | Gene by Tun et al6 . | Gene by Rubenstein et al7 . |
| A_23_P140190 | FAM30A | .001 | 2 | A_32_P167239 | AFAP1L1 | .029 | 56 | CX3CR1 | ZFP36 |
| A_24_P95723 | KIAA0125 | .001 | 3 | A_23_P43283 | GPR124 | .030 | 57 | AK091178 | MLL |
| A_24_P70183 | MYH11 | .002 | 5 | A_32_P76137 | Not found | .030 | 58 | D42043 | CHES1 |
| A_24_P76868 | Not found | .002 | 6 | A_24_P375761 | STGC3 | .030 | 59 | SQLE | EPHB3 |
| A_23_P206920 | MYH11 | .006 | 9 | A_24_P110618 | Not found | .030 | 60 | P2RY8 | RPS6KA3 |
| A_23_P76992 | PGF | .008 | 11 | A_23_P90710 | DES | .030 | 62 | LXN | DDR2 |
| A_24_P52697 | H19 | .008 | 12 | A_23_P218858 | ABI3BP | .031 | 63 | LPP | ADAM12 |
| A_23_P123853 | CCL19 | .008 | 13 | A_23_P310956 | COL6A2 | .031 | 64 | S56205 | MMP2 |
| A_24_P183128 | PLAC8 | .008 | 15 | A_23_P382065 | EMCN | .031 | 65 | Z24727 | COL4A2 |
| A_32_P172320 | CCND3 | .009 | 16 | A_24_P270044 | ASAM | .031 | 66 | STEAP | COL5A1 |
| A_32_P145968 | Not found | .011 | 18 | A_24_P99963 | CSNK1G2 | .032 | 68 | COL6A1 | VCAM1 |
| A_23_P78980 | B3GNT3 | .011 | 19 | A_23_P112470 | CCL21 | .032 | 69 | COL12A1 | CDH1 |
| A_23_P200741 | DPT | .012 | 21 | A_24_P86968 | CD3E | .032 | 70 | AK022110 | ITGAX |
| A_32_P444104 | LOC284454 | .015 | 24 | A_23_P158096 | COL27A1 | .032 | 71 | NR2F2 | |
| A_23_P45821 | Not found | .015 | 25 | A_24_P15062 | ZNF490 | .033 | 72 | NNMT | |
| A_23_P12686 | GFRA1 | .015 | 27 | A_24_P257224 | TPO | .033 | 74 | AK091153 | |
| A_23_P81219 | PLAC8 | .016 | 28 | A_23_P150064 | MMRN2 | .035 | 75 | BC040354 | |
| A_23_P149281 | EPHA2 | .016 | 30 | A_24_P361816 | IGLV6–57 | .038 | 76 | VEGFC | |
| A_23_P345581 | FOSL2 | .018 | 32 | A_23_P407840 | FNDC1 | .038 | 77 | LAMA4 | |
| A_23_P155900 | NPFFR2 | .018 | 33 | A_24_P101642 | LOC401847 | .042 | 80 | NM_015714 | |
| A_23_P381261 | ADCY4 | .020 | 34 | A_23_P215634 | IGFBP3 | .042 | 81 | CAMKK2 | |
| A_23_P96955 | FAM167B | .022 | 35 | A_23_P43276 | GPR124 | .043 | 83 | FBN1 | |
| A_23_P362694 | C4orf7 | .022 | 36 | A_32_P12454 | Not found | .043 | 84 | AJ001381 | |
| A_24_P788878 | FAM148B | .023 | 37 | A_24_P324787 | KANK2 | .043 | 85 | LUM | |
| A_23_P48936 | SMAD3 | .024 | 39 | A_23_P52161 | NUAK2 | .043 | 86 | LOXL1 | |
| A_24_P940288 | PGS1 | .024 | 40 | A_23_P208595 | LDLR | .043 | 87 | COL1A2 | |
| A_23_P215634 | IGFBP3 | .024 | 41 | A_23_P48936 | SMAD3 | .043 | 88 | ||
| A_23_P48936 | SMAD3 | .025 | 43 | A_23_P214168 | COL12A1 | .045 | 90 | ||
| A_24_P71280 | GPR157 | .026 | 44 | A_24_P216456 | MAP1LC3C | .045 | 91 | ||
| A_24_P207907 | R3HDM2 | .026 | 47 | A_23_P209799 | MYO7B | .046 | 92 | ||
| A_23_P156289 | OSMR | .026 | 49 | A_23_P127627 | DGKZ | .047 | 93 | ||
| A_23_P407695 | FAM151A | .027 | 51 | A_32_P210642 | EGFL7 | .047 | 94 | ||
| A_23_P1083 | GJA4 | .027 | 53 | A_23_P141055 | TGFB1I1 | .047 | 95 | ||
| A_32_P217750 | IL3RA | .027 | 54 | A_23_P256948 | MSC | .048 | 96 | ||
| A_23_P58588 | SLIT3 | .028 | 55 | ||||||
| PCNSL versus normal LN (present study) . | PCNSL versus non-CNS DLBCL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe ID . | Gene . | P (FWER) . | Rank . | Probe ID . | Gene . | P (FWER) . | Rank . | Gene by Tun et al6 . | Gene by Rubenstein et al7 . |
| A_23_P140190 | FAM30A | .001 | 2 | A_32_P167239 | AFAP1L1 | .029 | 56 | CX3CR1 | ZFP36 |
| A_24_P95723 | KIAA0125 | .001 | 3 | A_23_P43283 | GPR124 | .030 | 57 | AK091178 | MLL |
| A_24_P70183 | MYH11 | .002 | 5 | A_32_P76137 | Not found | .030 | 58 | D42043 | CHES1 |
| A_24_P76868 | Not found | .002 | 6 | A_24_P375761 | STGC3 | .030 | 59 | SQLE | EPHB3 |
| A_23_P206920 | MYH11 | .006 | 9 | A_24_P110618 | Not found | .030 | 60 | P2RY8 | RPS6KA3 |
| A_23_P76992 | PGF | .008 | 11 | A_23_P90710 | DES | .030 | 62 | LXN | DDR2 |
| A_24_P52697 | H19 | .008 | 12 | A_23_P218858 | ABI3BP | .031 | 63 | LPP | ADAM12 |
| A_23_P123853 | CCL19 | .008 | 13 | A_23_P310956 | COL6A2 | .031 | 64 | S56205 | MMP2 |
| A_24_P183128 | PLAC8 | .008 | 15 | A_23_P382065 | EMCN | .031 | 65 | Z24727 | COL4A2 |
| A_32_P172320 | CCND3 | .009 | 16 | A_24_P270044 | ASAM | .031 | 66 | STEAP | COL5A1 |
| A_32_P145968 | Not found | .011 | 18 | A_24_P99963 | CSNK1G2 | .032 | 68 | COL6A1 | VCAM1 |
| A_23_P78980 | B3GNT3 | .011 | 19 | A_23_P112470 | CCL21 | .032 | 69 | COL12A1 | CDH1 |
| A_23_P200741 | DPT | .012 | 21 | A_24_P86968 | CD3E | .032 | 70 | AK022110 | ITGAX |
| A_32_P444104 | LOC284454 | .015 | 24 | A_23_P158096 | COL27A1 | .032 | 71 | NR2F2 | |
| A_23_P45821 | Not found | .015 | 25 | A_24_P15062 | ZNF490 | .033 | 72 | NNMT | |
| A_23_P12686 | GFRA1 | .015 | 27 | A_24_P257224 | TPO | .033 | 74 | AK091153 | |
| A_23_P81219 | PLAC8 | .016 | 28 | A_23_P150064 | MMRN2 | .035 | 75 | BC040354 | |
| A_23_P149281 | EPHA2 | .016 | 30 | A_24_P361816 | IGLV6–57 | .038 | 76 | VEGFC | |
| A_23_P345581 | FOSL2 | .018 | 32 | A_23_P407840 | FNDC1 | .038 | 77 | LAMA4 | |
| A_23_P155900 | NPFFR2 | .018 | 33 | A_24_P101642 | LOC401847 | .042 | 80 | NM_015714 | |
| A_23_P381261 | ADCY4 | .020 | 34 | A_23_P215634 | IGFBP3 | .042 | 81 | CAMKK2 | |
| A_23_P96955 | FAM167B | .022 | 35 | A_23_P43276 | GPR124 | .043 | 83 | FBN1 | |
| A_23_P362694 | C4orf7 | .022 | 36 | A_32_P12454 | Not found | .043 | 84 | AJ001381 | |
| A_24_P788878 | FAM148B | .023 | 37 | A_24_P324787 | KANK2 | .043 | 85 | LUM | |
| A_23_P48936 | SMAD3 | .024 | 39 | A_23_P52161 | NUAK2 | .043 | 86 | LOXL1 | |
| A_24_P940288 | PGS1 | .024 | 40 | A_23_P208595 | LDLR | .043 | 87 | COL1A2 | |
| A_23_P215634 | IGFBP3 | .024 | 41 | A_23_P48936 | SMAD3 | .043 | 88 | ||
| A_23_P48936 | SMAD3 | .025 | 43 | A_23_P214168 | COL12A1 | .045 | 90 | ||
| A_24_P71280 | GPR157 | .026 | 44 | A_24_P216456 | MAP1LC3C | .045 | 91 | ||
| A_24_P207907 | R3HDM2 | .026 | 47 | A_23_P209799 | MYO7B | .046 | 92 | ||
| A_23_P156289 | OSMR | .026 | 49 | A_23_P127627 | DGKZ | .047 | 93 | ||
| A_23_P407695 | FAM151A | .027 | 51 | A_32_P210642 | EGFL7 | .047 | 94 | ||
| A_23_P1083 | GJA4 | .027 | 53 | A_23_P141055 | TGFB1I1 | .047 | 95 | ||
| A_32_P217750 | IL3RA | .027 | 54 | A_23_P256948 | MSC | .048 | 96 | ||
| A_23_P58588 | SLIT3 | .028 | 55 | ||||||
LN indicates lymph node.
Enriched functional annotation terms associated with top 96 genes
| Category . | Term . | % . | P* . | Genes . | Bonferroni* . |
|---|---|---|---|---|---|
| SP_PIR_KEYWORDS | Signal | 35.6 | < .0001 | OSMR, MMRN2, EMCN, C4orf7, DLAT, IL3RA, TACSTD1, SLIT3, ABI3BP, COL6A2, COL27A1, EGFL7, EPHA2, CD3E, DPT, PDIA2, CCL21, LDLR, COL12A1, ASAM, GFRA1, GPR124, CCL19, IGFBP3, PGF, TPO | 0.0018 |
| UP_SEQ_FEATURE | Signal peptide | 32.9 | < .0001 | ABI3BP, MMRN2, EMCN, COL6A2, COL27A1, C4orf7, EPHA2, EGFL7, DLAT, IL3RA, TACSTD1, CD3E, DPT, PDIA2, LDLR, CCL21, GFRA1, COL12A1, CCL19, GPR124, PGF, SLIT3, IGFBP3, TPO | 0.3724 |
| IPR008957 | Fibronectin, type III-like fold | 8.2 | .0004 | ABI3BP, OSMR, FNDC1, COL12A1, EPHA2, IL3RA | 0.9106 |
| GO:0009987 | Cellular process | 69.9 | .0018 | EXOC5, MAP1LC3C, DLAT, ADCY4, MSC, FOSL2, DGKZ, SLIT3, COL6A2, RASGRF1, MIOX, PIGA, NUAK2, GPR157, CCL21, LDLR, COL12A1, GPR124, IGFBP3, GFAP, OSMR, EMCN, IL3RA, PGS1, SORD, B3GNT3, DES, KIF21A, HOOK3, GOT2, MYH11, CSNK1G2, COL27A1, EGFL7, EPHA2, FOXG1, NPFFR2, CYB5R2, CD3E, DPT, CCND3, PDIA2, TGFB1I1, GJA4, SMAD3, GFRA1, CCL19, ZNF490, PGF, TPO, MRPL20 | 0.9999 |
| GO:0048513 | Organ development | 16.4 | .0028 | CD3E, MYH11, EMCN, PDLIM5, SMAD3, GJA4, COL12A1, PGF, FOXG1, EGFL7, IGFBP3, SLIT3 | 1.0000 |
| IPR003961 | Fibronectin, type III | 6.9 | .0037 | ABI3BP, OSMR, FNDC1, COL12A1, EPHA2 | 1.0000 |
| GO:0048731 | System development | 19.2 | .0041 | MYH11, EMCN, PDLIM5, EGFL7, EPHA2, FOXG1, CD3E, GJA4, SMAD3, COL12A1, GFRA1, PGF, SLIT3, IGFBP3 | 1.0000 |
| IPR001881 | EGF-like calcium-binding | 5.5 | .0055 | LDLR, EGFL7, SLIT3, TPO | 1.0000 |
| SM00060 | FN3 | 6.9 | .0059 | ABI3BP, OSMR, FNDC1, COL12A1, EPHA2 | 0.9731 |
| SP_PIR_KEYWORDS | Secreted | 16.4 | .0067 | DPT, MMRN2, EMCN, COL6A2, CCL21, COL12A1, C4orf7, CCL19, PGF, EGFL7, IGFBP3, SLIT3 | 0.9992 |
| GO:0044421 | Extracellular region part | 12.3 | .0077 | DPT, MMRN2, COL6A2, CCL21, COL27A1, COL12A1, CCL19, EGFL7, SLIT3 | 0.9988 |
| GO:0006519 | Amino acid and derivative Metabolic process | 8.2 | .0080 | GOT2, CD3E, CCND3, IGFBP3, DLAT, TPO | 1.0000 |
| SM00179 | EGF_CA | 5.5 | .0085 | LDLR, EGFL7, SLIT3, TPO | 0.9944 |
| GO:0005581 | Collagen | 4.1 | .0096 | COL6A2, COL27A1, COL12A1 | 0.9998 |
| Category . | Term . | % . | P* . | Genes . | Bonferroni* . |
|---|---|---|---|---|---|
| SP_PIR_KEYWORDS | Signal | 35.6 | < .0001 | OSMR, MMRN2, EMCN, C4orf7, DLAT, IL3RA, TACSTD1, SLIT3, ABI3BP, COL6A2, COL27A1, EGFL7, EPHA2, CD3E, DPT, PDIA2, CCL21, LDLR, COL12A1, ASAM, GFRA1, GPR124, CCL19, IGFBP3, PGF, TPO | 0.0018 |
| UP_SEQ_FEATURE | Signal peptide | 32.9 | < .0001 | ABI3BP, MMRN2, EMCN, COL6A2, COL27A1, C4orf7, EPHA2, EGFL7, DLAT, IL3RA, TACSTD1, CD3E, DPT, PDIA2, LDLR, CCL21, GFRA1, COL12A1, CCL19, GPR124, PGF, SLIT3, IGFBP3, TPO | 0.3724 |
| IPR008957 | Fibronectin, type III-like fold | 8.2 | .0004 | ABI3BP, OSMR, FNDC1, COL12A1, EPHA2, IL3RA | 0.9106 |
| GO:0009987 | Cellular process | 69.9 | .0018 | EXOC5, MAP1LC3C, DLAT, ADCY4, MSC, FOSL2, DGKZ, SLIT3, COL6A2, RASGRF1, MIOX, PIGA, NUAK2, GPR157, CCL21, LDLR, COL12A1, GPR124, IGFBP3, GFAP, OSMR, EMCN, IL3RA, PGS1, SORD, B3GNT3, DES, KIF21A, HOOK3, GOT2, MYH11, CSNK1G2, COL27A1, EGFL7, EPHA2, FOXG1, NPFFR2, CYB5R2, CD3E, DPT, CCND3, PDIA2, TGFB1I1, GJA4, SMAD3, GFRA1, CCL19, ZNF490, PGF, TPO, MRPL20 | 0.9999 |
| GO:0048513 | Organ development | 16.4 | .0028 | CD3E, MYH11, EMCN, PDLIM5, SMAD3, GJA4, COL12A1, PGF, FOXG1, EGFL7, IGFBP3, SLIT3 | 1.0000 |
| IPR003961 | Fibronectin, type III | 6.9 | .0037 | ABI3BP, OSMR, FNDC1, COL12A1, EPHA2 | 1.0000 |
| GO:0048731 | System development | 19.2 | .0041 | MYH11, EMCN, PDLIM5, EGFL7, EPHA2, FOXG1, CD3E, GJA4, SMAD3, COL12A1, GFRA1, PGF, SLIT3, IGFBP3 | 1.0000 |
| IPR001881 | EGF-like calcium-binding | 5.5 | .0055 | LDLR, EGFL7, SLIT3, TPO | 1.0000 |
| SM00060 | FN3 | 6.9 | .0059 | ABI3BP, OSMR, FNDC1, COL12A1, EPHA2 | 0.9731 |
| SP_PIR_KEYWORDS | Secreted | 16.4 | .0067 | DPT, MMRN2, EMCN, COL6A2, CCL21, COL12A1, C4orf7, CCL19, PGF, EGFL7, IGFBP3, SLIT3 | 0.9992 |
| GO:0044421 | Extracellular region part | 12.3 | .0077 | DPT, MMRN2, COL6A2, CCL21, COL27A1, COL12A1, CCL19, EGFL7, SLIT3 | 0.9988 |
| GO:0006519 | Amino acid and derivative Metabolic process | 8.2 | .0080 | GOT2, CD3E, CCND3, IGFBP3, DLAT, TPO | 1.0000 |
| SM00179 | EGF_CA | 5.5 | .0085 | LDLR, EGFL7, SLIT3, TPO | 0.9944 |
| GO:0005581 | Collagen | 4.1 | .0096 | COL6A2, COL27A1, COL12A1 | 0.9998 |
P value and Bonferroni were provided by DAVID bioinformatics resources.
Pathways specific to PCNSL
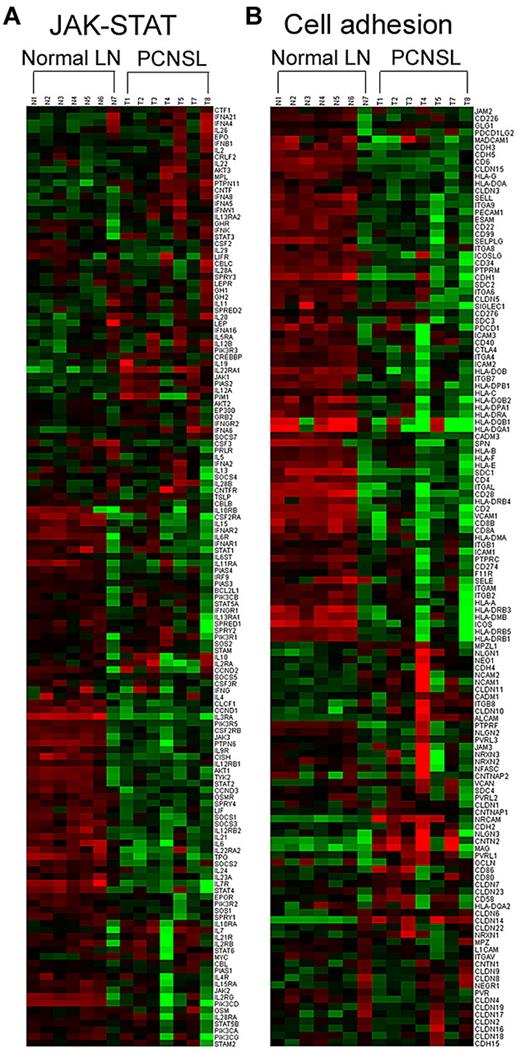
The pathway activities that most clearly distinguished PCNSL from normal lymph nodes (false discovery rate [FDR] Q value < 0.05) mediate functions, such as cytokine-receptor interaction, cell adhesion and migration, apoptosis, and signal transduction via Bad, Notch, JAK-STAT, and NF-κB pathways (Table 4). Expression signature analysis revealed that some component genes in the JAK-STAT pathway, including OSMR, CCND3, and TPO, were down-regulated in PCNSL. Other genes in the JAK-STAT path-way, such as STAT3, JAK1, AKT3, IL22RA1, and IL12A, were up-regulated in PCNSL, whereas SOCS1 and SOCS3, which inhibit JAK-STAT signaling, were down-regulated (Figure 3A). Gene expression changes that influence cell adhesion clearly distinguished the normal and tumor groups (Figure 3B).
Selection of pathways differentially expressed in PCNSL tissues compared with normal lymph node tissues
| Pathway name . | Nominal P . | FDR Q . |
|---|---|---|
| Cytokine cytokine receptor interaction | < .001 | <0.001 |
| Cell adhesion molecules | < .001 | 0.002 |
| ECM receptor interaction | < .001 | 0.014 |
| Leukocyte transendothelial migration | < .001 | 0.016 |
| Focal adhesion | < .001 | 0.017 |
| Apoptosis | .002 | 0.027 |
| Signal transduction pathway | ||
| Bad pathway | < .001 | 0.008 |
| Notch signaling pathway | < .001 | 0.010 |
| JAK-STAT signaling pathway | < .001 | 0.015 |
| NF-κB pathway | .007 | 0.026 |
| Pathway name . | Nominal P . | FDR Q . |
|---|---|---|
| Cytokine cytokine receptor interaction | < .001 | <0.001 |
| Cell adhesion molecules | < .001 | 0.002 |
| ECM receptor interaction | < .001 | 0.014 |
| Leukocyte transendothelial migration | < .001 | 0.016 |
| Focal adhesion | < .001 | 0.017 |
| Apoptosis | .002 | 0.027 |
| Signal transduction pathway | ||
| Bad pathway | < .001 | 0.008 |
| Notch signaling pathway | < .001 | 0.010 |
| JAK-STAT signaling pathway | < .001 | 0.015 |
| NF-κB pathway | .007 | 0.026 |
Representative molecular pathways differentially expressed in PCNSL compared with normal lymph node (LN). The genes involved in JAK-STAT (A) and cell adhesion (B) pathways were clustered according to phenotype (ie, as normal LN or PCNSL).
Representative molecular pathways differentially expressed in PCNSL compared with normal lymph node (LN). The genes involved in JAK-STAT (A) and cell adhesion (B) pathways were clustered according to phenotype (ie, as normal LN or PCNSL).
Gene expression changes associated with copy number aberrations
The regions that contained significant common aberrations in copy number were correlated to a dataset of the top 96 differentially expressed genes (FWER, P < .05) obtained from expression array. Eight genes (NPFFR2, C4orf7, OSMR, EMCN, TPO, FNDC1, COL12A1, and MSC) were identified in this correlational analysis. All 8 genes showed both down-regulation in expression microarray and deletion in aCGH in 2 or more patients (> 15%). The same 8 genes showed neither deletion nor amplification in all 13 normal samples from healthy men. The FNDC1 and COL12A1 genes most frequently showed deletions, which were identified in 5 patients (41.7%). The data for each patient are summarized in Table 5.
Deletion (Del) status in individual patients of 8 genes showing down-regulation by expression array
| Case no. . | NPFFR2 . | C4orf7 . | OSMR . | EMCN . | TPO . | FNDC1 . | COL12A1 . | MSC . |
|---|---|---|---|---|---|---|---|---|
| 1 | Del | Del | Del | Del | Del | — | Del | Del |
| 2 | — | — | — | — | — | Del | — | — |
| 3 | Del | Del | — | — | — | — | — | — |
| 4 | — | — | — | — | Del | — | — | — |
| 5 | — | — | — | — | — | — | Del | — |
| 6 | Del | Del | Del | Del | — | Del | Del | Del |
| 8 | — | — | — | — | — | — | — | — |
| 9 | — | — | — | — | — | — | — | — |
| 10 | — | — | — | — | — | — | — | — |
| 11 | — | — | Del | — | — | Del | Del | — |
| 12 | — | — | — | — | — | Del | Del | — |
| 13 | — | — | — | — | — | Del | — | — |
| Normal (n = 13) | — | — | — | — | — | — | — | — |
| Case no. . | NPFFR2 . | C4orf7 . | OSMR . | EMCN . | TPO . | FNDC1 . | COL12A1 . | MSC . |
|---|---|---|---|---|---|---|---|---|
| 1 | Del | Del | Del | Del | Del | — | Del | Del |
| 2 | — | — | — | — | — | Del | — | — |
| 3 | Del | Del | — | — | — | — | — | — |
| 4 | — | — | — | — | Del | — | — | — |
| 5 | — | — | — | — | — | — | Del | — |
| 6 | Del | Del | Del | Del | — | Del | Del | Del |
| 8 | — | — | — | — | — | — | — | — |
| 9 | — | — | — | — | — | — | — | — |
| 10 | — | — | — | — | — | — | — | — |
| 11 | — | — | Del | — | — | Del | Del | — |
| 12 | — | — | — | — | — | Del | Del | — |
| 13 | — | — | — | — | — | Del | — | — |
| Normal (n = 13) | — | — | — | — | — | — | — | — |
— indicates no Del or amplification.
Quantitative real-time RT-PCR for NPFFR2, C4orf7, OSMR, EMCN, TPO, FNDC1, COL12A1, and MSC
Eleven fresh-frozen tissue samples (6 tumors and 5 normal lymph nodes) were assayed for RNA expression levels, and the Ct was calculated for all 8 genes. The mRNA expression levels for each of the 8 genes were normalized to the reference gene (GAPDH). The concentration ratios of each of the 8 genes over GAPDH were calculated.
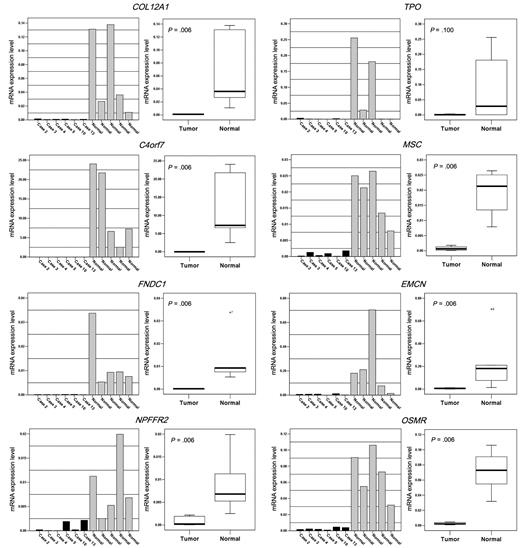
All 8 genes, except for TPO, showed significantly lower mRNA expression in the PCNSL group than in the normal lymph node group (Mann-Whitney U test, P < .05; Figure 4). The TPO mRNA expression level in the tumor group was not significantly lower than that in the normal group (mean concentration ratio = 0.001 [tumor] and 0.093 [normal], P = .1). The C4orf7 showed the most striking difference (mean concentration ratio = 0.007 [tumor] and 12.433 [normal], P = .006).
Significantly lower mRNA expression was detected for NPFFR2, C4orf7, OSMR, EMCN, FNDC1, COL12A1, and MSC in the PCNSL group than in the normal lymph node group (P < .05, Mann-Whitney U test) by quantitative real-time RT-PCR. TPO expression did not differ statistically between groups.
Significantly lower mRNA expression was detected for NPFFR2, C4orf7, OSMR, EMCN, FNDC1, COL12A1, and MSC in the PCNSL group than in the normal lymph node group (P < .05, Mann-Whitney U test) by quantitative real-time RT-PCR. TPO expression did not differ statistically between groups.
Prognostic significance of NPFFR2, C4orf7, OSMR, EMCN, TPO, FNDC1, COL12A1, and MSC
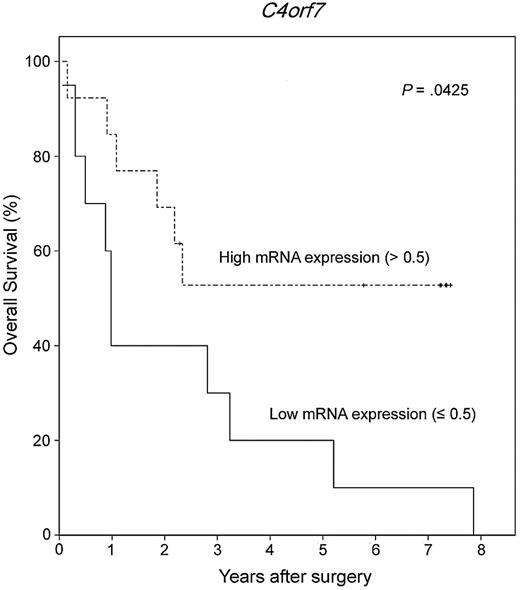
To estimate the prognostic significance of the 8 differentially expressed genes, we investigated an independent set of 23 FFPE tissue samples from primary CNS DLBCL patients. All patients received chemotherapy regimens based on high-dose methotrexate after diagnosis. Among them, 22 patients received additional whole-brain radiation therapy. From these 23 FFPE samples, we extracted total RNA and synthesized the corresponding cDNA. Quantitative real-time RT-PCR was performed as described in “Methods,” and concentration ratios were calculated for each gene. To identify a cut-off point for the mRNA concentration ratio that is most meaningful with respect to prognosis, we tested various levels of expression using the Kaplan-Meier method with log-rank test. Among the 8 genes, low mRNA expression of C4orf7, when the cut-off point was set as less than or equal to 0.5, was significantly associated with poor overall survival (P = .0425, Figure 5). The median and mean values of concentration ratio of C4orf7 in 23 FFPE tumor samples were 0.58 and 0.71, respectively (range, 0.11-2.49). Significant associations with survival were not found for the 7 other genes (P > .05).
Low mRNA expression of C4orf7 was associated with poor overall survival in 23 patients with PCNSL.
Low mRNA expression of C4orf7 was associated with poor overall survival in 23 patients with PCNSL.
Discussion
In this study, we observed 96 genes showing the greatest difference in expression between PCNSL and normal lymphoid tissue. From these, we selected 8 candidate genes based on gene expression changes associated with copy number aberrations. Quantitative real-time RT-PCR confirmed down-regulation of all 8 genes in tumor tissue compared with normal lymphoid tissue. FNDC1, EMCN, and COL12A1 are associated with ECM and cell adhesion. Our results suggest that ECM and cell adhesion contribute to the pathogenesis of PCNSL. This finding converges with prior studies that have found that the gene set that is most significantly affected in PCNSL encodes components of the ECM-receptor pathway.6 These results support the model that an interaction between the CNS microenvironment and lymphoma cells strongly influences PCNSL development.
The other genes (ie, OSMR, C4orf7, NPFFR2, TPO, and MSC) mediate cell signaling. MSC, also called activated B-cell factor-1, is a transcriptional repressor that inhibits the transactivational capacity of TCF3/E47 and may participate in antigen-dependent B-cell differentiation. MSC is frequently silenced by promoter methylation in follicular lymphoma, DLBCL, and Burkitt lymphoma.21 NPFFR2 is a receptor protein for NPAF and NPFF neuropeptides and acts through association with G proteins that activate a phosphatidylinositol-calcium second messenger system. C4orf7 is expressed by follicular dendritic cells and activates leukocytes during an ongoing immune response. These leukocytes may then bind to the surface of B-lymphoma cells, but not T-lymphoma cells, which is consistent with a function for C4orf7 as a secreted B cell-specific mediator. In this study, low expression of C4orf7 mRNA was significantly associated with poor survival in 23 patients with primary CNS DLBCL. C4orf7 has been previously identified as playing a role in the lymphatic metastasis of human breast cancer.22 However, the specific mechanism for the prognostic effect of C4orf7 in PCNSL remains unclear, and further studies with a larger sample are needed.
OSMR is activated by oncostatin M (OSM), which is a member of the IL6 family of cytokines. OSM inhibits the growth of many tumor cell types, such as neuroblastoma, fibrosarcoma, and ovarian, lung, stomach, and breast carcinoma cell lines.23 Thus, OSMR, as the OSM receptor, may act as a tumor suppressor. In the present study, we found that OSMR was significantly down-regulated in PCNSL with deletion, which is consistent with the idea of a tumor suppressor role for OSMR in the pathogenesis of PCNSL. On the other hand, binding of OSM to OSMR triggers the activation of multiple members of the JAK family,24 which in turn leads to the activation of transcription factors known as STATs.25 Moreover, OSMR participates in STAT3 (and possibly STAT1 and STAT5) activation and mediates OSM-specific signaling events. Aberrant STAT3 and STAT5 expression in lymphoma may promote tumor progression.26 In this study, STAT1 was generally down-regulated in PCNSL, which correlates with OSMR down-regulation. STAT3, however, was up-regulated in PCNSL. Thus, factors other than OSMR may influence STAT3 expression in this type of tumor. For example, we also observed down-regulation of SOCS1 and SOCS3 expression. SOCS is known as inhibitor of JAK-STAT pathway, and a study has explored the negative relationship between SOCS3 and the STAT3 pathway.27
In the aCGH analysis, we identified 9p21.3 containing CDKN2A as the region most frequently affected by deletion. CDKN2A is frequently mutated or deleted in a wide variety of tumors, which is evidence of its role as a tumor suppressor gene. Sulong et al28 observed CDKN2A deletion as a significant secondary abnormality in childhood acute lymphoblastic leukemia strongly correlated with phenotype and genotype. Schwindt et al29 reported frequent deletion at the CDKN2A locus in PCNSL, and they suggest that both deletion and DNA methylation may be considered as mechanisms of CDKN2A inactivation in PCNSL. Collectively, these findings indicate the importance of CDKN2A deletion in the pathogenesis of PCNSL.
Deletion of 6q also appears to be a common and important finding in the pathogenesis of PCNSL as well as in systemic DLBCL. Weber et al15 reported 6q as the region most frequently affected by losses of genomic material and mapped a commonly deleted region to 6q21-q22. Boonstra et al30 also described frequent 6q deletion in PCNSL, and they suggested that one or more suppressor genes involved in the pathogenesis of PCNSL may be localized in the region 6q16–22. Recently, Cady et al12 reported that del 6q22 is common in PCNSL, and it has been associated with inferior overall survival. In the present study, aCGH analysis of PCNSL frequently detected deletions in the region of chromosome 6q22, but no significantly down-regulated genes matched in this region. Among several genes showing deletion in this region, RSPO3, which is located at 6q22, showed deletion in 50% of the cases and also demonstrated decreased expression with borderline statistical significance (unadjusted P < .001 but FWER P > .05) in this study.
Using the expression array, we detected 96 significant genes (FWER, P < .05) from approximately 44 000 genes. To select significant genes, there are 2 standard statistical measurements, FWER and FDR, in multiple testing problems. FWER is one of the most conservative methods and provides much better protection against false positives than FDR, although this method has the disadvantage that some biologically significant genes could be missed because of its statistical conservatism. For example, SPP1 (osteopontin) has been reported as the gene most significantly up-regulated in PCNSL.6 In our study, SPP1 was not included in the top up-regulated genes; however, we identified that SPP1 was significantly up-regulated in PCNSL when the FDR method was applied (FDR Q < 0.05). Although we may have missed identifying a few biologically significant genes using an FWER correction, we wanted to protect against identifying false positives.
In the expression array analysis, we found that TACSTD1 (EPCAM), a pan-epithelial differentiation antigen commonly expressed on carcinomas, was significantly up-regulated. Tun et al6 also present this gene as a potentially important contributor to PCNSL. Another interesting up-regulated gene is HOOK3. This microtubule-binding protein interacts with scavenger receptor A, which is implicated in cell adhesion, innate immunity, and atherogenesis.31 Fusion of RET to the HOOK3 gene may lead to formation of an oncogene.32 Among the down-regulated genes, a few additional tumor suppressor genes were identified. Kank was found as a candidate tumor suppressor gene for renal cell carcinoma.33 Loss of Smad expression may be associated with development of some T-cell lymphomas.34 Smad3 expression may have a critical role in tumor suppression in the early stages of gastric carcinogenesis.35 ABI3BP (TARSH) mRNA expression is significantly down-regulated in all lung cancer cells and in follicular thyroid carcinoma.36,37 IGFBP-3 acts as tumor suppressor; IGFBP3 down-regulation is associated with shorter disease-specific survival in early non-small cell lung cancer and is an early event during head and neck carcinogenesis.38,39
In this study, we used normal peripheral blood and normal lymph node tissue samples as the control group to identify genomic alterations in PCNSL. Until now, there have been only 3 studies of gene expression profiling PCNSL, which have all used systemic DLBCL as a control comparison5-7 because one of major issues in studying PCNSL is characterizing how it differs from systemic DLBCL. These previous studies suggest that PCNSL is indeed significantly different from DLBCL.6,7 Compared with systemic DLBCL, PCNSL appears to have a specific signature in the differential expression of ECM- and adhesion-related pathways, including candidate genes, such as SPP1, DDR1, CXCL13, MUM1, TCL1A, and CHI3L1.6 PCNSL also has a higher expression of c-MYC and Pim-1, and it may be that disruption of the IL-4 signaling pathway involving JAK1 and STAT6 could potentially facilitate apoptosis in PCNSL.7 Furthermore, several genes are differentially expressed between PCNSL and systemic DLBCL, including IGM, FN, SERPINA3, SPP1, TF, and HBA1.5,40 In this study, we identified a few overlapped genes, including TACSTD1, FOXG1, SPP1, COL12A1, COL6A, FN, and JAK1, despite using normal lymphoid tissue as control. These genes may be key genes to understand PCNSL biology. This study suggests that JAK-STAT pathway may have a significant role in pathogenesis of PCNSL, in addition to an ECM and cell-adhesion related pathway. The results are in agreement with those of Rubenstein et al,7 who also identified increased expression of JAK-1, suggesting a significant role of the JAK-STAT pathway in the pathogenesis of PCNSL.
Recently, Richter et al41 reported that PCNSL and DLBCL do not differ in their DNA methylation pattern; however, they identified 194 genes differentially methylated between PCNSL and normal hematopoietic controls. Among 194 genes, there were several genes, specifically MYH11, IGFBP3, and DES, which overlapped with findings from our results examining differentially expressed genes. These genes were unmethylated in normal controls but methylated in PCNSL,41 and these genes were significantly down-regulated in our study.
However, there are many genes that do not overlap, which may be the result of the methodologic differences among studies. Montesinos-Rongen et al40 described several reasons for this, including different platforms used, different algorithms applied, different methods to eliminate the background signature, low number of cases, and sample-induced bias (resection vs biopsy). Besides these, different control samples used and heterogeneity of patients may also result in different genes found among the studies. For example, some patients with PCNSL were given steroid before biopsy because their condition was very serious and rapidly progressed, or malignant lymphoma was not suspected from either clinical or radiologic standpoints. Such glucocorticoid treatment could affect gene expression, especially of apoptosis-related genes associated with glucocorticoid. In the expression array analysis of the current study, 2 of 7 patients received steroid treatment before surgery. Although the whole gene expression pattern of these 2 patients showed no clustering distinct from the non–glucocorticoid-treated group, there may have been potential influence in gene expression, which will need to be evaluated in larger studies.
In conclusion, we identified pathways involved in signal transduction and cell adhesion that may contribute to PCNSL pathogenesis. By combining aCGH with expression array analysis, we identified several candidate genes involved in cell signaling, ECM activity, and cell adhesion, which may perform tumor suppressor gene functions in the pathogenesis of PCNSL. Although the molecular and cellular pathways that lead to PCNSL still remain largely unknown and existing evidence appears to be mixed, this study identified novel tumor suppressor genes that may serve as therapeutic targets of PCNSL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kyusam Choi of Samsung Cancer Research Institute in Seoul for his excellent technical support in real-time RT-PCR from formalin-fixed paraffin-embedded tissues.
This work was supported by the Samsung Biomedical Research Institute (grant SBRI C-A8-202), Korea Healthcare Technology R&D Project, and Ministry for Health & Welfare, Republic of Korea (A092255).
Authorship
Contribution: C.O.S. performed a pathology review and research, analyzed data, collected clinical data, and wrote the manuscript; S.C.K. analyzed data; S.K., K.K., and M.S. performed research and obtained samples; H.J.S. and D.-H.N. obtained samples and collected clinical data; S.J.K. and W.S.K. collected clinical data; Y.-L.S. and J.Y.K. performed a pathology review; S.-H.K. performed research; and Y.-H.K. designed research, performed a pathology review and research, analyzed data, obtained samples, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Young-Hyeh Ko, Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, 50 Irwon-dong Gangnam-gu, Seoul 135-710, South Korea; e-mail: yhko310@skku.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal