Abstract
Ontogenesis of T cells in the thymus is a complex process whose molecular control is poorly understood. The present study investigated microRNAs involved in human thymocyte differentiation by comparing the microRNA expression profiles of thymocytes at the double-positive, single-positive CD4+ and single-positive CD8+ maturation stages. Microarray analysis showed that each thymocyte population displays a distinct microRNA expression profile that reflects their developmental relationships. Moreover, analysis of small-RNA libraries generated from human unsorted and double-positive thymocytes and from mature peripheral CD4+ and CD8+ T lymphocytes, together with the microarray data, indicated a trend toward up-regulation of microRNA expression during T-cell maturation after the double-positive stage and revealed a group of microRNAs regulated during normal T-cell development, including miR-150, which is strongly up-regulated as maturation progresses. We showed that miR-150 targets NOTCH3, a member of the Notch receptor family that plays important roles both in T-cell differentiation and leukemogenesis. Forced expression of miR-150 reduces NOTCH3 levels in T-cell lines and has adverse effects on their proliferation and survival. Overall, these findings suggest that control of the Notch pathway through miR-150 may have an important impact on T-cell development and physiology.
Introduction
The development of T cells in the thymus is controlled by a complex signaling network, which has been the object of extensive studies.1,2 Based on the differential expression of the CD4 and CD8 coreceptors and their developmental stage, thymocytes can be subdivided into double-negative (DN), double-positive (DP), and single-positive (SP) CD4+ or CD8+ cells. During this maturation process, T-cell progenitors undergo rearrangement of T-cell receptor genes and alternate phases of intense selection and proliferation.3
MicroRNAs (miRs) are a class of small noncoding RNAs (∼ 22 nucleotides) that control gene expression at the post-transcriptional level by impairing translation or by promoting degradation of the target messenger RNA (mRNA).4–7 Given that the human genome codes for more than 1000 miRs (miRBase Release 16, September 2010), each with the potential to regulate the expression of multiple genes, the impact of the miR regulatory network on normal cell physiology is enormous. miR expression is dynamically regulated during hematopoiesis and the immune response and several miRs are already known to play essential roles in these processes.8–11 Furthermore, deregulated expression of specific miRs is associated with various diseases, including solid and hematopoietic tumors.12,13 So far, studies on the expression of miRs during T-cell maturation have been performed exclusively in mouse models.14,15 Nielson et al15 reported that specific miRs are enriched at distinct stages of thymocyte development in the mouse; this correlated with stage-specific depletion of transcripts harboring seed matches to these miRs.
Because little is known regarding the possible role of miRs during the physiologic development of human T cells, we investigated the miR expression profiles of human thymocytes at different stages of maturation: DP (CD4+CD8+), SP CD4+ (CD4+CD8−), and SP CD8+ (CD4−CD8+). In parallel, small-RNA libraries were generated from unsorted and DP thymocytes, as well as from mature CD4+ and CD8+ peripheral blood T lymphocytes. Comparison of the array and sequencing data allowed us to identify a group of miRs consistently regulated during normal T-cell maturation. miR-150, in particular, represents one of the top up-regulated miRs during human T-cell differentiation. In the mouse, miR-150 was demonstrated to be selectively expressed in mature naive B and T cells, being down-regulated in their progenitors and on lymphocyte activation.16–19 Although miR-150 has been extensively studied in B cells, its role in T cells is still poorly understood. In this work, we validated NOTCH3, a member of the Notch receptor family, as a new target of miR-150, confirmed this relationship in T acute lymphoblastic leukemia (T-ALL) cell lines and provided initial evidence that miR-150 levels can modulate proliferation and death of T-ALL cells.
Methods
Cell lines and in vitro culture conditions
The T-ALL cell lines MOLT-3, Jurkat, and CCRF-HSB-2 were purchased from ATCC. DND41 and TALL-1 cell lines were kindly provided by A. Ferrando (Columbia University). All T-ALL cell lines were cultured in RPMI 1640 (EuroClone) supplemented with 10% FCS (Invitrogen), 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (Cambrex Bioscience), and 2mM L-glutamine (complete RPMI). For serum starvation experiments, DND41 cells were cultured in RPMI 1640 supplemented with 1% FCS, 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, and 2mM L-glutamine. The human embryonic kidney epithelium cell line 293T was purchased from ATCC and cultured in Dulbecco modified Eagle medium (Sigma-Aldrich), supplemented with 10% FCS and 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid.
Purification of thymocyte subsets and mature peripheral T lymphocytes
After informed consent, thymus samples were obtained as surgical tissue discards from 35 pediatric patients, ranging in age from 2 days to 5 years, undergoing cardiac surgery at the Universitary Hospital of Padova. Thymocytes were isolated by cutting the thymic lobes into small pieces and forcing them through a plastic mesh. CD4+ SP (CD2+/CD4+/CD8−), CD8+ SP (CD2+/CD4−/CD8+), and CD4+/CD8+ DP (CD2+/CD4+/CD8+) thymocyte subsets were purified (> 95%) by cytofluorimetric cell sorting after triple labeling with anti-CD2 (CD2-PC5; Beckman Coulter), anti-CD4 (CD4-FITC; Beckman Coulter), and anti-CD8 (CD8-phycoerythrin; Beckman Coulter) antibodies. Cell sorting was performed on a FACSVantage Cell Sorter (BD Biosciences).
Peripheral blood mononuclear cells were isolated from the peripheral blood of healthy volunteers by Ficoll-Paque (GE Healthcare) density centrifugation. CD4+ and CD8+ T lymphocytes were isolated from peripheral blood mononuclear cell by immunomagnetic separation (Miltenyi Biotec) to a purity > 85%.
RNA extraction and quantitative RT-PCR
Total RNA was extracted using TRIzol Reagent (Invitrogen). Quantitative RT-PCR analysis of miR-150, miR-146b-5p, miR-146a, miR-151–5p, miR-23a, miR-342–3p, miR-155, miR-29b, miR-196b, miR-128, miR-17, miR-92, and RNU44 was performed using TaqMan MicroRNA Assays (Applied Biosystems). For NOTCH3 mRNA quantification, complementary deoxyribonucleic acid was synthesized from 0.5 to 1 μg of total RNA using Superscript II first-strand system (Invitrogen) for reverse transcription followed by quantitative PCR using SYBR Green (Invitrogen). miR and mRNA PCRs were performed in an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Results were analyzed using the ΔΔCt method with normalization against RNU44 and β2-microglobulin expression for miR and mRNA, respectively.
miR and mRNA expression profiling
miR expression profiles in normal human thymus tissue, purified DP, SP CD4+, and SP CD8+ human thymocytes were investigated with the Human miRNA Microarray kit Version 2.0 (Agilent Technologies), which allows detection of 723 known human and 76 human viral miRNAs (miRBase Version 10.1), as previously described.20 Six biologic replicates for each T-cell population were hybridized on the arrays. Total RNA was labeled with the miRNA Complete Labeling and Hyb Kit (Agilent Technologies); fluorescent signals were extracted and analyzed with Feature Extraction Software (Version 9.5.3.1; Agilent Technologies).
Gene expression profiles of DP and SP CD4+ human thymocytes were obtained by hybridization on Human genome U-133 PLUS microarrays (Affymetrix), as described in the supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Three to 6 biologic replicates for each sample were hybridized on the arrays. Gene expression data were extracted by GeneChip software (Affymetrix Microarray Suite 5.0 [MAS 5.0] software).
Microarray normalization and the identification of differentially expressed features were performed as detailed in the supplemental Methods. All microarray data are available on the Gene Expression Omnibus (National Center for Biotechnology Information) public database under accession number GSE26158.
Generation of small-RNA libraries
Small-RNA libraries were prepared from human DP thymocytes, total unsorted thymocytes, and peripheral mature CD4+ and CD8+ T lymphocytes. As detailed in supplemental Methods, the small-RNA fraction (18-26 nucleotides) was separated from total RNA, modified by the addition of 3′ and 5′ sequences, and converted to cDNAs using reverse transcriptase. Resulting cDNAs were PCR-amplified and sequenced according to the 454 massive sequencing-by-synthesis protocol (Roche Diagnostics, 454 Life Sciences). Bioinformatic analysis of sequence reads for identification and quantification of known miR sequences was performed with in-house developed procedures, as previously described,21 and are detailed in supplemental Methods.
Western blot analysis
Cell lysates were run on 4% to 12% gradient polyacrylamide gels; separated proteins were then blotted for 2 hours at 400 mA onto a nitrocellulose membrane. Blots were probed with rabbit polyclonal antibodies against human NOTCH3 (1:1000, Abcam), poly(adenosine 5′-diphosphate-ribose) polymerase (1:1000; Cell Signaling Technology), and β-actin (1:500; Sigma-Aldrich) or with a mouse monoclonal antibody recognizing α-tubilin (1:2000; Sigma-Aldrich), followed by hybridization with a horseradish peroxidase-conjugated anti–rabbit or anti–mouse antibody (1:5000; GE Healthcare). Blots were developed using a chemiluminescence detection kit (SuperSignal West Femto; Thermo Scientific). Signal intensities were measured using a Bio-Rad XRS imaging system.
Expression constructs and stable transduction
The hsa-miR-150 expression plasmid pCDNA_miR-150 was constructed by cloning a PCR-amplified fragment spanning the hsa-miR-150 hairpin (chromosome 19: 50003857-500043 49, minus strand; 493 bp) into pCRBlunt plasmid (Invitrogen) followed by subcloning into the cytomegalovirus promoter-driven plasmid pCDNA3.1 (Invitrogen). Primer sequences used for cloning are listed in supplemental Methods.
Lentiviral vectors expressing hsa-miR-150 and an anti-miR-150 shRNA (referred to in the text as LV-pre-miR-150 and LV-miRZip-150, respectively) and corresponding control vectors (pCDH-cytomegalovirus-MCS-EF1-copGFP and pGreenPuro Scramble Hairpin Control-Construct, referred to in the text as LV-control and LV-miRZiP-control, respectively) were purchased from SBI (System BioScience). All the vectors express the green fluorescent protein reporter gene under the control of the human EF1α gene promoter (LV-pre-miR-150 and LV-control) or the cytomegalovirus promoter (LV-miRZip-150 and LV-miRZip-control). T-ALL cell lines were seeded in a 6-well plate at 106 cells/mL in 1 mL of complete RPMI and transduced overnight with 1 × 109 transducing units of the lentiviral vectors produced in 293T cells as previously described.22
Cloning of 3′-UTR reporter constructs and reporter assays
NOTCH3 3′-untranslated region (UTR) segments (human NOTCH3, NM_000435; chromosome 19: 15270445-15271472; 1028 bp) were amplified by PCR from human genomic deoxyribonucleic acid and inserted into the pMIR-REPORT vector (Ambion) between the HindIII and SpeI sites, resulting in the pMIR-Notch3 plasmid. A derivative of pMIR-Notch3 containing a 4-nucleotide deletion in positions 3 to 6 of the miR-150 seed match complementary site was generated by PCR using the QuikChange II Site-Directed Mutagenesis Kit (Agilent); mutations were confirmed by deoxyribonucleic acid sequencing. The primers used for cloning, mutagenesis, and sequencing are listed in supplemental Methods.
293T cells were plated in 24-well plates at 1.5 × 105 cells/well and cotransfected using Lipofectamine 2000 (Invitrogen) with the pMIR-REPORT constructs (300 ng) along with 550 ng of pCDNA_miR-150 or control plasmid (pCDNA3.1, referred to in the text as pCDNA_empty). The pSV-β-galactosidase control plasmid (150 ng; Promega) was used to normalize transfection efficiency. In the experiments performed with the lentiviral vectors, 293T cells were transduced in vitro with the LV-pre-miR-150 or LV-control virus (108 transducing units) 48 hours before transfection with the pMIR-REPORT constructs. Thirty hours after transfection, luciferase and β-galactosidase activities were measured in triplicate using the Britelite Plus Kit (PerkinElmer) and Beta-Glo assay system (Promega), respectively.
To test the effect of miR-150 knockdown on expression of pMIR-REPORT constructs or on the endogenous levels of NOTCH3 protein, 293T and T-ALL cells were first transduced with LV-miRZip-150 or the LV-miRZip-control and then retransduced 48 hours later with the lentiviral vector expressing miR-150 or control vector (supplemental Figures 3-4).
Methods to measure cellular proliferation and apoptosis, and computational analysis are reported as supplemental Methods.
Statistical analysis
Results were expressed as mean value ± SD. Statistically significant differences were investigated by Student t test or Mann-Whitney Rank-Sum test, as indicated in the text. Differences were considered statistically significant when P < .05.
Results
Dynamic regulation of miRs during human thymocyte differentiation
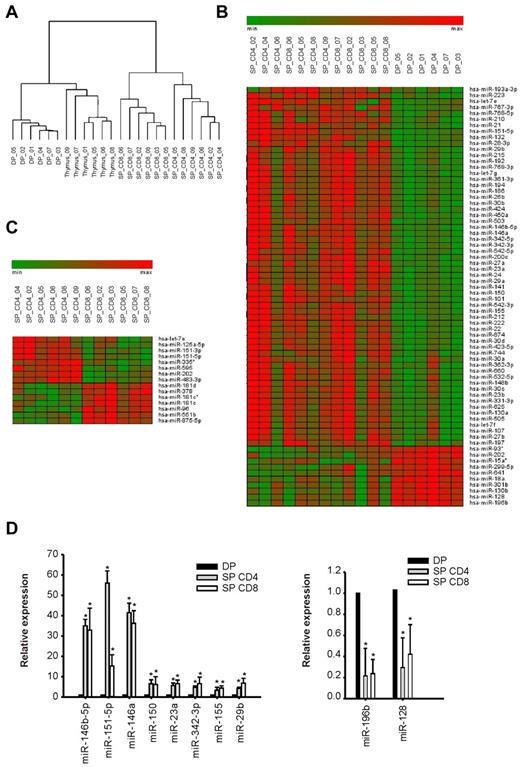
The miR expression profile of unsorted human thymocytes and purified thymocyte populations at 3 different stages of maturation (DP, SP CD4+, SP CD8+) was analyzed by hybridization of total RNA to microarrays (Agilent Human miRNA microarray kit Version 2.0). Hierarchical clustering (Figure 1A) indicated that each thymic population was characterized by a distinct miR expression pattern, which reflected the developmental relationships across maturation stages in T precursors. In particular, DP thymocytes showed a miR profile quite distinct from those of CD4+ and CD8+ SP cells; on the other hand, expression profiles of the 2 SP populations showed important similarities. In accordance with the heterogeneous cellular composition of the thymus, unsorted thymocytes also presented a characteristic miR expression profile (Figure 1A).
miR expression during human thymocyte development. miR expression profiles were generated as described in “miR and mRNA expression profiling” from a dataset consisting of 6 samples each of normal thymus tissue, DP thymocytes, CD4+ SP thymocytes (SP_ CD4), and CD8+ SP thymocytes (SP_CD8). (A) Dendrogram based on miR expression profiles of thymic populations showing the hierarchical clustering of biologic samples. (B-C) Heatmap representations of feature expression, limiting to the subsets of miRs that were differentially expressed in the following comparisons: DP versus SP thymocytes (B) and SP CD4+ versus SP CD8+ thymocytes (C). Differentially expressed features were identified by Significance Analysis of Microarray analysis23 with false discovery rate = 0.001; the heatmaps were generated with the Genesis software package (BRB array tools ver. 3.6 Genesis 1.7.2).24 (D) Validation of miR expression profiles in thymocyte subpopulations by quantitative RT-PCR (P < .05; t test). Reported are results of TaqMan quantitative RT-PCR to detect miRs chosen among the top-regulated miRs during thymocyte transition from DP to SP stage. The level of expression of was normalized to that of the small noncoding RNA RNU44. Plotted are the average values ± SD of 3 independent experiments from 3 biologic samples for each population.
miR expression during human thymocyte development. miR expression profiles were generated as described in “miR and mRNA expression profiling” from a dataset consisting of 6 samples each of normal thymus tissue, DP thymocytes, CD4+ SP thymocytes (SP_ CD4), and CD8+ SP thymocytes (SP_CD8). (A) Dendrogram based on miR expression profiles of thymic populations showing the hierarchical clustering of biologic samples. (B-C) Heatmap representations of feature expression, limiting to the subsets of miRs that were differentially expressed in the following comparisons: DP versus SP thymocytes (B) and SP CD4+ versus SP CD8+ thymocytes (C). Differentially expressed features were identified by Significance Analysis of Microarray analysis23 with false discovery rate = 0.001; the heatmaps were generated with the Genesis software package (BRB array tools ver. 3.6 Genesis 1.7.2).24 (D) Validation of miR expression profiles in thymocyte subpopulations by quantitative RT-PCR (P < .05; t test). Reported are results of TaqMan quantitative RT-PCR to detect miRs chosen among the top-regulated miRs during thymocyte transition from DP to SP stage. The level of expression of was normalized to that of the small noncoding RNA RNU44. Plotted are the average values ± SD of 3 independent experiments from 3 biologic samples for each population.
Significance Analysis of Microarray analysis23 of the miR expression levels (supplemental Methods) revealed that 70 miRs were differentially regulated in SP compared with DP thymocytes (60 up-regulated and 10 down-regulated). Interestingly, a general up-regulation was observed in the maturation of thymocytes from the DP to the SP stage (Figure 1B). As suggested by the array data, the 5 most up-regulated miRs in SP thymocytes were miR-146b, miR-151–5p, miR-146a, miR-132, and miR-150, whereas the 5 most down-regulated were miR-196b, miR-128, miR-15a*, miR-301b, and miR-130b (supplemental Figure 1A). Comparison of the CD4+ and CD8+ SP populations revealed 15 differentially regulated miRs (Figure 1C); in particular, 8 miRs were up-regulated in CD4+ cells compared with CD8+ (miR-151–3p, miR-151–5p, let-7e, miR-595, miR-125a-5p, miR-335*, miR-202, miR-483–3p), and 7 were down-regulated (miR-181d, miR-378, miR-181c, miR-96, miR-181c*, miR-551b, miR-875–5p) (Figure 1C; supplemental Figure 1B).
For experimental validation, we selected 10 miRs among those whose expression changed most dramatically in the transition from the DP to SP stage: miR-146b, miR-151–5p, miR-146a, miR-150, miR-23a, miR-342–3p, miR-155, miR-29b (up-regulated in SP vs DP), and miR-196b, miR-128 (down-regulated in SP vs DP). RT-PCR measurements confirmed significant differences (P < .05, t test) in the expression levels of these miRs comparing DP with SP CD4+ and SP CD8+ thymocytes (Figure 1D). Because fold changes calculated from RT-PCR measurements were in close agreement with those derived from the expression levels monitored by microarrays and Pearson analysis test revealed a very high correlation (r = 0.987, P < .01; supplemental Figure 2), we could trust that chips reflected the real ranking of miR expression levels.
Analysis of small-RNA libraries from T lymphoid subpopulations
To further confirm the consistency of the data from miR microarrays findings and obtain absolute quantification of miR frequencies in T-cell populations, we generated and analyzed small-RNA libraries from unsorted human thymocytes, DP thymocytes, and mature peripheral CD4+ and CD8+ T lymphocytes. Unfortunately, the large amount of biologic material required for the generation of small-RNA libraries precluded the analysis of the human SP thymocyte populations. Nevertheless, the inclusion in this study of human mature circulating T lymphocytes allowed us to identify a group of individual miRs, which are consistently up-regulated/down-regulated in T cells committed to the periphery, and also to compare our data with the results obtained by other groups in the mouse model from analogous T-cell populations using similar techniques.25–27
Mass sequencing of the libraries yielded 23 006 sequences ranging in length from 18 to 26 nucleotides (Table 1). Overall, the estimate of library complexity, performed as described by Basso et al,20 exceeded 80% for all the libraries (Figure 2A). A total of 139 known miRs were identified among the sequence reads, as described in supplemental Methods. Some “dominant” miRs were expressed ubiquitously and at high frequencies in all the T-cell subsets analyzed (Figure 2A). These included miR-142–3p and miR-16, whose frequencies exceeded, respectively, 40% and 5% of the entire pool of known miRs in the 4 libraries. Three miRs (ie, miR-150, miR-21, and let-7a) showed remarkably high frequencies only in mature peripheral CD4+ and CD8+ T lymphocytes while being expressed at very low levels in both unsorted and DP thymocytes (Figure 2A).
miRs identified by sequencing of small-RNA libraries
| . | CD4+ T lymphocytes . | CD8+ T lymphocytes . | DP thymocytes . | Unsorted thymocytes . |
|---|---|---|---|---|
| Total no. of sequences | 7709 | 5323 | 9974 | 6738 |
| Total no. of mature miR sequences | 144 | 132 | 147 | 120 |
| Total no. of known miRs (≥ 1 count) | 99 | 88 | 86 | 89 |
| . | CD4+ T lymphocytes . | CD8+ T lymphocytes . | DP thymocytes . | Unsorted thymocytes . |
|---|---|---|---|---|
| Total no. of sequences | 7709 | 5323 | 9974 | 6738 |
| Total no. of mature miR sequences | 144 | 132 | 147 | 120 |
| Total no. of known miRs (≥ 1 count) | 99 | 88 | 86 | 89 |
Results of mass sequencing and bioinformatic analysis of the 4 small-RNA libraries generated in this study.
Small-RNA libraries. (A) Distribution of the 5 most abundant miRs in the T lymphoid subsets. The sequencing frequency of each miR is shown as the percentage of the total number of known miRs isolated from each T-cell population. Percentages in parentheses on the x-axis report the coverage of each library, estimated by bootstrap analysis as described by Basso et al.20 (B) Top 15 up-regulated and 10 down-regulated miRs in peripheral CD4+ and CD8+ T lymphocytes compared with DP thymocytes, based on the sequencing data (P < .05; Fisher Exact Test). Histograms report the relative frequency of each miR in the 3 libraries. The value of miR frequency in the DP population was set to 1. (C) miRs consistently regulated during T-cell maturation after the DP stage, identified by comparing data from miR microarrays and small-RNA libraries. The table reports the 18 miRs found to be concordantly regulated in the comparison of SP vs DP thymocytes (from microarrays) and mature T lymphocytes vs DP (from small-RNA libraries).
Small-RNA libraries. (A) Distribution of the 5 most abundant miRs in the T lymphoid subsets. The sequencing frequency of each miR is shown as the percentage of the total number of known miRs isolated from each T-cell population. Percentages in parentheses on the x-axis report the coverage of each library, estimated by bootstrap analysis as described by Basso et al.20 (B) Top 15 up-regulated and 10 down-regulated miRs in peripheral CD4+ and CD8+ T lymphocytes compared with DP thymocytes, based on the sequencing data (P < .05; Fisher Exact Test). Histograms report the relative frequency of each miR in the 3 libraries. The value of miR frequency in the DP population was set to 1. (C) miRs consistently regulated during T-cell maturation after the DP stage, identified by comparing data from miR microarrays and small-RNA libraries. The table reports the 18 miRs found to be concordantly regulated in the comparison of SP vs DP thymocytes (from microarrays) and mature T lymphocytes vs DP (from small-RNA libraries).
Statistically significant differences in miR expression between mature T lymphocytes and DP T cells were assessed by applying the Fisher Exact test to the sequence read counts of known miRs. Comparison of mature CD4+ and CD8+ T cells to DP cells yielded 50 and 48 miRs characterized by significant differences in counts, respectively (P < .05). Forty-one of these miRs were concordantly regulated (30 up-regulated and 11 down-regulated) in both mature lymphocyte populations compared with DP thymocytes, confirming the general up-regulation of miR expression during T-cell maturation previously observed by microarray analysis (Figure 2B).
Eighteen of these significantly up- and down-regulated miRs matched those identified as differentially expressed in SP compared with DP thymocytes on the basis of microarray expression data. This group of miRs was differentially expressed in the thymus during the DP to SP transition, and based on library results appeared to be modulated in the same direction in peripheral T lymphocytes (Figure 2C). The list of differentially expressed miRs identified by both approaches included miR-150, miR-146a, miR-146b (up-regulated), and miR128 (down-regulated), which were among the 5 miRs whose expression showed the greatest change in the transition from the DP to SP stage of T-cell development according to the microarray data. These results suggested that this group of miRs could play an important role in the maturation of T-cell precursors from the DP stage to subsequent stages of development.
Combined analysis of differentially expressed miRs and mRNAs
Because miR-mRNA interactions may lead to degradation of the mRNA, integration of gene expression data with miR expression profiles can facilitate identification of genuine mRNA targets from the long list of potential targets provided by prediction algorithms.15,28 To exploit this approach, we analyzed the mRNA expression profiles of T cells during their maturation from DP to SP CD4+ cells using the HG-U133 Plus 2.0 platform (Affimetrix): 702 genes were identified as up-regulated in SP compared with DP, whereas 351 were down-regulated (supplemental Table 1). The Functional Annotation Clustering tool implemented in the DAVID website (www.david.abcc.ncifcrf.gov) confirmed that a high number of these differentially expressed genes were involved in T-cell activation and differentiation (Table 2).
Functional Annotation Clustering of differentially expressed genes
| Annotation term . | P . | False discovery rate . |
|---|---|---|
| Annotation cluster 1 Enrichment score: 9.08 | ||
| Cell activation | 3.3 × 10−13 | 6.0 × 10−10 |
| Leukocyte activation | 8.6 × 10−13 | 1.5 × 10−9 |
| Lymphocyte activation | 1.1 × 10−11 | 2.0 × 10−8 |
| Lymphocyte differentiation | 2.3 × 10−11 | 4.1 × 10−8 |
| Leukocyte differentiation | 3.8 × 10−11 | 6.9 × 10−8 |
| Hematopoietic or lymphoid organ development | 1.4 × 10−10 | 2.6 × 10−7 |
| Immune system development | 9.0 × 10−10 | 1.6 × 10−6 |
| Hemopoiesis | 1.6 × 9−13 | 2.8 × 10−6 |
| T-cell activation | 1.1 × 10−8 | 2.0 × 10−5 |
| T-cell differentiation | 1.1 × 10−8 | 2.0 × 10−5 |
| B-cell activation | 1.9 × 10−5 | 3.4 × 10−2 |
| B-cell differentiation | 9.9 × 10−5 | 1.8 × 10−1 |
| Annotation cluster 2 Enrichment score: 8.09 | ||
| Immunoglobulin-like | 2.5 × 10−15 | 3.9 × 10−12 |
| Immunoglobulin-like fold | 3.0 × 10−15 | 4.8 × 10−12 |
| Immunoglobulin domain | 5.6 × 10−11 | 8.1 × 10−8 |
| Immunoglobulin V-set | 2.1 × 10−4 | 3.4 × 10−1 |
| Immunoglobulin subtype | 3.0 × 10−4 | 4.8 × 10−1 |
| Immunoglobulin | 1.1 × 10−2 | 1.3 × 10 |
| Annotation term . | P . | False discovery rate . |
|---|---|---|
| Annotation cluster 1 Enrichment score: 9.08 | ||
| Cell activation | 3.3 × 10−13 | 6.0 × 10−10 |
| Leukocyte activation | 8.6 × 10−13 | 1.5 × 10−9 |
| Lymphocyte activation | 1.1 × 10−11 | 2.0 × 10−8 |
| Lymphocyte differentiation | 2.3 × 10−11 | 4.1 × 10−8 |
| Leukocyte differentiation | 3.8 × 10−11 | 6.9 × 10−8 |
| Hematopoietic or lymphoid organ development | 1.4 × 10−10 | 2.6 × 10−7 |
| Immune system development | 9.0 × 10−10 | 1.6 × 10−6 |
| Hemopoiesis | 1.6 × 9−13 | 2.8 × 10−6 |
| T-cell activation | 1.1 × 10−8 | 2.0 × 10−5 |
| T-cell differentiation | 1.1 × 10−8 | 2.0 × 10−5 |
| B-cell activation | 1.9 × 10−5 | 3.4 × 10−2 |
| B-cell differentiation | 9.9 × 10−5 | 1.8 × 10−1 |
| Annotation cluster 2 Enrichment score: 8.09 | ||
| Immunoglobulin-like | 2.5 × 10−15 | 3.9 × 10−12 |
| Immunoglobulin-like fold | 3.0 × 10−15 | 4.8 × 10−12 |
| Immunoglobulin domain | 5.6 × 10−11 | 8.1 × 10−8 |
| Immunoglobulin V-set | 2.1 × 10−4 | 3.4 × 10−1 |
| Immunoglobulin subtype | 3.0 × 10−4 | 4.8 × 10−1 |
| Immunoglobulin | 1.1 × 10−2 | 1.3 × 10 |
The list of differentially expressed genes was used as input for the Functional Annotation Clustering function of DAVID Bioinformatics Resources 6.7.29 The 2 most relevant clusters are reported.
We then used the miRanda30 and TargetScan31 software packages to predict targets of miRs that were differentially expressed in SP CD4+ versus DP cells. Comparison of the list of targets identified by both algorithms with the subset of genes observed to be differentially expressed during T-cell maturation yielded a group of candidate targets whose expression was inversely correlated to that of related miRs (supplemental Table 2). Functional Annotation Clustering performed on this restricted subset of targets revealed an enrichment in genes involved in regulation of the cell cycle and mitosis (supplemental Table 3). Several of these targets were of potential interest because of their known role in the control of T-cell survival, proliferation, and differentiation as well as neoplastic transformation, including NOTCH3,32 BCL2L1,33 CDKN2D,34 and NFKB1.35
Identification of NOTCH3 as a new target of miR-150
Combined analysis of differentially expressed miRs and mRNAs and predictions from MiRanda and TargetScan identified NOTCH3 as a potential target of miR-150, one of the miRs that showed the strongest up-regulation during human T-cell development in our screening (Figure 3A). NOTCH3 is a member of the Notch family of receptors, which plays an important role in both T-cell differentiation and leukemogenesis.32,36 Data from gene expression arrays indicating that NOTCH3 mRNA decreased in abundance with T-cell maturation from DP to SP CD4+ (ie, in a trend opposite to that of miR-150; Figure 3B) were confirmed by quantitative RT-PCR, which revealed down-regulation of the mRNA in both SP CD4+ and SP CD8+ thymocytes (P < .05; Figure 3C). As expected, NOTCH3 protein levels also decreased in the transition from DP to SP human thymocytes (Figure 3D).
NOTCH3 is a target of miR-150. (A) Sequence alignment between miR-150 and the 3′-UTR of NOTCH3 in human and mouse, as predicted by miRanda algorithm.30 Solid line represents seed match region; and dashed line, deleted region. (B) Relative expression of miR-150 (left) and NOTCH3 mRNA (right) in DP vs SP CD4+ thymocytes, based on microarray data. The histogram reports the relative fold change (FC) in miR-150 and NOTCH3 in SP CD4+ compared with DP thymocytes. (C) RT-PCR for NOTCH3 in human thymocyte subsets DP, SP CD4+, and SP CD8+ (P < .05; t test). The level of expression was normalized to that of the housekeeping gene β2-microglobulin. Shown are the average values ± SD of 2 independent experiments. (D) Western blot to detect NOTCH3 protein in human DP, SP CD4+, and SP CD8+ thymocytes. β-actin was used as a loading control. (E) Luciferase assay using LV-pre-miR-150/LV-control vectors. 293T cells were transduced with a lentiviral vector expressing pre-miR-150 (LV-pre-miR-150) or with a control vector (LV-control). After 48 hours, cells were cotransfected with a β-galactosidase expression plasmid and a luciferase reporter plasmid containing the human NOTCH3 3′-UTR (pMIR-NOTCH3) or with the parental pMIR-REPORT plasmid (pMIR-empty). (F) Luciferase assay using pCDNA_miR-150/pCDNA_empty. The 293T cells were cotransfected with the pCDNA_miR-150 expression plasmid, a β-galactosidase expression plasmid, and a luciferase reporter plasmid containing the human NOTCH3 3′-UTR (pMIR-NOTCH3), no 3′-UTR (pMIR-empty), or the miR-150-binding site-deleted human NOTCH3 3′-UTR (pMIR-NOTCH3DEL). PCDNA3.1 plasmid lacking the miR-150 fragment (pCDNA_empty) was used as a negative control. Luciferase activities were measured 30 hours after transfection and normalized to the corresponding β-galactosidase activity. Each experiment was repeated at least 3 times. *P < .05 (t test). **P < .01 (t test). ***P < .001 (t test).
NOTCH3 is a target of miR-150. (A) Sequence alignment between miR-150 and the 3′-UTR of NOTCH3 in human and mouse, as predicted by miRanda algorithm.30 Solid line represents seed match region; and dashed line, deleted region. (B) Relative expression of miR-150 (left) and NOTCH3 mRNA (right) in DP vs SP CD4+ thymocytes, based on microarray data. The histogram reports the relative fold change (FC) in miR-150 and NOTCH3 in SP CD4+ compared with DP thymocytes. (C) RT-PCR for NOTCH3 in human thymocyte subsets DP, SP CD4+, and SP CD8+ (P < .05; t test). The level of expression was normalized to that of the housekeeping gene β2-microglobulin. Shown are the average values ± SD of 2 independent experiments. (D) Western blot to detect NOTCH3 protein in human DP, SP CD4+, and SP CD8+ thymocytes. β-actin was used as a loading control. (E) Luciferase assay using LV-pre-miR-150/LV-control vectors. 293T cells were transduced with a lentiviral vector expressing pre-miR-150 (LV-pre-miR-150) or with a control vector (LV-control). After 48 hours, cells were cotransfected with a β-galactosidase expression plasmid and a luciferase reporter plasmid containing the human NOTCH3 3′-UTR (pMIR-NOTCH3) or with the parental pMIR-REPORT plasmid (pMIR-empty). (F) Luciferase assay using pCDNA_miR-150/pCDNA_empty. The 293T cells were cotransfected with the pCDNA_miR-150 expression plasmid, a β-galactosidase expression plasmid, and a luciferase reporter plasmid containing the human NOTCH3 3′-UTR (pMIR-NOTCH3), no 3′-UTR (pMIR-empty), or the miR-150-binding site-deleted human NOTCH3 3′-UTR (pMIR-NOTCH3DEL). PCDNA3.1 plasmid lacking the miR-150 fragment (pCDNA_empty) was used as a negative control. Luciferase activities were measured 30 hours after transfection and normalized to the corresponding β-galactosidase activity. Each experiment was repeated at least 3 times. *P < .05 (t test). **P < .01 (t test). ***P < .001 (t test).
The interaction between miR-150 and NOTCH3 was further investigated in 293T cells transfected with constructs containing the firefly luciferase gene fused to the 3′-UTR of NOTCH3 (pMIR-NOTCH3). The 293T cell line was chosen in view of its low expression of endogenous miR-150. Figure 3E shows results of assays in which the reporter plasmids were transiently transfected into 293T cells previously transduced with a lentiviral vector overexpressing miR-150 (LV-pre-miR-150) or with a control vector (LV-control). Cells transduced with the LV-pre-miR-150 vector showed a 6-fold reduction in luciferase expression compared with cells transduced with LV-control (P < .001), whereas expression of luciferase from the parental pMIR-REPORT plasmid lacking the NOTCH3 3′-UTR (pMIR-empty) was not affected. The inhibitory effect of miR-150 on NOTCH3 3′-UTR was reversed by cotransducing a lentiviral vector producing a shRNA against miR-150 (LV-miRZip-150; supplemental Figure 3).
Similar results were obtained after transfection of 293T cells with the luciferase reporter plasmid pMIR-Notch3 and either a miR-150-expression plasmid (pCDNA_miR-150) or a control plasmid (pCDNA_empty). Suppression of luciferase expression was relieved using the luciferase construct lacking the NOTCH3 3′-UTR (pMIR-empty; P < .01) or with a deletion of the miR-150 binding site in the NOTCH3 3′-UTR (pMIR-Notch3DEL; P < .05), confirming the specific interaction between this target and the miR (Figure 3F).
miR-150 regulates NOTCH3 expression in T-ALL cell lines
To investigate whether the NOTCH3/miR-150 interaction was also operative in a T-cell system, we analyzed miR-150 expression levels in the T-ALL cell lines Jurkat, MOLT-3, DND41, CCRF-HSB-2, and TALL-1. RT-PCR assays revealed that miR-150 was almost undetectable in Jurkat, MOLT-3, and DND41 and was present at very low levels in CCRF-HSB-2 and TALL-1 (supplemental Figure 4). To address the effect of miR-150 on NOTCH3, we transduced Jurkat, MOLT-3, and DND41 T-ALL cells with the lentiviral vector expressing miR-150 (LV-pre-miR-150) or with the control vector (LV-control). RT-PCR assays confirmed that expression of miR-150 was increased in all 3 cell lines on transduction with LV-pre-miR-150, although fold induction levels varied depending on the cell line (Figure 4A). Increased expression of miR-150 was accompanied by a 20% to 40% decrease in the levels of full-length NOTCH3 protein in all 3 cell lines (Figure 4B). A control experiment performed in DND41 cells confirmed restoration of NOTCH3 protein expression when the cells were cotransduced with the anti-miR-150 shRNA vector (LV-miRZip-150; Figure 4C). As shown in Figure 4D, forced expression of miR-150 was not associated with a significant decrease in the expression of the NOTCH3 transcript (Figure 4D). This observation suggests that miR-150 specifically represses NOTCH3 expression at the level of translation rather than mRNA stability in the context of these T-ALL cell lines.
The miR 150/NOTCH3 interaction is operative in T-ALL cell lines. (A) RT-PCR to detect miR-150 in the T-ALL cell lines Jurkat, MOLT-3, and DND41 72 hours after transduction with the lentiviral vector LV-pre-miR-150 or LV-control. The level of expression was normalized to that of the small noncoding RNA RNU44. The figure shows the average of 3 independent experiments plus or minus SD. (B) Western blot showing that miR-150 overexpression induces a decrease in the levels of endogenous full-length (FL) NOTCH3 protein in the T-ALL cell lines. α-tubulin was used as a loading control. The numbers below the bands indicate the relative ratio between the NOTCH3 and the α-tubulin bands, normalized by setting the value obtained for the controls at 1. (C) Western blot showing that miRZip anti-miR-150 vector (LV-miRZip-150) prevents attenuation of NOTCH3 protein levels in DND41 cells cotransduced with LV-pre-miR-150. β-actin was used as a loading control. The numbers below the bands indicate the relative ratio between the NOTCH3 and the β-actin bands, normalized by setting the value obtained for the untreated cells at 1. (D) RT-PCR to detect NOTCH3 mRNA in the T-ALL cell lines Jurkat, MOLT-3, and DND41 72 hours after transduction with the lentiviral vector LV-pre-miR-150 or LV-control. The level of expression was normalized to that of the housekeeping gene β2-microglobulin. The figure shows the average of 3 independent experiments plus or minus SD. *P < .05 (t test). **P < .001 (t test).
The miR 150/NOTCH3 interaction is operative in T-ALL cell lines. (A) RT-PCR to detect miR-150 in the T-ALL cell lines Jurkat, MOLT-3, and DND41 72 hours after transduction with the lentiviral vector LV-pre-miR-150 or LV-control. The level of expression was normalized to that of the small noncoding RNA RNU44. The figure shows the average of 3 independent experiments plus or minus SD. (B) Western blot showing that miR-150 overexpression induces a decrease in the levels of endogenous full-length (FL) NOTCH3 protein in the T-ALL cell lines. α-tubulin was used as a loading control. The numbers below the bands indicate the relative ratio between the NOTCH3 and the α-tubulin bands, normalized by setting the value obtained for the controls at 1. (C) Western blot showing that miRZip anti-miR-150 vector (LV-miRZip-150) prevents attenuation of NOTCH3 protein levels in DND41 cells cotransduced with LV-pre-miR-150. β-actin was used as a loading control. The numbers below the bands indicate the relative ratio between the NOTCH3 and the β-actin bands, normalized by setting the value obtained for the untreated cells at 1. (D) RT-PCR to detect NOTCH3 mRNA in the T-ALL cell lines Jurkat, MOLT-3, and DND41 72 hours after transduction with the lentiviral vector LV-pre-miR-150 or LV-control. The level of expression was normalized to that of the housekeeping gene β2-microglobulin. The figure shows the average of 3 independent experiments plus or minus SD. *P < .05 (t test). **P < .001 (t test).
miR-150 expression exerts proapoptotic and antiproliferative effects in T-ALL cells
Having confirmed that miR-150 targets NOTCH3 in T-ALL cells, we next examined the impact of forced expression of this miR on cell proliferation and death. Transduction of Jurkat, MOLT-3, and DND41 cells with the LV-pre-miR-150 vector caused a significant reduction in cell numbers, measured 72 hours after infection (P < .05; t test; Figure 5A). In Jurkat and MOLT-3 cells, this effect was associated with the induction of apoptosis, measured as an increase in caspase 3/7 activity (Figure 5B left and middle). Although forced expression of miR-150 did not induce apoptosis of DND41 cells under standard culture conditions (10% FCS; Figure 5B right), it indeed reduced the proliferation rate of the cells, as detected by thymidine incorporation assay (supplemental Figure 5). Cultivation of DND41 cells under stress conditions (ie, serum starvation; 1% FCS) revealed a clear increase in apoptosis in miR-150-transduced cells (Figure 5B right). In this set of experiments, anisomycin-treated cells were used as a positive control for apoptosis induction. These findings were further confirmed on both Jurkat and DND41 cell lines by the increase of the cleaved form of poly(adenosine 5′-diphosphate-ribose) polymerase, detected by Western blot, in LV-pre-miR-150 transduced cells compared with controls (Figure 5C). Taken together, these experiments suggest that the up-regulation of miR-150 can exert proapoptotic and antiproliferative effects in T-ALL cells.
Functional effects of miR-150 expression in T-ALL cells. (A) Relative cell numbers of Jurkat, MOLT-3, and DND41 cells 72 hours after transduction with the lentiviral vector LV-pre-miR-150 or LV-control (P < .05; t test). The relative cell numbers were calculated by setting the value of the control at 1. The figure shows the average of 4 independent experiments ± SD. (B) Caspase 3/7 Activity Test on Jurkat, MOLT-3, and DND41 cells 72 hours after transduction with the lentiviral vector LV-pre-miR-150 or LV-control. As a positive control for activated caspase activity, we treated T-ALL cells with the protein synthesis inhibitor anisomycin (2 μg/mL) for 24 hours. In standard culture conditions (10% FCS), miR-150 overexpression induces a significant increase of caspase activity in Jurkat and MOLT-3 T-ALL cell lines. In the DND41 cell line, miR-150-forced expression induces an increase in the caspase activity only when the cells are cultured in serum deprivation (1% FCS), although it is not detectable in normal serum conditions (10% FCS). The level of caspase activity was expressed in terms of absolute intensity of luminescence measured in counts per second (cps). The figure shows the result of one representative experiment of 2 for each cell line. (C) Western blot analysis to detect uncleaved and cleaved poly(adenosine 5′-diphosphate-ribose) polymerase in lysates of Jurkat and DND41 cells treated as described in panel B.
Functional effects of miR-150 expression in T-ALL cells. (A) Relative cell numbers of Jurkat, MOLT-3, and DND41 cells 72 hours after transduction with the lentiviral vector LV-pre-miR-150 or LV-control (P < .05; t test). The relative cell numbers were calculated by setting the value of the control at 1. The figure shows the average of 4 independent experiments ± SD. (B) Caspase 3/7 Activity Test on Jurkat, MOLT-3, and DND41 cells 72 hours after transduction with the lentiviral vector LV-pre-miR-150 or LV-control. As a positive control for activated caspase activity, we treated T-ALL cells with the protein synthesis inhibitor anisomycin (2 μg/mL) for 24 hours. In standard culture conditions (10% FCS), miR-150 overexpression induces a significant increase of caspase activity in Jurkat and MOLT-3 T-ALL cell lines. In the DND41 cell line, miR-150-forced expression induces an increase in the caspase activity only when the cells are cultured in serum deprivation (1% FCS), although it is not detectable in normal serum conditions (10% FCS). The level of caspase activity was expressed in terms of absolute intensity of luminescence measured in counts per second (cps). The figure shows the result of one representative experiment of 2 for each cell line. (C) Western blot analysis to detect uncleaved and cleaved poly(adenosine 5′-diphosphate-ribose) polymerase in lysates of Jurkat and DND41 cells treated as described in panel B.
Discussion
The present study profiled miR expression in human T-cell populations from sequential stages of maturation using microarrays and small-RNA libraries. To our knowledge, this is the first comprehensive analysis of miR regulation performed in human thymocytes as all previous studies were conducted either in mouse models or in peripheral T lymphocytes at later stages of development.
Our results indicate that each human thymocyte subpopulation (DP, SP CD4+, SP CD8+) is characterized by a distinct miR expression profile, with the pattern of miR expression reflecting the developmental relationship between maturation stages (Figure 1). We detected a general trend of up-regulated miR expression in the maturation from the DP to SP stage of thymocyte development, a property that was also observed in the comparison between DP thymocytes and peripheral T lymphocytes and was common to both SP subsets. Furthermore, the level of up-regulation of miRs generally exceeded that of down-regulation in terms of fold change. This result is consistent with previous studies that correlated miR levels to cell differentiation37 and suggests that miR up-regulation in this context could be important for down-regulating genes associated with an immature phenotype. Moreover, because immature thymocytes at the DP stage are subjected to both positive and negative selection through T-cell receptor-activated apoptotic/survival signals, the up-regulation of miRs during the maturation to SP cells could also play a role in suppressing genes involved in this selection process.38 In support of these considerations, among the miRs that are highly modulated in the transition from DP to SP T cells, we indeed found some that had previously been reported to play an important role in the differentiation of the lymphoid (miR-150, miR-155, miR-23a) or myeloid (miR-146, miR-223) hematopoietic lineages,13,18,39 and in the regulation of cellular proliferation or apoptosis (miR-150, miR-23a, miR-27a, miR-24).40,41
Besides some similarities, we observed some important differences in the regulation of miRs during thymocyte maturation in humans compared with the mouse model. In particular, Neilson et al15 reported the miR-181 family to be highly expressed in the thymus and specifically enriched in mouse DP thymocytes. Surprisingly, we did not find differential expression of any member of the miR-181 family in the transition from DP to SP thymocytes. However, our analysis of small-RNA libraries identified 2 members of this family (miR-181a and miR-181b) among the most down-regulated miRs in human mature peripheral T lymphocytes compared with DP thymocytes; moreover, both miRs were also detected at high levels in unsorted human thymocytes. These observations suggest that human miR-181 is highly expressed both in DP and SP thymocyte subpopulation, being strongly down-regulated only when mature T cells egress the thymus and migrate to the periphery.
Our small-RNA libraries highlighted the existence of some “dominant” miRs (ie, miR-142–3p and miR-16) abundant in thymocytes as well as mature peripheral T cells and others that are up-regulated in the mature peripheral populations (ie, miR-150, miR-21, and let-7a; Figure 2A). This is consistent with previous studies showing that some miRs are dominantly expressed in specific cell types and tissues.26,27
By comparing the array and sequencing data, we identified a group of 18 miRs, including miR-150, which are consistently up- or down-regulated during the DP-to-SP transition. Their continued up- or down-modulation in T cells that have migrated from the thymus to the periphery suggests that these miRs could play a particularly important role in T development starting from the DP stage through subsequent stages of T-cell maturation.
Our integrated analysis of candidate miR targets and miR versus gene expression data facilitated identification of NOTCH3 as a bona fide target of miR-150 (Figure 3). NOTCH3 was recently shown to be a target of miR-206 in HeLa cells; in this system, forced expression of miR-206 resulted in a 3-fold increase of apoptosis, which, however, was not rescued by NOTCH3 overexpression.42 Unlike miR-150, we did not observe differential expression of miR-206 during human T-cell development, suggesting that different miRs may regulate NOTCH3 expression depending on the cell type considered.
One of the top up-regulated miRs during T-cell maturation (Figures 1, 2), miR-150, is a lymphoid-specific miR that is known to be highly expressed in mature T and B lymphocytes and involved in B-cell development and antigen-induced lymphocyte activation.16,43,44 Notch receptors control cell cycle progression and apoptosis in different cell types and are essential regulators of T-cell progenitor differentiation. In particular, NOTCH3 expression levels are significantly higher in DN and DP thymocytes with respect to mature SP cells. Of note, NOTCH3 has been shown to play a role in the intrathymic pre-T-cell receptor selection of T cells.32,45
Although we do not have direct evidence that miR-150 influences maturation of human thymocytes through suppression of NOTCH3, this hypothesis is supported by the observed inverse regulation of NOTCH3 and miR-150 expression and reduction in the levels of NOTCH3 protein measured in SP vs DP thymocytes (Figure 3B-D). Further indirect evidence for a miR-150-NOTCH3 interaction in T-cell physiology and development is provided by studies in mice demonstrating that specific up-regulation of NOTCH3 occurs in thymocytes in the late DN phase and that forced expression of miR-150 in T-cell progenitors interferes with the DN3 to DN4 transition.18
Mice expressing a constitutively activated form of NOTCH3 develop aggressive T-cell leukemia/lymphomas with high penetrance.46 Deregulated NOTCH3 signaling has likewise been proposed to be important in T-ALL pathogenesis, although activating mutations of this gene have not been described.45 This property, together with the identification of miR-150 as one of the most conspicuously down-regulated miRs in ALL compared with their normal counterparts,47 suggests a role for miR-150/NOTCH3 deregulation in this disease. The presence of very low levels of miR-150 in the 5 T-ALL cell lines examined in the present study (supplemental Figure 3) led us to investigate in this T-cell system the impact of forced miR-150 expression, which demonstrated functional effects ranging from the inhibition of cell proliferation to the induction of cell apoptosis, depending on the cell line (Figure 5). Nevertheless, additional experiments will be required to verify whether these effects depend on the suppression of NOTCH3 or on the silencing of other targets of miR-150. One interesting known target of miR-150 is C-MYB, a transcription factor that plays an essential role in the hematopoietic process and, in particular, in T-cell differentiation and leukemogenesis.48,49 Further studies of these and other miR-target interactions during normal T-cell differentiation will thus probably elucidate key miRs involved in the regulation of this process and contribute to the identification of novel therapeutics for T-ALL and other T-cell malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Elisabetta Rossi for help and advice with cytofluorimetric analyses, Anna Merlo for discussion of results, and Paola Dalla Pria for technical assistance.
This work was supported by the Italian Ministry of Health, the Italian Association for Cancer Research (AIRC), the European Union (“The role of chronic infections in the development of cancer”; contract no. 2005-018704), Istituto Superiore Sanità -Alleanza Contro il Cancro, and Fondazione Cariparo-Progetti d'Eccellenza.
Authorship
Contribution: M.G. designed and performed experiments, analyzed and interpreted data, and wrote the manuscript; A.C., S.M., A.G., and G.D.B. performed bioinformatic analysis; K.B. and L.M. performed microarray experiments; C.F. performed fluorescence-activated cell sorter sorting and flow cytometry; V.S., K.R., and L.B. performed experiments; G.S. and G.G. provided thymic surgical specimen; D.M.D., G.B., and V.B. discussed and analyzed the results; A.A. and S.I. discussed and analyzed the results and wrote the manuscript; and P.Z. discussed and analyzed the results, wrote the manuscript, and supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of A.G. is Genomnia srl, Lainate, Italy.
Correspondence: Paola Zanovello, Oncology Section, Department of Oncology and Surgical Sciences, University of Padova, Via Gattamelata, 64, 35128 Padova, Italy; e-mail: paola.zanovello@unipd.it.
References
Author notes
M.G. and A.C. contributed equally to this study.
Supplemental data
Upregulated miRNAs (in red, in the header line) and downregulated miRNAs (in green, in the header line) in the SP versus DP comparison are predicted to target the genes whose symbols are reported in the corresponding columns. Only the targets predicted by both miRanda and TargetScan algorithms were considered and reported. Complete algorithms' predictions are freely downloadable from http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/download.pl and http://www.targetscan.org/cgi-bin/targetscan/data_download.cgi?db=vert_50, respectively. Genes identified as up- or down-regulated on microarrays are highlighted in red and green, respectively.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal