Abstract
Chromosomal translocations of the mixed lineage leukemia (MLL) gene are a common cause of acute leukemias. The oncogenic function of MLL fusion proteins is, in part, mediated through aberrant activation of Hoxa genes and Meis1, among others. Here we demonstrate using a tamoxifen-inducible Cre-mediated loss of function mouse model that DOT1L, an H3K79 methyltransferase, is required for both initiation and maintenance of MLL-AF9–induced leukemogenesis in vitro and in vivo. Through gene expression and chromatin immunoprecipitation analysis we demonstrate that mistargeting of DOT1L, subsequent H3K79 methylation, and up-regulation of Hoxa and Meis1 genes underlie the molecular mechanism of how DOT1L contributes to MLL-AF9–mediated leukemogenesis. Our study not only provides the first in vivo evidence for the function of DOT1L in leukemia, but also reveals the molecular mechanism for DOT1L in MLL-AF9 mediated leukemia. Thus, DOT1L may serve as a potential therapeutic target for the treatment of leukemia caused by MLL translocations.
Introduction
The mixed lineage leukemia 1 (MLL1) protein contains an evolutionarily conserved SET domain and possesses intrinsic methyltransferase activity toward histone H3 lysine 4 (H3K4), whose methylation generally correlates with transcriptional activation.1,2 The full-length MLL1 precursor protein is proteolytically cleaved by Taspase1 to generate N-terminal 300 kDa (MLL1N) and carboxyl-terminal 180 kDa (MLL1C) proteins that form a heterodimer as part of a multi-subunit protein complex.3 Previous studies indicated that MLL dimerization is required for its stability as well as proper spatio-temporal activation of homeobox (Hox) gene expression during embryonic development and hematopoiesis.3,4
The gene that encodes human MLL1 protein is located at 11q23. One unique feature of this genomic locus is that it harbors an 8.3 kb breakpoint cluster region which is known to be involved in translocation with > 50 different genes.5,6 Chromosomal translocation of MLL is a common cause of acute leukemias, accounting for 5%-10% of adult acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL). Strikingly, MLL rearrangements account for ∼ 80% of ALL and ∼ 60% of AML infant leukemias.5,6 In MLL-related leukemias, genes of the Hoxa cluster are frequently up-regulated, and their sustained expression is required for leukemic stem cell (LSC) maintenance.7 Because the oncogenic fusion proteins lack the SET domain of MLL, it is believed that the translocation partner protein is responsible for maintaining Hox gene expression either through its intrinsic transcription activation activity or recruitment of other effector molecules.8 Among the ∼ 50 different MLL fusion partners, AF4, AF9, AF10, and ENL account for more than two-thirds of all MLL-associated leukemias and have been reported to associate with each other through direct or indirect protein-protein interactions.8-10 AF4, AF9, and ENL have been recently shown to be components of a large protein complex named AEP11 or EAP,12 which also contain the transcription elongation factor P-TEFb.
Yeast disruptor of telomeric silencing (DOT1) and its mammalian homolog DOT1L methylate Lysine 79 within the globular domain of histone H3 (H3K79).13-15 Although DOT1 was originally identified as a regulator of telomeric silencing,16 data suggest that DOT1-mediated H3K79 methylation is linked to euchromatic gene transcription17,18 and transcription elongation. A recent study indicates that DOT1L exists in a large protein complex that regulates expression of wingless target genes.19 In addition, DOT1L has been shown to regulate erythroid differentiation during embryonic hematopoiesis.20 Interestingly, DOT1L was also shown to be part of the EAP complex,12,21 suggesting that DOT1L may interact with MLL-fusion oncogenes and play a role in leukemic transcription.
Using in vitro methylcellulose replating assays, we have previously shown that mistargeting of DOT1L and dysregulation of MLL-target genes is probably the mechanism underlying how DOT1L contributes to leukemogenesis in MLL-AF10 and CALM-AF10.22,23 Similar in vitro studies were also reported for leukemia caused by MLL-ENL,24 MLL-AF4,25 and MLL-AF9.26 To further substantiate the role of DOT1L in leukemogenesis, we generated a conditional DOT1L knockout mouse model and evaluated the role of DOT1L in MLL-AF9–mediated leukemogenesis. Our studies not only establish a critical function of DOT1L in the initiation and maintenance of leukemia, but also revealed a role for DOT1L in the activation of leukemia genes such as Meis1 and Hoxa genes.
Methods
Mice and in vivo tamoxifen treatment
The DOT1L conditional mouse was previously described.27 The R26-Cre-ERT2 mice were generated by Tyler Jacks laboratory28 and were obtained from the NCI Mouse Models of Human Cancers Consortium (Strain 01XAB). DOT1L2lox/+ and DOT1L1lox/+ mice were intercrossed to generate DOT1L2lox/1lox;Cre-ERT2 and DOT1L+/+;Cre-ERT2 mice. Mice were kept on a 129Sv/Jae and C57/B6 Ly5.2 background. Wild-type C57/B6 Ly5.1 and Ly5.2 mice were purchased from The Jackson Laboratory and were intercrossed to generate Ly5.1/Ly5.2 mixed-background bone marrow cells (BMCs). For in vivo Cre-recombination, tamoxifen (TAM; Sigma-Aldrich) was administered via intraperitoneal injection every 2 days (100 μL of 10 mg/mL in corn oil) for a period of 3 weeks. All animal protocols adhere to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the UNC Institutional Animal Care and Use Committee.
Retrovirus production, transduction, methylcellulose colony replating assay, and cell culture
All retroviral constructs, pMSCV, used in the manuscript contain a neomycin selection marker and are abbreviated as pMSCN. The pMSCN-MLL-AF9 construct was a gift from Jay Hess (University of Michigan). The pMSCN-MLL-AFX construct was a gift from Michael Cleary (Stanford University). Retroviral production was performed as previously described.22 Briefly, pMSCN-MLL-AF9 and pMSCN-MLL-AFX were cotransfected with pGag-pol and pVSVG into 293T cells using Superfect (QIAGEN). At 48 hours after transfection, viral supernatant were collected for transduction of c-Kit-positive HPCs purified as follows: Mice 4 to 10 weeks old were injected intravenously with 5-fluorouracil (150 mg/kg). Bone marrow was harvested from both femurs 5 days after injection and stained with c-Kit-APC (BD Biosciences). Positive stained HPCs were FACS sorted using a BD FACSAria II Flow Cytometer. Sorted HPCs were maintained in mFTOC (20% FBS in RPMI1640, 1mM MEM sodium pyruvate, 1% MEM nonessential amino acid, 10mM HEPES at pH 7.3, 50 μm 2-mercaptoethanol) with 50 ng/mL mSCF, 50 ng/mL mIL-6, and 10 ng/mL mIL-3 (PeproTech). Viral supernatants were used to transduce the sorted cells via spinoculation, and cells plated onto methylcellulose 24 hours after infection. For methylcellulose colony formation assays, 1 × 104 transduced cells were plated into 0.9% Methocult M3534 (StemCell Technologies; Iscove MDM plus FBS, BSA, rh insulin, human transferrin, 2-mercaptoethanol, mSCF, mIL-3, and mIL-6) supplemented with 10 ng/mL GM-CSF and 1 mg/mL G418 (Gibco) for selection. After 7 days of culture, G418-resistant colonies were collected and single-cell suspensions replated into M3534 with GM-CSF. Replating was performed every 7 to 10 days. Liquid cultures of transformed hematopoietic progenitor cells (HPCs) and LSCs were maintained in mFTOC with 5 ng/mL mIL-3. For in vitro recombination, 4-OHT (Sigma-Aldrich) was resuspended in ethanol and added to cell culture media at final concentration of 250nM. Media was changed daily.
Bone marrow transplantation
Recipient C57/B6 Ly5.1 mice (6-12 weeks old) began a prophylactic regimen to prevent gastrointestinal infections through administration of sulfamethoxazole/trimethoprim (SMZ; TW Medical) at a concentration of 6 mg/1.2 mg per mL water 1 week before bone marrow transplantation (BMT) procedures and continued for 4 weeks after transplantation. SMZ containing water was changed every other day. On the day of transplantation, recipient mice received 2 doses, 4.8 Gy each, of total body irradiation (TBI) 4 hours apart using a cesium radiation source. Four hours after the second irradiation dose, recipient mice were anesthetized through intraperitoneal injection of 300 μL/10 g body weight of avertin (0.0125 g/mL 2,2,2, tribromoethanol and 1.25% tert-amyl alcohol in PBS). After mice were anesthetized, 0.5 × 105 LCs plus 2.5 × 105 radio-protector Ly5.1/Ly5.2 cells resuspended in 100 μL of PBS (1:5 ratio) were transplanted via retro-orbital injection.
Histology
Tissues were harvested from mice and fixed in 4% paraformaldehyde. Livers were paraffin embedded and tissue sections, 5 μm thick, were used for H&E staining. The samples were examined using an Axio Observer Z1 microscope (Carl Zeiss) with a Zeiss EC Plan-NeoFluar 10×/0.3 numeric aperture Ph1 DIC objective lens (Carl Zeiss). The images were captured by an AxioCam MRC5 camera (Carl Zeiss) and processed by AxioVision40 Version 4.8.2.0 image software (Carl Zeiss).
FACS analysis and cell sorting
Bone marrow cells were flushed from both femurs of mice using a 25-G needle and syringe. Red blood cells were lysed using ammonium chloride (StemCell Technologies). Cells were resuspended in PBS + 2% FBS at a concentration of 106 cells per 100 μL. Cells were stained for 30 minutes at 4°C, washed 3 times with PBS, and resuspended in PBS + 2% FBS. Antibodies used for FACS analysis and sorting are: Rat antimouse c-Kit-APC, Ly5.1-PE-Cy7, and Ly5.2-PerCP-Cy5.5 plus corresponding Rat IgG2a Isotype controls (eBioscience). All analysis and sorting was performed using BD FACSAria II Flow Cytometer.
PI staining for DNA content
Cells grown in liquid culture were washed twice with ice-cold PBS and resuspended in 500 μL of PBS. While vortexing gently, 4.5 mL of ice-cold 70% ethanol was slowly added and then stored at 4°C overnight. On the following day, fixed cells were pelleted by centrifugation at 4°C for 5 minutes at 1500 rpm. Cell pellet was washed with PBS, centrifuged, and resuspended in 250 μL of PBS. Propidium iodide (PI) staining mix (RNase A 100 μg/mL, PI 40 μg/mL, and 0.1% Triton X-100 in PBS) was prepared fresh, and 750 μL added to cells. Cells were incubated for 15 minutes at 37°C and then analyzed by FACS using a Beckman Coulter CyAn Flow Cytometer. Data were analyzed using ModFit LT Version 3.2 software.
RT-qPCR and chromatin immunoprecipitation
For gene-expression analysis, total RNA was isolated from cells using RNeasy Kit (QIAGEN) and reverse transcribed using Improm II (Promega). SYBR GreenER qPCR SuperMix (Invitrogen) was used for qPCR. qPCR was performed using Applied Biosystems 7300 Real-Time PCR system. Relative expression was normalized to Gapdh. Primer sequences are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). ChIP experiments were carried out as previously described22,29 with the following modifications. DNA was fragmented into 300-500 bp in length by sonication. Immunoprecipitation was performed using anti-H3K79me2/3 (Abcam) and antiRabbit IgG (Santa Cruz; sc-2027) and DynaBeads (Invitrogen). ChIPed samples were washed twice with low-salt (140mM NaCl) RIPA buffer, once with high-salt (500mM NaCl) RIPA buffer, and twice with TE buffer. DNA was purified using Chelex-100. Quantitative real-time PCR of ChIPed DNA was analyzed using SYBR GreenER qPCR SuperMix (Invitrogen) and Applied Biosystems 7300 Real-Time PCR system. Primer sequences are listed in supplemental Table 2.
Results
DOT1L is required for MLL-AF9–induced transformation in vitro
Previous studies suggest that DOT1L plays an important role in leukemogenesis involving MLL fusion proteins. Most of the studies analyzed the effect of DOT1L in MLL-fusion mediated transformation in vitro using siRNA/shRNA-mediated knockdown approaches22-25 or DOT1L knockout cells.26 To further establish a role for DOT1L in leukemogenesis in vivo, we generated an inducible knockout system by crossing mice carrying DOT1L conditional allele (DOT1l2lox; supplemental Figure 127,29 with mice harboring a Cre-recombinase-Estrogen-Receptor-T2 allele (Cre-ERT2) at the ubiquitous ROSA26 locus.28 On TAM administration, the Cre-ERT2 system allows for translocation of Cre-recombinase into the nucleus resulting in efficient recombination of genomic loxP sites.28 Previous studies have confirmed that Cre-recombination of the Dot1l allele removes exons 5 and 6, which encode part of the S-adenosyl methionine-binding motif required for enzymatic activity, to generate DOT1l1lox allele (supplemental Figure 1), resulting in complete loss of H3K79 mono-, di-, and trimethylation.27,29
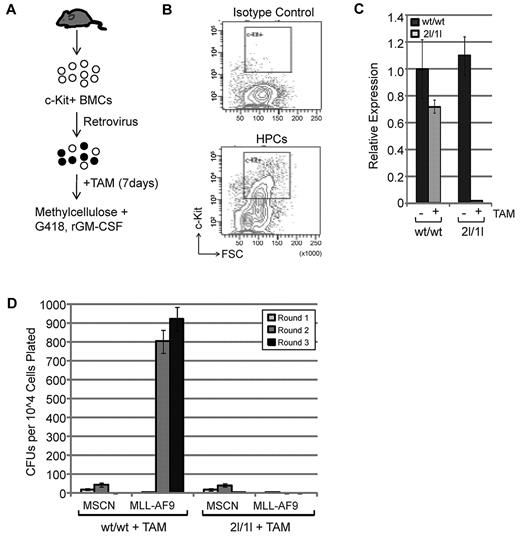
Using this inducible system, we first investigated the role of DOT1L in MLL-AF9–mediated transformation of HPCs. To this end, we FACS sorted the c-Kit+ HPCs from bone marrow of conditional DOT1L2lox/1lox;Cre-ERT2 (2l/1l) and control DOT1Lwt/wt;Cre-ERT2 (wt/wt) mice (Figure 1B) and cultured in the presence of IL-3, IL-6, and SCF. The HPCs were then infected with retroviruses expressing MLL-AF9 or empty vector control. After infection, cells were administered TAM for 7 days and efficient deletion of Dot1l was confirmed by RT-qPCR (Figure 1C, supplemental Figure 2A). To evaluate the effect of DOT1L depletion on MLL-AF9–mediated transformation, infected 2l/1l and wt/wt cells, treated and untreated, were plated in methylcellulose in the presence of G418 and GM-CSF. Serial methylcellulose replating assay demonstrates that DOT1L is required for transformation by MLL-AF9 as no colonies were observed in the second and third rounds of methylcellulose replating when DOT1l is depleted in HPCs harboring a conditional Dot1l allele (Figure 1D, supplemental Figure 2B-C). The observed effect of DOT1L deletion on the initial transformation capability of MLL-AF9 is not because of virus transduction efficiency of the two different HPC genotypes as a similar number of colonies was observed at the end of the first-round plating (supplemental Figure 2A).
DOT1L is required for MLL-AF9-induced transformation in vitro. (A) Diagram of the procedure used for in vitro analysis. (B) Bone marrow cells were stained with rat IgG2a-APC isotype control to identify c-Kit-negative population or rat antimouse c-Kit-APC. c-Kit+ cells were sorted using a BD FACSAria II instrument. (C) RT-qPCR analysis of DOT1L expression level, normalized to Gapdh, after 7 days of TAM treatment (250nM final concentration) demonstrates efficient recombination. Mouse genotype is indicated. (D) Serial methylcellulose colony replating assay shows that MLL-AF9 fails to transform HPCs in the absence of DOT1L. An equal number of cells transduced with empty vector MSCN or MLL-AF9 were plated at each round and colony forming units (CFUs) counted after 7 to10 days. Experiment was performed 3 times and presented as average number of CFUs with standard deviation (SD).
DOT1L is required for MLL-AF9-induced transformation in vitro. (A) Diagram of the procedure used for in vitro analysis. (B) Bone marrow cells were stained with rat IgG2a-APC isotype control to identify c-Kit-negative population or rat antimouse c-Kit-APC. c-Kit+ cells were sorted using a BD FACSAria II instrument. (C) RT-qPCR analysis of DOT1L expression level, normalized to Gapdh, after 7 days of TAM treatment (250nM final concentration) demonstrates efficient recombination. Mouse genotype is indicated. (D) Serial methylcellulose colony replating assay shows that MLL-AF9 fails to transform HPCs in the absence of DOT1L. An equal number of cells transduced with empty vector MSCN or MLL-AF9 were plated at each round and colony forming units (CFUs) counted after 7 to10 days. Experiment was performed 3 times and presented as average number of CFUs with standard deviation (SD).
DOT1L is required for MLL-AF9–mediated leukemogenesis in vivo
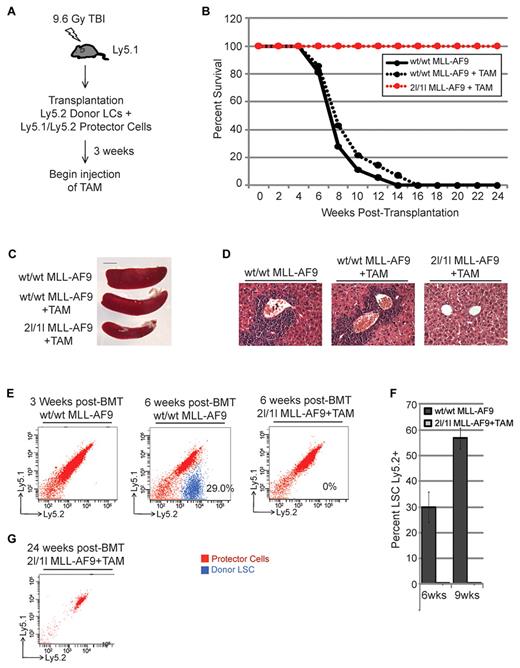
Taking advantage of our inducible system, we next asked whether DOT1L is required for MLL-AF9–mediated leukemogenesis in the mouse via BMT. To this end, donor 2l/1l and wt/wt mice are maintained on a 129Sv/Jae and C57/B6 Ly5.2 background, while recipient mice are maintained in a C57/B6 Ly5.1 background. Thus, donor LCs can be distinguished from wild-type recipient and radio-protector cells, which do not contain the Dot1L conditional allele or Cre-ERT2 allele, by FACS. Donor MLL-AF9–transformed cells, collected from the third round of colonies grown on methylcellulose, were mixed with radio-protector Ly5.1/Ly5.2 BMCs at a 1:5 ratio and transplanted into recipient Ly5.1 mice. TAM administration was initiated 3 weeks after BMT, before the detection of LCs in bone marrow (Figure 2E), to induce Dot1L recombination in donor cells. RT-qPCR analysis confirmed efficient deletion of DOT1L by the TAM treatment (supplemental Figure 1). As expected, all mice receiving wt/wt MLL-AF9 transplantation died by 14 weeks after BMT (Figure 2B). However, TAM treatment increased survival of mice transplanted with 2l/1l MLL-AF9 cells (Figure 2B). The extended life span of the TAM treated group (red dotted line) is not because of other effects of TAM as only a 1- to 2-week increase in survival was observed between treated and untreated wt/wt MLL-AF9 (Figure 2B, dotted and solid black lines). Consistent with the improvement in survival, TAM treatment prevented splenomegaly (Figure 2C) and leukemia infiltration of liver (Figure 2D) in mice that received 2l/1l MLL-AF9 transplantation, but not to those receiving wt/wt MLL-AF9. Furthermore, FACS analysis demonstrates that donor wt/wt MLL-AF9 LCs constituted one-third (58%) of the bone marrow population at 6 weeks (or 9 weeks) after BMT, whereas no 2l/1l MLL-AF9+TAM donor cells were detectable when analyzed at the same time (Figure 2E-F), indicating that the ability of transplanted MLL-AF9–transformed cells to repopulate the recipient bone marrow requires functional DOT1L. Strikingly, by 24 weeks after BMT no 2l/1l MLL-AF9 cells were detected in the peripheral blood of TAM-treated mice (Figure 2G), demonstrating that MLL-AF9–transformed cells fail to establish AML in the absence of DOT1L. Collectively, these results demonstrate a critical function for DOT1L in the initiation of leukemia in vivo.
DOT1L is required for MLL-AF9–mediated leukemogenesis in vivo. (A) Diagram of the BMT procedure to examine the effect of DOT1L depletion on the establishment of AML by MLL-AF9–transformed cells in vivo. TAM treatment began at 3 weeks after transplantation. (B-G) In vivo deletion of DOT1L prevents MLL-AF9–mediated acute leukemia development. (B) Kaplan-Meier plot showing survival of transplanted mice treated and untreated with TAM. Median survival of wt/wt MLL-AF9–transplanted mice without TAM = 7 weeks (n = 17) and wt/wt MLL-AF9 transplanted mice treated with TAM = 7 weeks (n = 14). All 2lox/1lox MLL-AF9–transplanted mice treated with TAM (n = 18) survive at least 24 weeks after transplantation. (C) Representative image of spleens harvested from transplanted mice at 6 weeks after transplantation show splenomegaly in wt/wt MLL-AF9 transplanted mice (n = 3 per group). (D) Representative H&E staining of liver tissue sections at 6 weeks after transplantation show leukemic infiltration in wt/wt MLL-AF9 transplanted mice (n = 3 per group). (E) Donor cells depleted of DOT1L are absent in recipient bone marrow. Representative images showing FACS analysis of bone marrow cells isolated from transplanted mice (n = 3 per group). Donor MLL-AF9 cells are Ly5.2+ (blue) while recipient/protector cells are Ly5.1+/Ly5.2+ (red). (F) Percentage of MLL-AF9 LCs present in recipient bone marrow at 6 and 9 weeks after transplantation as determined by FACS analysis (n = 3 per group). (G) No transformed donor cells are detected by FACS analysis of peripheral blood obtained from 2lox/1lox MLL-AF9+TAM mice at 24 weeks after transplantation. Representative image from 1 of 6 mice analyzed.
DOT1L is required for MLL-AF9–mediated leukemogenesis in vivo. (A) Diagram of the BMT procedure to examine the effect of DOT1L depletion on the establishment of AML by MLL-AF9–transformed cells in vivo. TAM treatment began at 3 weeks after transplantation. (B-G) In vivo deletion of DOT1L prevents MLL-AF9–mediated acute leukemia development. (B) Kaplan-Meier plot showing survival of transplanted mice treated and untreated with TAM. Median survival of wt/wt MLL-AF9–transplanted mice without TAM = 7 weeks (n = 17) and wt/wt MLL-AF9 transplanted mice treated with TAM = 7 weeks (n = 14). All 2lox/1lox MLL-AF9–transplanted mice treated with TAM (n = 18) survive at least 24 weeks after transplantation. (C) Representative image of spleens harvested from transplanted mice at 6 weeks after transplantation show splenomegaly in wt/wt MLL-AF9 transplanted mice (n = 3 per group). (D) Representative H&E staining of liver tissue sections at 6 weeks after transplantation show leukemic infiltration in wt/wt MLL-AF9 transplanted mice (n = 3 per group). (E) Donor cells depleted of DOT1L are absent in recipient bone marrow. Representative images showing FACS analysis of bone marrow cells isolated from transplanted mice (n = 3 per group). Donor MLL-AF9 cells are Ly5.2+ (blue) while recipient/protector cells are Ly5.1+/Ly5.2+ (red). (F) Percentage of MLL-AF9 LCs present in recipient bone marrow at 6 and 9 weeks after transplantation as determined by FACS analysis (n = 3 per group). (G) No transformed donor cells are detected by FACS analysis of peripheral blood obtained from 2lox/1lox MLL-AF9+TAM mice at 24 weeks after transplantation. Representative image from 1 of 6 mice analyzed.
DOT1L is required for maintaining MLL-AF9 transformation in vitro
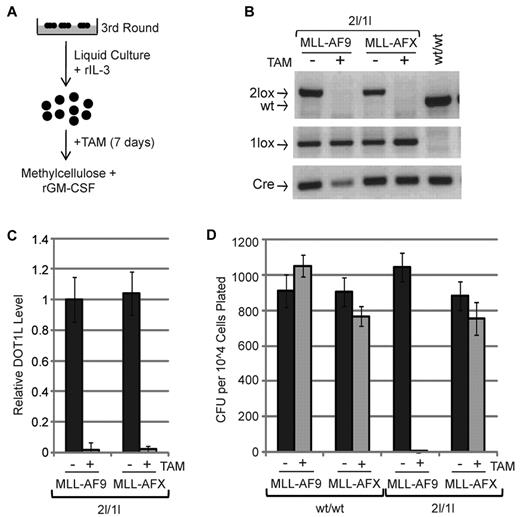
Having established the function for DOT1L in the initiation of leukemia, we next asked whether DOT1L is also required for maintaining the transformed state induced by MLL-AF9. To this end, MLL-AF9 transformed cells, after the third round of methylcellulose, were propagated in liquid culture containing IL-3 in the presence or absence of TAM for a period of 7 days. After verification of efficient TAM-induced Dot1l recombination by genotyping (Figure 3B) and RT-qPCR (Figure 3C), an equal number of cells were plated onto methylcellulose. MLL-AFX–transformed cells were used as a negative control because DOT1L was previously shown not required for maintaining the leukemia state induced by MLL-AFX.22 Results shown in Figure 3D demonstrate that 2l/1l MLL-AF9–transformed cells failed to produce colonies on TAM-induced depletion of DOT1L. In contrast, similar treatment exhibited no effect on MLL-AFX–transformed cells, indicating that depletion of DOT1L did not cause a general cell toxic effect. The lack of an effect on MLL-AFX is not because of inefficient recombination as efficient recombination is observed in the MLL-AFX transformed cells on TAM treatment (supplemental Figure 3A-B). Consistent with the notion that DOT1L is not involved in leukemogenesis by MLL-AFX, loss of DOT1L activity did not affect up-regulation of Hoxa5, Hoxa9, and Hoxa10 in MLL-AFX–transformed cells (supplemental Figure 3C). Moreover, TAM treatment by itself had no effect on wt/wt colony formation potential (Figure 3D). This study demonstrates that MLL-AF9–transformed cells require DOT1L for maintaining clonal growth in vitro.
MLL-AF9 cells require DOT1L to maintain transformation in vitro. (A) Diagram of the procedure analyzing role of DOT1L in maintaining transformation in vitro. (B) Genotyping to verify efficient Cre-mediated recombination of DOT1L2lox conditional allele. (C) RT-qPCR analysis of DOT1L expression level in 2lox/1lox MLL-AF9 cells, normalized to Gapdh, after 7 days of TAM treatment (250nM final concentration) demonstrates efficient recombination. (D) DOT1L is required for maintaining MLL-AF9 transformation but not for MLL-AFX. Methylcellulose colony formation assay after TAM treatment of transformed cells. Equal number of cells were plated and average numbers of CFUs from 3 independent experiments are shown. Error bars represent SD.
MLL-AF9 cells require DOT1L to maintain transformation in vitro. (A) Diagram of the procedure analyzing role of DOT1L in maintaining transformation in vitro. (B) Genotyping to verify efficient Cre-mediated recombination of DOT1L2lox conditional allele. (C) RT-qPCR analysis of DOT1L expression level in 2lox/1lox MLL-AF9 cells, normalized to Gapdh, after 7 days of TAM treatment (250nM final concentration) demonstrates efficient recombination. (D) DOT1L is required for maintaining MLL-AF9 transformation but not for MLL-AFX. Methylcellulose colony formation assay after TAM treatment of transformed cells. Equal number of cells were plated and average numbers of CFUs from 3 independent experiments are shown. Error bars represent SD.
DOT1L is required for MLL-AF9–induced leukemia progression in vivo
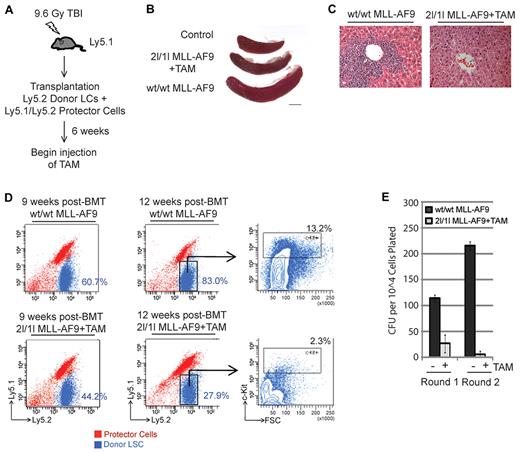
Our in vivo results shown in Figure 1E through J suggest that DOT1L is required for leukemogenesis by MLL-AF9. In those experiments, TAM administration was initiated at 3 weeks after BMT. At this stage, acute leukemia had yet to develop because donor cells in the bone marrow were undetectable by FACS (Figure 2E). To investigate the importance of DOT1L in the maintenance of MLL-AF9–induced leukemia in vivo, we administrated TAM at 6 weeks after BMT when donor MLL-AF9 cells account for ∼ 30% of the bone marrow population (Figure 2F). At 9 weeks after BMT, TAM-induced DOT1L depletion greatly reduced splenomegaly (Figure 4B) and liver infiltration (Figure 4C) in 2l/1l MLL-AF9 mice. FACS analysis of recipient bone marrow revealed that 2l/1l MLL-AF9 cells represented a lower percentage (44.2% vs 60.7% at 9 weeks; 27.9% vs 83.0% at 12 weeks) of total BMCs in treated mice compared with untreated controls (Figure 4D). In TAM-treated mice, we also observed a decrease in 2l/1l MLL-AF9 cells at 12 weeks compared with 9 weeks after BMT (27.9% vs 44.2%; Figure 4D). In contrast, the percentage of wt/wt MLL-AF9 cells increased from 60.7% to 83.0% in the same period (Figure 4D). Moreover, FACS analysis indicated that the remaining DOT1L-deficient donor cells expressed lower levels of stem cell/progenitor cell marker c-Kit (Figure 4D), indicating a depletion of MLL-AF9 clonogenic leukemic progenitor/stem cells30 in the absence of DOT1L.
DOT1L is required for MLL-AF9–induced acute leukemia progression in vivo. (A) Diagram of BMT procedure to examine the requirement of DOT1L in MLL-AF9 AML progression. TAM treatment began at 6 weeks after transplantation after leukemia development. (B-E) In vivo deletion of DOT1L prevents MLL-AF9 AML progression. (B) Reduced spleen size in TAM-treated 2lox/1lox MLL-AF9 recipient mice. Representative images of spleens harvested from transplanted mice at 9 weeks after transplantation (n = 3 per group). (C) TAM-induced DOT1L deletion prevents leukemic infiltration of the liver. Representative H&E staining of liver tissue sections at 9 weeks after transplantation (n = 3 per group). (D) On DOT1L depletion, the percentage of transformed donor cells in bone marrow diminishes over time. FACS analysis of bone marrow cells isolated from transplanted mice (n = 3 per group, per time point). Donor MLL-AF9 cells are Ly5.2+ (blue) and recipient/protector cells are Ly5.1+/Ly5.2+ (red). Percentage of donor Ly5.2 cells in the total bone marrow cell population at 9 and 12 weeks after transplantation are indicated. At 12 weeks after transplantation, cells were also stained with the stem cell/progenitor marker c-Kit. Transformed donor Ly5.2 population is gated and percentage of c-Kit+ cells indicated. Shown are representative images from 3 independent experiments. (E) Loss of clonogenic leukemic stem cells after in vivo DOT1L depletion. Methylcellulose colony formation assay of donor cells sorted from mice at 12 weeks after transplantation. Average CFUs are shown (n = 3). Error bars represent SD.
DOT1L is required for MLL-AF9–induced acute leukemia progression in vivo. (A) Diagram of BMT procedure to examine the requirement of DOT1L in MLL-AF9 AML progression. TAM treatment began at 6 weeks after transplantation after leukemia development. (B-E) In vivo deletion of DOT1L prevents MLL-AF9 AML progression. (B) Reduced spleen size in TAM-treated 2lox/1lox MLL-AF9 recipient mice. Representative images of spleens harvested from transplanted mice at 9 weeks after transplantation (n = 3 per group). (C) TAM-induced DOT1L deletion prevents leukemic infiltration of the liver. Representative H&E staining of liver tissue sections at 9 weeks after transplantation (n = 3 per group). (D) On DOT1L depletion, the percentage of transformed donor cells in bone marrow diminishes over time. FACS analysis of bone marrow cells isolated from transplanted mice (n = 3 per group, per time point). Donor MLL-AF9 cells are Ly5.2+ (blue) and recipient/protector cells are Ly5.1+/Ly5.2+ (red). Percentage of donor Ly5.2 cells in the total bone marrow cell population at 9 and 12 weeks after transplantation are indicated. At 12 weeks after transplantation, cells were also stained with the stem cell/progenitor marker c-Kit. Transformed donor Ly5.2 population is gated and percentage of c-Kit+ cells indicated. Shown are representative images from 3 independent experiments. (E) Loss of clonogenic leukemic stem cells after in vivo DOT1L depletion. Methylcellulose colony formation assay of donor cells sorted from mice at 12 weeks after transplantation. Average CFUs are shown (n = 3). Error bars represent SD.
To evaluate the clonal growth capacity of the remaining DOT1L deleted MLL-AF9 cells, we sorted donor cells at 12 weeks after BMT from bone marrow aspirates and plated equal numbers onto methylcellulose. Results shown in Figure 4E demonstrate that the colony formation capacity of 2l/1l MLL-AF9 cells from TAM-treated mice is greatly reduced after the first round of plating. Although colonies were observed, those cells failed to give rise to more colonies after the second round of plating. In contrast, wt/wt MLL-AF9 cells derived from untreated mice have increased colony formation capacity. These data suggest that the block in AML progression on in vivo TAM-induced DOT1L depletion is because of inhibition of LC clonal growth, which is consistent with the decreased c-Kit expression (Figure 4D).
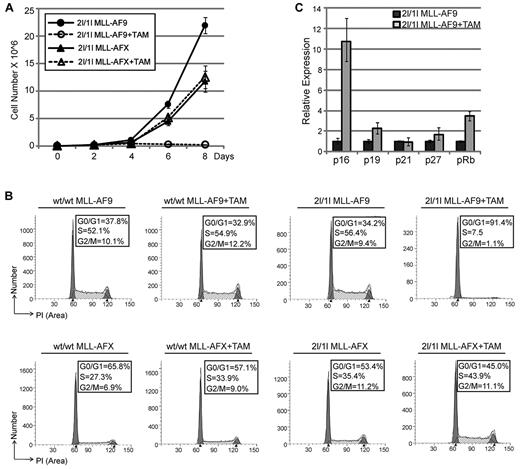
Loss of DOT1L function causes cell-cycle arrest in MLL-AF9–transformed cells
Previous studies have shown that loss of DOT1L induces cell-cycle arrest in embryonic stem cells27,31 and erythroid cells.32 To analyze the effect of DOT1L on cell cycle, we cultured transformed cells in liquid media supplemented with IL-3 in the presence or absence of TAM. Results shown in Figure 5A and supplemental Figure 4 demonstrate that loss of DOT1L inhibits proliferation of cells transformed by MLL-AF9, but not cells transformed by MLL-AFX, indicating that depletion of DOT1L did not cause a general cell toxic effect. PI staining and FACS analysis indicate that TAM-mediated depletion of DOT1L induced cell-cycle arrest at G0/G1 in MLL-AF9–transformed cells, but has no effect on MLL-AFX–transformed cells (Figure 5B). The general anticancer effect of TAM is not responsible for the observed cell-cycle effect as TAM treatment has no effect on wt/wt MLL-AF9 cells (Figure 5A-B). Consistent with a G0/G1 cell-cycle arrest, we observed substantial up-regulation of 2 regulators of G1 progression, pRb and the CDK inhibitor p16,33,34 in 2l/1l MLL-AF9 cells on loss of DOT1L (Figure 5C). Given that DOT1L-mediated H3K79 methylation is generally linked to transcription activation, lost function of DOT1L is expected to down-regulate DOT1L direct targets. Therefore, the effect of DOT1L on p16 and pRb expression is probably to be indirect.
Loss of DOT1L inhibits cell proliferation because of a G0/G1 cell-cycle arrest. (A) In vitro TAM-induced DOT1L deletion inhibits proliferation of MLL-AF9–transformed cells in liquid culture but not MLL-AFX. Total cell numbers were counted every 2 days. Shown is the average of 3 independent experiments with error bars. (B) In vitro TAM-induced DOT1L deletion causes cell-cycle arrest at G0/G1 phase for MLL-AF9–transformed cells. Cell-cycle analysis was performed by PI staining. Experiment was repeated twice. Data were analyzed using ModFit software. (C) Gene-expression analysis by RT-qPCR shows up-regulation of CDK inhibitors after DOTL depletion, normalized to Gapdh.
Loss of DOT1L inhibits cell proliferation because of a G0/G1 cell-cycle arrest. (A) In vitro TAM-induced DOT1L deletion inhibits proliferation of MLL-AF9–transformed cells in liquid culture but not MLL-AFX. Total cell numbers were counted every 2 days. Shown is the average of 3 independent experiments with error bars. (B) In vitro TAM-induced DOT1L deletion causes cell-cycle arrest at G0/G1 phase for MLL-AF9–transformed cells. Cell-cycle analysis was performed by PI staining. Experiment was repeated twice. Data were analyzed using ModFit software. (C) Gene-expression analysis by RT-qPCR shows up-regulation of CDK inhibitors after DOTL depletion, normalized to Gapdh.
DOT1L regulates expression of Hoxa cluster and Meis1
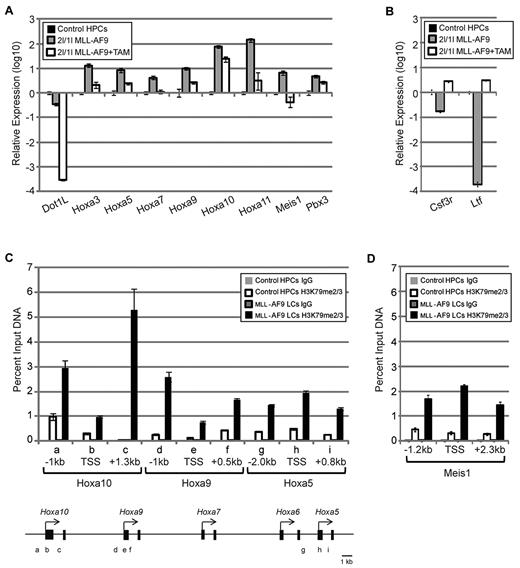
We have presented evidence supporting a function for DOT1L in the initiation and maintenance of transformation by the leukemic oncoprotein MLL-AF9, including clonal growth and proliferation. Previous studies have shown that the Hoxa gene cluster is up-regulated in MLL-AF9 LSCs.30,35 To determine whether DOT1L is involved in Hoxa gene up-regulation, we analyzed the expression by RT-qPCR. We found that all analyzed Hoxa genes are up-regulated in MLL-AF9–transformed cells compared with normal HPCs (Figure 6A). Importantly, DOT1L plays a crucial role in activation of these Hoxa genes as TAM-induced DOT1L depletion caused their down-regulation (Figure 6A). In addition, 2 Hoxa cofactors, Meis1 and Pbx3, required for maintaining MLL-AF9 leukemic stem cell potential,36 are also down-regulated in response to TAM-induced DOT1L depletion (Figure 6A). Previous studies have shown that in addition to their role in maintaining LSC identity, continued Hoxa and Meis1 expression also help block differentiation.30,37 Accordingly, Csf3r and Ltf genes associated with myeloid differentiation and down-regulated in MLL-AF9 LCs are also reactivated on TAM-induced DOT1L deletion (Figure 6B). These data support the notion that DOT1L is required for maintaining a transcriptional program that supports LSC self-renewal and blocks differentiation.
DOT1L directly regulates expression of Hoxa and Meis1 genes in MLL-AF9–transformed cells. (A) RT-qPCR analysis shows up-regulation of Hoxa cluster and their cofactors Meis1 and Pbx3, normalized to Gapdh, in MLL-AF9–transformed cells, which become down-regulated on DOT1L deletion. (B) RT-qPCR analysis shows down-regulation of myeloid differentiation markers, normalized to Gapdh, in MLL-AF9–transformed cells, which become up-regulated on DOT1L deletion. (C) ChIP analysis demonstrates that H3K79me2/3 is specifically enriched at Hoxa loci in MLL-AF9–transformed cells compared with control HPCs. IgG was used for control ChIP and amplicon positions are indicated (TSS indicates transcription start site). (D) ChIP analysis demonstrates that H3K79me2/3 is enriched in the Meis1 gene in MLL-AF9 LCs compared with that in the control HPCs. IgG was used for control ChIP and amplicon positions are indicated (TSS indicates transcription start site).
DOT1L directly regulates expression of Hoxa and Meis1 genes in MLL-AF9–transformed cells. (A) RT-qPCR analysis shows up-regulation of Hoxa cluster and their cofactors Meis1 and Pbx3, normalized to Gapdh, in MLL-AF9–transformed cells, which become down-regulated on DOT1L deletion. (B) RT-qPCR analysis shows down-regulation of myeloid differentiation markers, normalized to Gapdh, in MLL-AF9–transformed cells, which become up-regulated on DOT1L deletion. (C) ChIP analysis demonstrates that H3K79me2/3 is specifically enriched at Hoxa loci in MLL-AF9–transformed cells compared with control HPCs. IgG was used for control ChIP and amplicon positions are indicated (TSS indicates transcription start site). (D) ChIP analysis demonstrates that H3K79me2/3 is enriched in the Meis1 gene in MLL-AF9 LCs compared with that in the control HPCs. IgG was used for control ChIP and amplicon positions are indicated (TSS indicates transcription start site).
To determine whether DOT1L is directly involved in the regulation of Hoxa and Meis1 expression, we performed ChIP analysis using MLL-AF9–transformed cells. Because no ChIP-grade DOT1L antibody is available, we used an antibody that recognizes H3K79 di- and tri-methylation (H3K79me2/3; Abcam), modifications deposited by DOT1L.13 We chose to analyze Hoxa5 and Hoxa9 loci because they have been shown to be required for leukemogenesis of various MLL-fusion proteins.22,23,38 We also included Hoxa10 because its overexpression alone has been reported to induce transplantable AML in mice.39 ChIP analysis verified that Hoxa5, Hoxa9, and Hoxa10 are preferentially enriched for H3K79me2/3 in MLL-AF9–transformed cells compared with control HPCs (Figure 6C). Similarly, H3K79me2/3 is also enriched at the Meis1 locus in MLL-AF9–transformed cells compared with control HPCs (Figure 6C). These data suggest that DOT1L-mediated H3K79 methylation contributes directly to Hoxa and Meis1 gene activation in MLL-AF9–induced leukemogenesis.
Discussion
MLL-AF9 fusion caused leukemia represents approximately one-third of all infant acute leukemias and approximate one-third of adult AML.8 Recent studies demonstrating the interaction between MLL-AF9 and several functional partners such as Menin,40-42 wild-type MLL,43 and the PAF complex37,44 have shed some light on the underlying mechanism of how MLL-AF9 causes leukemia. However, a complete understanding of the molecular mechanism underlying MLL-AF9–induced leukemia requires the identification of all the important players. DOT1L has been previously shown to directly interact with the C-terminus of AF945 and has been identified in AF9-associated protein complexes.9,12,21 In addition, H2B ubiquitination mediated by the PAF complex is required for both DOT1L methyltransferase activity46,47 and for MLL-AF9 transformation.37,44 These data suggest a possible link between DOT1L and MLL-AF9–mediated leukemogenesis.
Using a TAM-inducible DOT1L deletion mouse system, we demonstrate that DOT1L is required for the initiation of transformation by MLL-AF9 both in vitro by methylcellulose colony formation assays and in vivo by bone marrow transplantation assays (Figures 1–2). In addition, we demonstrate that DOT1L is required for maintaining the transformed state of MLL-AF9–induced leukemia in vitro and in vivo as loss of DOT1L abrogated colony formation in methylcellulose colony replating assay as well as AML progression in mice (Figures 3–4). Further analysis revealed that DOT1L is required for maintaining expression of c-Kit, a marker of MLL-AF9 clonogenic leukemic progenitor/stem cells (Figure 4D). In addition, in vitro clonal growth capacity and cell proliferation of MLL-AF9, but not MLL-AFX, transformed cells (Figures 3,Figure 4–5). Consistent with the phenotypic effect, gene-expression analysis demonstrates that activation of a group of genes, including Hoxa cluster, Meis1, and Pbx3, important for LSCs is compromised on the loss of DOT1L function (Figure 4). Importantly, ChIP assay supports that DOT1L-mediated H3K79 methylation directly contributes to activation of these genes.
We note that while this paper was in preparation, 2 papers were published that address the role of DOT1L in MLL-AF9 transformation.26,48 Hess et al6 demonstrated that di- and tri-methyl H3K79 marks are selectively enriched at the Hoxa9 locus in MLL-AF9–transformed cells by ChIP,48 which is consistent with our ChIP analysis. Although Zhang et al45 did not perform ChIP, they did use a knockout approach to show that DOT1L is required for MLL-AF9 transformation in vitro. However, these studies did not evaluate the role of DOT1L in vivo, which leaves open the question of the in vivo relevance of their studies. Through bone marrow transplantation, here we demonstrate that DOT1L plays a critical role in the initiation and maintenance of MLL-AF9–mediated leukemia, thus firmly establishing the function of DOT1L in leukemia.
In contrast to our findings, Chang et al26 found that DOT1L is required for maintaining MLL-AFX transformation in vitro. One possible explanation is that the 2 studies used different mouse models. On Cre-recombination of the Dot1L conditional allele in our mouse model, exons 5 and 6 are excised. Given that exons 4 and 7 will still be transcribed and translated in-frame, a truncated mutant DOT1L missing 108 residues within the catalytic domain can still be generated (supplemental Figure 1). However, recombination of the conditional allele used in Chang et al only removes exon 5, which results in a frame-shift and the expression of a truncated protein containing only the first 87 residues.26 Their deletion may have a more severe phenotype because of the loss of other DOT1L functions aside from its catalytic activity.
We and others have previously shown that mistargeting of DOT1L and subsequent methylation on H3K79 plays an important role in the leukemogenesis process involving MLL-AF10, MLL-ENL, and MLL-AF4 transformation in vitro.22,24,25 Consistently, DOT1L has been shown to associate with all 3 of the aforementioned MLL fusion partners through direct or indirect interactions 9,10,12,21,22,24 . It is important to note that 2 reports have failed to identify DOT1L in an AF4-associated complex,11,49 putting into question the role of DOT1L in MLL-AF4 leukemia. However, it is indisputable that AF4 interacts with AF9 and that AF9 interacts with DOT1L; thus, DOT1L may be mistargeted to genomic loci by MLL-AF4 indirectly through its association with AF9. In support of this possibility, it has been shown that disruption of the AF4-AF9 interaction by the synthetic peptide PFWT inhibits MLL-AF4 cell proliferation,50 which is similar to the results we observe with MLL-AF9 in the absence of DOT1L.
This study, together with previous studies, strongly suggests a common mechanism by which DOT1L contributes to leukemogenesis of MLL-fusion proteins. We believe that mistargeting of DOT1L to Hoxa and Meis1 genes, subsequent H3K79 methylation, and up-regulation of these genes is a common mechanism underlying the involvement of DOT1L in leukemogenesis. Although a similar mechanism for MLL-AF10 and MLL-AF4 has been previously proposed,22,25,51 all previous studies were performed using in vitro systems with DOT1L knockdown or knockout. Using MLL-AF9 as a model, here we provide the first in vivo evidence demonstrating an essential role for DOT1L in leukemogenesis. Our study thus solidifies DOT1L's role in leukemias involving MLL rearrangements and makes DOT1L a prime candidate target for therapeutic intervention.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jay Hess for providing the pMSCN-MLL-AF9 construct and Michael Cleary for providing the pMSCN-MLL-AFX construct.
The work is supported by an NIH grant (CA119133). A.T.N. is a recipient of the Pre-doctoral Fellowship from the American Heart Association. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
National Institutes of Health
Authorship
Contribution: A.T.N. and Y.Z. designed research and wrote the paper; A.T.N. performed research; O.T. generated tamoxifen-inducible mice; and J.H. helped with bone marrow transplantation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yi Zhang, 450 West Dr, LCCC CB 7295, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599; e-mail: yi_zhang@med.unc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal