Abstract
Variant Philadelphia (Ph) chromosome translocations have been reported in 5%-10% of patients with newly diagnosed chronic myeloid leukemia (CML). Variant translocations may involve one or more chromosomes in addition to 9 and 22, and can be generated by 2 different mechanisms, 1-step and 2-step rearrangements, as revealed by fluorescence in situ hybridization. The prognostic significance of the occurrence of variant translocations has been discussed in previous studies. The European LeukemiaNet recommendations do not provide a “warning” for patients with variant translocations, but there is limited information about their outcome after therapy with tyrosine kinase inhibitors. To identify the role of variant translocations in early chronic phase (CP) CML patients treated with imatinib mesylate, we performed an analysis in a large series of 559 patients enrolled in 3 prospective imatinib trials of the Gruppo Italiano Malattie EMatologiche dell'Adulto (GIMEMA) Working Party on CML. Variant translocations occurred in 30 patients (5%). Our data show that the presence of variant translocations has no impact on the cytogenetic and molecular response or on outcome, regardless of the involvement of different mechanisms, the number of involved chromosomes, or the presence of deletions. Therefore, we suggest that patients with variant translocations do not constitute a “warning” category in the imatinib era. This study is registered at www.clinicaltrials.gov as NCT00514488 and NCT00510926.
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disorder characterized by the presence of the Philadelphia (Ph) chromosome resulting from the reciprocal translocation t(9;22)(q34;q11).1,2 The molecular consequence of this translocation is the generation of the BCR-ABL fusion gene, which encodes a constitutively active protein tyrosine kinase. The oncogenic protein tyrosine kinase, which is located in the cytoplasm, is responsible for the leukemia phenotype through the constitutive activation of multiple signaling pathways involved in the cell cycle and in adhesion and apoptosis.3
In 5%-10% of newly diagnosed CML cases, one or more additional chromosomes are added to 9 and 22 and are involved in the translocation, and this is termed variant translocation.4,5 Occasionally, the chromosome changes are submicroscopic so the translocation can be masked and revealed only by fluorescence in situ hybridization (FISH) or by molecular analysis.3
The mechanisms of the generation of the variant translocations are not fully clear; some authors have suggested 2 different mechanisms: a 1-step mechanism in which chromosome breakage occurs simultaneously on 3 or 4 different chromosomes in 3-way or 4-way translocation, respectively, and a 2-step mechanism involving 2 sequential translocations in which a standard t(9;22) translocation is followed by a second translocation involving additional chromosomes.4-6 The 2-step mechanism may suggest that the formation of a variant translocation is similar to clonal evolution, and therefore this mechanism might be associated with a poorer prognosis.4-6 FISH is the technique used at present to determine the mechanism of variant translocations.
Some previous studies have suggested that patients with variant Ph translocations may have a worse outcome than those with classic translocations, whereas other studies have reported no different impact on prognosis in imatinib mesylate (IM)–treated patients in either the chronic phase (CP) or the accelerated phase.6,8 In the pre-imatinib era, some studies reported a strong association of variant translocations with the presence of der(9) deletions,9-11 suggesting that the poor prognosis may be due to the increased frequency of the latter change. No systematic studies of early CP patients in large prospective clinical trials with imatinib have been reported previously.
We report an analysis of the Gruppo Italiano Malattie EMatologiche dell'Adulto (GIMEMA) Working Party (formerly the Italian Cooperative Study Group) on CML aiming to clarify the role of variant Ph translocations in newly diagnosed CML patients in CP treated with IM as a first-line therapy. To our knowledge, this is the first large, systematic study performed within prospective studies.
Methods
Patients
The study involved 559 patients, all of whom were at least 18 years old, had Ph-positive and BCR-ABL–positive CML in early CP, and were enrolled in 3 concurrent studies sponsored by the GIMEMA working party on CML that opened for enrollment in 2004: (1) CML/021 (www.clinicaltrials.gov NCT00514488), a phase 2 trial exploring IM 800 mg in intermediate Sokal risk CP CML; (2) CML/022 (www.clinicaltrials.gov NCT00510926), a phase 3 trial comparing IM 400 vs 800 mg in high Sokal risk CP CML; and (3) CML/023, an observational trial of IM 400 mg in CP CML. All patients provided written informed consent before the enrollment.
The studies were reviewed and approved by the institutional review boards of all participating institutions, and were performed in accordance with Good Clinical Practice and the Declaration of Helsinki.
Treatment monitoring and definition of response
Blood count and serum chemistry were performed at enrollment, monthly until the 12th month of treatment, and every 3 months thereafter. A complete hematologic response (CHR) was defined as a white blood cell count of < 10 × 109/L, a platelet count of < 450 × 109/L, no immature cells (blasts, promyelocytes, or myelocytes) in the peripheral blood, and the disappearance of all signs and symptoms related to leukemia (including palpable splenomegaly).
All study protocols required conventional cytogenetics and FISH performed on bone marrow cells at baseline, after 6 and 12 months of treatment, and every 6 months thereafter or in case of failure or disease progression. Cytogenetic response, based on the results of conventional banding analysis, was identified as: complete (CCgR), 0% Ph-positive metaphases; partial (PCgR), 1%-35% Ph-positive metaphases; major (MCgR), 0%-35% Ph-positive metaphases; minor, 36%-65% Ph-positive metaphases; minimal, 66%-95% Ph-positive metaphases; or no response, > 95% Ph-positive metaphases, as described previously.12,13
Real-time quantitative PCR was performed on peripheral blood and bone marrow samples at baseline and after 3, 6, and 12 months; and on peripheral blood every 6 months thereafter. Qualitative RT-PCR for the BCR-ABL transcript was routinely performed at enrollment on bone marrow for determining the type of transcript. The molecular response was defined as major (MMolR) if the BCR-ABL/ABL ratio was < 0.10% on the International Scale.14-16
Cytogenetic analysis
The GIMEMA Working Party on CML has established a network of cytogenetic laboratories covering all the country. Fourteen of these laboratories performed cytogenetic studies for more than one clinical center (reference laboratories), and 24 laboratories performed cytogenetic studies for their respective clinical center (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Chromosome-banding analysis was performed on bone marrow cells after short-term culture (24 and/or 48 hours). The cells were treated with a colchicine and hypotonic solution, the pellet was fixed and washed in methanol-acetic acid (3:1), and the cells were resuspended in fixative and dropped onto slides. Karyotypes were examined after G- or Q-banding techniques according to the International System for Human Cytogenetic Nomenclature (ISCN 2009)17 and were not centrally reviewed. At least 20 metaphases were analyzed in 87% of samples and 10-19 metaphases in 13% of samples.
FISH analysis
FISH was performed on bone marrow cells prepared according to standard cytogenetic techniques and using DNA probes that hybridize at the BCR and ABL regions. The mostly frequently used probes were: the Locus Specific Identifier BCR/ABL Dual-Color, Dual-Fusion Translocation Probe (Abbott Molecular-Vysis) or the Double-Fusion Signal D-FISH BCR/ABL Probe (Oncor-QBiogene). The dual-fusion strategy uses probes that span the common breakpoints in the ABL and BCR gene regions. A normal nucleus will exhibit 2 orange and 2 green signal patterns; a nucleus with a classic t(9;22) translocation will display one orange (normal 9 chromosome), one green (normal 22 chromosome), and 2 yellow fusion signals on the derivative chromosomes der(9) and der(22).18,19
In one case, an extra signal system was applied: the Locus Specific Identifier BCR/ABL Extra-Signal Dual-Color Translocation Probe (Abbott Molecular-Vysis). Using this probe, a normal nucleus will exhibit 2 orange and 2 green signal patterns; a nucleus with a classic t(9;22) translocation will display 2 orange signals on normal 9 chromosome and der(9), and one yellow fusion signal on der(22) chromosomes.
In cases with a variant Ph translocation or with structural changes involving chromosomes 9 and 22, the hybridization pattern may vary. Hybridization procedures were done according to the manufacturer's recommendation with slight modifications. Only one laboratory used a PAC/BAC system: a pool of PAC, RP5-1132H12, and RP5-835J22 for the ABL gene and BAC and RP11-164n13 for the BCR gene.20 FISH analysis was performed on 200-300 nuclei and on metaphases to verify the interphase FISH pattern and to localize the rearrangements.
Molecular analysis
All samples and tests were centralized in Bologna, Italy. Whole buffy-coat cells were used. RNA extraction, RT-PCR, and real-time quantitative PCR were performed according to European recommendations, as described previously.21-23 Real-time quantitative PCR was performed on an ABI PRISM 7700 sequence detector (Perkin Elmer).24 ABL was used as housekeeping gene to correct differences in RNA quality and/or RT efficacy. BCR-ABL and ABL plasmid dilutions (Ipsogen) were used as standards. The final results were calculated as the ratios of BCR-ABL to ABL and are expressed as percentages. All experiments were performed in duplicate, and the results are expressed as the percent ratio to ABL. The BCR-ABL:ABL ratios were further multiplied by the conversion factor of the Bologna laboratory to set the results on an international scale.14-16 Samples yielding an ABL threshold cycle > 30, corresponding to less than 1000 ABL transcript copies, were considered to have degraded RNA and were discarded.
Definition of progression, failure, and events
Progression to the accelerated/blast phase was identified by a blood myeloblast or myeloblast and promyelocyte percentage of at least 10% or 30%, respectively, or by any extramedullary blast involvement, excluding spleen and liver. According to 2009 European LeukemiaNet recommendations, failures were defined as no CHR at 3 months, no CgR at 6 months, no PCgR at 12 months, no CCgR at 18 months, loss of CHR or CCgR, new BCR-ABL mutation insensitive to IM, or progression to the accelerated/blast phase.25 Events were defined as treatment failure or permanent discontinuation of IM for any reason, including toxicity, patient refusal, or loss to follow-up.
Statistical analysis
All comparisons between the 2 different groups of patients (those with and without variant translocations) were assessed with the Student t test and with Fisher exact test for categorical variables, as appropriate. Prism 4 software (GraphPad) was used throughout for statistical analysis. Times to CCgR and MMolR were calculated from the date of start of treatment until the achievement of the response. Overall survival (OS), progression-free survival (PFS), failure-free survival (FFS), and event-free survival (EFS) were calculated from the date of the first IM dose until death (OS), until progression to accelerated phase or blast phase or death (PFS), until failure or death (FFS), and until any event (EFS). Probabilities of CCgR, MMolR, OS, PFS, FFS, and EFS were calculated using the Kaplan-Meier method.26 Survival times were compared with the log-rank test.27 Multivariate logistic regression analysis was used to assess the relationship between various predictors of interest and response or outcome. All analyses were performed according to the intention-to-treat principle.
Results
In total, 559 patients were enrolled in the GIMEMA Working Party on CML trials, and were evaluable by cytogenetic analysis at diagnosis. Thirty patients (5%) showed variant Ph translocations and 529 patients (95%) showed classic t(9;22)(q34;q11) translocations.
Detailed baseline characteristics of the 2 groups are presented in Table 1. The 2 groups, those with and those without variant translocations, were similar in demographic and hematologic characteristics. The Sokal and Hasford risk score distributions were also comparable. The proportion of patients treated with high-dose IM was higher in the variant translocation groups, but the difference was not statistically significant. The presence of additional cytogenetic abnormalities was comparable in patients with and without variant translocations. Finally, the frequency of der(9) deletions was higher in patients with variant translocations, which is in agreement with previous studies,4,7 but the difference was not statistically significant.
Comparison of patient characteristics at diagnosis
| Characteristics . | Patients with variant translocations (n = 30) . | Patients without variant translocations (n = 529) . | P . |
|---|---|---|---|
| Median age, y (range) | 52 (33-84) | 52 (18-80) | .49 |
| Sex, male/female; n (%) | 19/11 (63/37) | 317/212 (60/40) | .85 |
| Spleen, cm; median (range) | 1.5 (0-18) | 1 (0-24) | .76 |
| Median hemoglobin level, g/L (range) | 11.8 (7.9-16.1) | 12.2 (6.4-17.5) | .41 |
| Median platelet count, 109/L (range) | 317 (124-1364) | 353 (100-4920) | .36 |
| Peripheral blasts, %; median (range) | 2 (0-5) | 1 (0-10) | .55 |
| Eosinophils, %; median (range) | 2 (0-6) | 2 (0-15) | .89 |
| Basophils, %; median (range) | 2 (0-5) | 2 (0-19) | .08 |
| Sokal risk score | |||
| low | 13 (43) | 206 (39) | .81 |
| intermediate | 7 (23) | 209 (40) | |
| high | 10 (33) | 114 (22) | |
| Hasford risk score | |||
| low | 10 (33) | 229 (43) | .69 |
| intermediate | 15 (50) | 266 (50) | |
| high | 5 (17) | 34 (6) | |
| Additional cytogenetic abnormalities | 1 (3) | 20 (4) | 1.00 |
| Deletion der(9) | 6 (20) | 54 (10) | .12 |
| Imatinib dose | |||
| 400 mg | 20 (67) | 403 (76) | .27 |
| 800 mg | 10 (33) | 126 (24) |
| Characteristics . | Patients with variant translocations (n = 30) . | Patients without variant translocations (n = 529) . | P . |
|---|---|---|---|
| Median age, y (range) | 52 (33-84) | 52 (18-80) | .49 |
| Sex, male/female; n (%) | 19/11 (63/37) | 317/212 (60/40) | .85 |
| Spleen, cm; median (range) | 1.5 (0-18) | 1 (0-24) | .76 |
| Median hemoglobin level, g/L (range) | 11.8 (7.9-16.1) | 12.2 (6.4-17.5) | .41 |
| Median platelet count, 109/L (range) | 317 (124-1364) | 353 (100-4920) | .36 |
| Peripheral blasts, %; median (range) | 2 (0-5) | 1 (0-10) | .55 |
| Eosinophils, %; median (range) | 2 (0-6) | 2 (0-15) | .89 |
| Basophils, %; median (range) | 2 (0-5) | 2 (0-19) | .08 |
| Sokal risk score | |||
| low | 13 (43) | 206 (39) | .81 |
| intermediate | 7 (23) | 209 (40) | |
| high | 10 (33) | 114 (22) | |
| Hasford risk score | |||
| low | 10 (33) | 229 (43) | .69 |
| intermediate | 15 (50) | 266 (50) | |
| high | 5 (17) | 34 (6) | |
| Additional cytogenetic abnormalities | 1 (3) | 20 (4) | 1.00 |
| Deletion der(9) | 6 (20) | 54 (10) | .12 |
| Imatinib dose | |||
| 400 mg | 20 (67) | 403 (76) | .27 |
| 800 mg | 10 (33) | 126 (24) |
Cytogenetic analysis
A total of 30 CML cases of variant translocations were identified and are described in Table 2. We did not find recurring breakpoints. In addition to chromosomes 9 and 22, the most involved chromosome was chromosome 17 (4 cases), followed by chromosomes 1, 9, 11, 12, and 15 (3 cases); chromosomes 3, 5, 8, 16 (2 cases); and chromosomes 10, 14, 19, 21, and 22 (1 case). The majority of cases (28 of 30; 93%) showed a 3-way translocation, and only 2 translocations (7%) involved 4 chromosomes (cases n.3 and n.29). One case (n.22) did not show the Ph chromosome by conventional cytogenetics (masked Ph chromosome), and only one case (n.19) carried an additional cytogenetic abnormality: t(7;19)(q21;p13) in all of the metaphases.
Cytogenetic results and treatment responses of 30 patients with variant translocations
| Case . | Sex . | Karyotype at diagnosis . | Der(9) deletion . | FISH pattern . | Mechanism . | CgR at 12 mo . | CgR at last contact . |
|---|---|---|---|---|---|---|---|
| 1 | F | 46,XX,t(3;9;22)(q27;q34;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 2 | F | 46,XX,t(5;9:22)(q13;q34;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 3 | M | 46,XY,t(5;9;22;17)(q12;q34;q11;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 4 | M | 46,XY,t(8;9;22)(q21;q34;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 5 | F | 46,XX,t(8,9,22)(q22;q34;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 6 | M | 46,XY,t(9;9;22)(p13;q34;q22) | 1F2R2G | 1-step | Dropout | Death | |
| 7 | M | 46,XY,t(9;22;10)(q34;q11;q21) | 1F2R2G | 1-step | CCgR | CCgR | |
| 8 | F | 46,XX,t(9;22;11)(q34;q11;q13) | 1F2R2G | 1-step | CCgR | CCgR | |
| 9 | M | 46,XY,t(9;22;11)(q34;q11;q13) | 1F2R2G | 1-step | CCgR | CCgR | |
| 10 | M | 46,XY,t(9;22;12)(q34;q11;q24) | 1F2R2G | 1-step | CCgR | CCgR | |
| 11 | M | 46,XY,t(9;22;14)(q34;q11;q32) | 1F2R2G | 1-step | NE | CCgR | |
| 12 | M | 46,XY,t(9;22;15)(q34;q11;q22) | 1F2R2G | 1-step | Dropout | Death | |
| 13 | M | 46,XY,t(9;22;15)(q34;q11;q21) | 1F2R2G | 1-step | Failure | Failure | |
| 14 | M | 46,XY,t(9;22;17)(q34;q11;p13) | 1F2R2G | 1-step | CCgR | CCgR | |
| 15 | M | 46,XY,t(9;22;17)(q34;q11;q21) | 1F2R2G | 1-step | PCgR | CCgR | |
| 16 | F | 46,XX,t(9;22;19)(q34;q11;p13) | 1F2R2G | 1-step | CCgR | CCgR | |
| 17 | M | 46,XY,t(9;9;22)(q34;q34;q11) | 2F1R1G | 2-step | CCgR | Failure | |
| 18 | F | 46,XX,t(9;22;16)(q34;q11;p12) | 2F1R1G | 2-step | CCgR | CCgR | |
| 19 | F | 46,XX,t(9;22;22)(q34;q11;q11), t(7;19)(q21;p13) | 2F1R | 1-step | CCgR | CCgR | |
| 20 | M | 46,XY,t(9;22;11)(q34;q11;q13) | BCR on 3rd chromosome | 1F2R1G | 2-step | CCgR | CCgR |
| 21 | F | 46,XX,t(9;22;15)(q34;q11;q15) | BCR on 3rd chromosome | 1F2R1G | 1-step | CCgR | Death |
| 22 | M | 46,XY,t(1;9;22)(q21;q34;q11) Ph masked | 1F2R1G | Multistep | Failure | Failure | |
| 23 | M | 46,XY,t(9;22;12)(q34;q11;q24) | ABL1/BCR on 3rd chromosome | 1F1R1G | 2-step | NE | CCgR |
| 24 | F | 46,XX,t(9;9;22)(p13;q34;q22) | ABL1 | 1F1R1G | Multistep | PCgR | CCgR |
| 25 | M | 46,XY,t(1;9;22)(?;q34;q11) | NE | NE | CCgR | CCgR | |
| 26 | F | 46,XX,t(1;9;22)(?;q34;q11) | ABL1 | NE | NE | Failure | Failure |
| 27 | F | 46,XX,t(9;22;12)(q34;q11;?) | NE | NE | CCgR | CCgR | |
| 28 | M | 46,XY,t(3;9;22)(p21;q34;q11) | ABL1 | NE | NE | CCgR | CCgR |
| 29 | M | 46,XY,t(9;22;16;17)(q34;q11;?;?) | NE | NE | CCgR | CCgR | |
| 30 | M | 46,XY,t(9;22;21)(q34;q11;?) | NE | NE | CCgR | CCgR |
| Case . | Sex . | Karyotype at diagnosis . | Der(9) deletion . | FISH pattern . | Mechanism . | CgR at 12 mo . | CgR at last contact . |
|---|---|---|---|---|---|---|---|
| 1 | F | 46,XX,t(3;9;22)(q27;q34;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 2 | F | 46,XX,t(5;9:22)(q13;q34;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 3 | M | 46,XY,t(5;9;22;17)(q12;q34;q11;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 4 | M | 46,XY,t(8;9;22)(q21;q34;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 5 | F | 46,XX,t(8,9,22)(q22;q34;q11) | 1F2R2G | 1-step | CCgR | CCgR | |
| 6 | M | 46,XY,t(9;9;22)(p13;q34;q22) | 1F2R2G | 1-step | Dropout | Death | |
| 7 | M | 46,XY,t(9;22;10)(q34;q11;q21) | 1F2R2G | 1-step | CCgR | CCgR | |
| 8 | F | 46,XX,t(9;22;11)(q34;q11;q13) | 1F2R2G | 1-step | CCgR | CCgR | |
| 9 | M | 46,XY,t(9;22;11)(q34;q11;q13) | 1F2R2G | 1-step | CCgR | CCgR | |
| 10 | M | 46,XY,t(9;22;12)(q34;q11;q24) | 1F2R2G | 1-step | CCgR | CCgR | |
| 11 | M | 46,XY,t(9;22;14)(q34;q11;q32) | 1F2R2G | 1-step | NE | CCgR | |
| 12 | M | 46,XY,t(9;22;15)(q34;q11;q22) | 1F2R2G | 1-step | Dropout | Death | |
| 13 | M | 46,XY,t(9;22;15)(q34;q11;q21) | 1F2R2G | 1-step | Failure | Failure | |
| 14 | M | 46,XY,t(9;22;17)(q34;q11;p13) | 1F2R2G | 1-step | CCgR | CCgR | |
| 15 | M | 46,XY,t(9;22;17)(q34;q11;q21) | 1F2R2G | 1-step | PCgR | CCgR | |
| 16 | F | 46,XX,t(9;22;19)(q34;q11;p13) | 1F2R2G | 1-step | CCgR | CCgR | |
| 17 | M | 46,XY,t(9;9;22)(q34;q34;q11) | 2F1R1G | 2-step | CCgR | Failure | |
| 18 | F | 46,XX,t(9;22;16)(q34;q11;p12) | 2F1R1G | 2-step | CCgR | CCgR | |
| 19 | F | 46,XX,t(9;22;22)(q34;q11;q11), t(7;19)(q21;p13) | 2F1R | 1-step | CCgR | CCgR | |
| 20 | M | 46,XY,t(9;22;11)(q34;q11;q13) | BCR on 3rd chromosome | 1F2R1G | 2-step | CCgR | CCgR |
| 21 | F | 46,XX,t(9;22;15)(q34;q11;q15) | BCR on 3rd chromosome | 1F2R1G | 1-step | CCgR | Death |
| 22 | M | 46,XY,t(1;9;22)(q21;q34;q11) Ph masked | 1F2R1G | Multistep | Failure | Failure | |
| 23 | M | 46,XY,t(9;22;12)(q34;q11;q24) | ABL1/BCR on 3rd chromosome | 1F1R1G | 2-step | NE | CCgR |
| 24 | F | 46,XX,t(9;9;22)(p13;q34;q22) | ABL1 | 1F1R1G | Multistep | PCgR | CCgR |
| 25 | M | 46,XY,t(1;9;22)(?;q34;q11) | NE | NE | CCgR | CCgR | |
| 26 | F | 46,XX,t(1;9;22)(?;q34;q11) | ABL1 | NE | NE | Failure | Failure |
| 27 | F | 46,XX,t(9;22;12)(q34;q11;?) | NE | NE | CCgR | CCgR | |
| 28 | M | 46,XY,t(3;9;22)(p21;q34;q11) | ABL1 | NE | NE | CCgR | CCgR |
| 29 | M | 46,XY,t(9;22;16;17)(q34;q11;?;?) | NE | NE | CCgR | CCgR | |
| 30 | M | 46,XY,t(9;22;21)(q34;q11;?) | NE | NE | CCgR | CCgR |
In the FISH patterns, R indicates red signal; G, green signal; and F, fusion signal.
NE indicates not evaluable; failure was according to the definition by the 2009 European LeukemiaNet recommendations; and dropout, IM permanent discontinuation for any reason other than failure.
FISH signal patterns: identification of different mechanisms
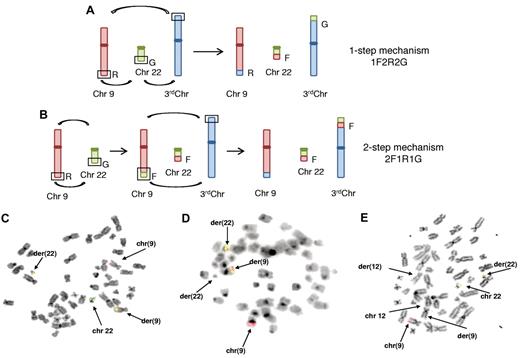
The FISH signal pattern was evaluable in 24 of 30 (80%) patients. The most common signal pattern (n.1-n.16; 53%) was one fusion signal on der(22), 2 red signals on normal and der(9) chromosomes, and 2 green signals on normal chromosome 22 and on the third involved chromosome, 1F2R2G (Figure 1A). This signal pattern was indicative of a 1-step mechanism in which 3 chromosomes broke and rejoined at the same time.
Mechanisms of genesis and FISH analysis of variant translocations. Shown are the 1-step mechanism (A) and the 2-step mechanism (B). The segments in the open boxes exchange material, as shown by the curved arrows. The letters indicate the probe colors used in FISH studies: R for red, labeling the ABL1 gene sequence region; G for green, labeling the BCR green sequence region; and F for fusion.4 (C) Case n.17, 46, XY, t(9;9;22)(q34;q34;q11). (D) Case n.19, 46, XX, t(9;22;22)(q34;q11;q11), t(7;19)(q21;p13). (E) Case n.23, 46, XY, t(9;22;12)(q34;q11;q24). BCR is shown by the green signal, ABL by the red signal, and fusion by the green/red signal (see “Results”).
Mechanisms of genesis and FISH analysis of variant translocations. Shown are the 1-step mechanism (A) and the 2-step mechanism (B). The segments in the open boxes exchange material, as shown by the curved arrows. The letters indicate the probe colors used in FISH studies: R for red, labeling the ABL1 gene sequence region; G for green, labeling the BCR green sequence region; and F for fusion.4 (C) Case n.17, 46, XY, t(9;9;22)(q34;q34;q11). (D) Case n.19, 46, XX, t(9;22;22)(q34;q11;q11), t(7;19)(q21;p13). (E) Case n.23, 46, XY, t(9;22;12)(q34;q11;q24). BCR is shown by the green signal, ABL by the red signal, and fusion by the green/red signal (see “Results”).
Two cases (n.17 and n.18; 7%) showed a different FISH pattern: 1 red, 1 green, 1 fusion on the Ph chromosome, and 1 fusion on the third chromosome, 2F1R1G. This can be explained by a 2-step mechanism: a classic translocation t(9;22) followed by a second translocation involving the ABL/BCR fusion from chromosome 9 to the third chromosome (Figure 1B-C).
We also found 4 other FISH patterns that could have originated from other mechanisms probably associated with deletions of the involved chromosomes. In the first, a 2F1R signal pattern was observed in case 19: 1 red signal on normal chromosome 9, 1 fusion signal on der(9), and 1 fusion signal on chromosome 22, apparently bigger than the classic Ph chromosome by conventional cytogenetics. Chromosome 22 without any signal was smaller than the normal one (Figure 1D). We have suggested a 1-step mechanism in which the signal fusion, normally located on der(22), moved to the third chromosome, which in this case was the other chromosome 22.
In the second FISH pattern, 3 cases (n.20-n.22) showed the same 1F2R1G pattern, a result of different events during the translocations. Case n.20 showed 1 fusion signal on der(22), 1 green signal on normal chromosome 22, and 2 red signals on normal chromosome 9 and on der(11), suggesting a 2-step mechanism with a loss of the BCR signal usually located on the third chromosome. Case n.21 showed 1 fusion signal on der(22), 1 green signal on normal chromosome 22, and 2 red signals on normal chromosome 9 and on der(9). In this pattern, a 1-step mechanism probably occurred, with loss of the BCR signal on the third chromosome. In case n.22, Ph was masked by conventional cytogenetics, chromosomes 1 and 9 showed evident abnormalities, whereas FISH analysis revealed 1 fusion signal on der(22), 1 red signal on der(9), and ABL1 and BCR signals on normal chromosomes 9 and 22, suggesting a multistep rearrangement.
In the third pattern, 1 case (n.23) displayed a 1F1R1G signal pattern: 1 fusion signal on der(22), 1 red signal on normal chromosome 9, and 1 green signal on normal chromosome 22. Chromosome analysis showed that this translocation involved chromosome 12 and could have originated via a 2-step mechanism, with deletion of material of der(9) moving to der(12) (Figure 1E).
In the fourth FISH pattern, 1 case (n.24) was analyzed only by Extra-Signal FISH (Abbott Molecular-Vysis) because of the scarcity of material for the analysis, and showed a 1F1R1G signal pattern. As described previously, FISH painting revealed a multistep mechanism that led to a fusion signal on the short arm of chromosome 9 and no signal on der(9q).28
In all cases except n.19 and n.24, the BCR-ABL fusion gene was located on der(22q). Deletions of der(9q) were present in 6 cases with variant translocations (20%). In 3 patients (n.24, n.26, and n.28), the translocation was associated with deletion of the ABL1 signal; in 2 cases (n.20 and n.21), it was associated with deletion of the BCR signal on the third chromosome, and in 1 case (n.23) there was deletion of the signal fusion (ABL/BCR) on the third chromosome.
Response to IM therapy and survival
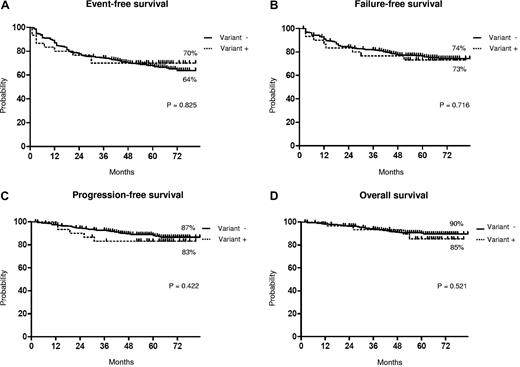
The median follow-up times were 61 months (range 2-83 months) and 64 months (range 30-81 months) in patients without and with variant translocations, respectively. The response rates to IM are shown in Table 3. No significant difference in CHR, CCgR, and MMolR was observed between patients without or with variant translocations. The median time to CCgR was 6 months in both groups; the overall estimated probability of CCgR was 90% and 84% in patients without and with variant translocations, respectively (P = .49). The median duration of cytogenetic response was 51 and 59 months in patients without and with variant translocations, respectively (P = .35). The median time to MMolR was 6 months and 9 months in patients without and with variant translocations, respectively; the overall estimated probability of MMolR was 90% and 84% in patients without and with variant translocations, respectively (P = .66; Figure 2). The number of patients alive without failure on IM was similar in the 2 groups: 23 of 30 patients with variant translocations (77%; Table 2) and 412 of 529 patients without variant translocations (78%; Figure 3). The estimated overall probabilities for EFS, FFS, PFS, and OS were 64% and 70% (P = .82), 74% and 73% (P = .72), 87% and 83% (P = .42), and 90% and 85% (P = .52) for patients without and with variant translocations, respectively (Figure 3). The 2 groups were not significantly different. We observed only 2 cases (n.3 and n.29) carrying 4-way translocations that showed CCgR and MMolR.
Response to imatinib
| Response . | Patients with variant translocations (n = 30) . | Patients without variant translocations (n = 529) . | P . |
|---|---|---|---|
| CHR | |||
| Overall | 28 (93%) | 518 (98%) | .150 |
| CCgR | |||
| 12-month | 21 (70%) | 413 (78%) | .366 |
| Overall | 25 (83%) | 466 (88%) | .394 |
| MMolR | |||
| 12-month | 17 (57%) | 312 (59%) | .850 |
| Overall | 25 (83%) | 450 (85%) | .793 |
| Response . | Patients with variant translocations (n = 30) . | Patients without variant translocations (n = 529) . | P . |
|---|---|---|---|
| CHR | |||
| Overall | 28 (93%) | 518 (98%) | .150 |
| CCgR | |||
| 12-month | 21 (70%) | 413 (78%) | .366 |
| Overall | 25 (83%) | 466 (88%) | .394 |
| MMolR | |||
| 12-month | 17 (57%) | 312 (59%) | .850 |
| Overall | 25 (83%) | 450 (85%) | .793 |
Patients were categorized by their response at 12 months and by their best overall response.
Cytogenetic and molecular response. Shown are the results of the Kaplan-Meier analysis of estimates of time to CCgR (A) and of time to MolR by variant translocations (B). The dotted line indicates the presence of variant translocations, and the solid line indicates the presence of classic translocations.
Cytogenetic and molecular response. Shown are the results of the Kaplan-Meier analysis of estimates of time to CCgR (A) and of time to MolR by variant translocations (B). The dotted line indicates the presence of variant translocations, and the solid line indicates the presence of classic translocations.
Long-term outcome. Shown are the results of the Kaplan-Meier analysis of estimates of EFS (A), FFS (B), PFS (C), and OS (D). The dotted line indicates the presence of variant translocations, and the solid line indicates the presence of classic translocations.
Long-term outcome. Shown are the results of the Kaplan-Meier analysis of estimates of EFS (A), FFS (B), PFS (C), and OS (D). The dotted line indicates the presence of variant translocations, and the solid line indicates the presence of classic translocations.
Regarding the type of rearrangement, 18 cases involved a 1-step mechanism, whereas in 4 cases, a 2-step mechanism was reported and in 2 cases a probable multistep mechanism. Of 18 cases involving a 1-step mechanism, 14 are actually in CCgR and MMolR, 3 went off trial, and the last failed to respond. Of 4 cases involving the 2-step mechanism, 3 are in CCgR and MMolR and 1 failed to respond after the first year of treatment, with the appearance of additional abnormalities described as der(3) t(3;?), del(7q), add(21q). One case involving a multistep mechanism maintained CCgR during the follow-up and the other one failed to achieve CHR. Because of the small number of cases with different variants of the BCR/ABL rearrangement, their association with outcome was not assessable.
Discussion
Usually, variant Ph translocations are observed at diagnosis in CP CML patients, and their occurrence is not associated with disease evolution. These translocations have been reported in 5%-10% of CML patients at diagnosis.5-8 The prognostic significance of the occurrence of variant translocations has been discussed in previous studies, but data about the outcome of patients with variant translocations after therapy with tyrosine kinase inhibitors are limited. The studies that are available are usually case reports, small series, or large series but involving patients in different phases of disease and treated with different drugs (eg, interferon and tyrosine kinase inhibitors). Therefore, the European LeukemiaNet recommendations do not provide a warning for patients with variant translocations, suggesting that variant translocations have no impact on the prognosis of CML.29 Further, some studies have suggested that patients with variant translocations do have adverse outcomes,4,30-33 whereas others have reported that such translocations have no influence on response or survival.6,8,34-37
A previous analysis reported 44 cases with variant translocations in a series of 721 patients (4%) who had failed prior interferon therapy and were either in CP or the accelerated phase. No differences in outcome were evident in that study. Multivariate analysis showed that the variant translocations had no impact in response rate, OS, or duration of response when the patients were treated with imatinib.8 Another more recent report described 10 CML patients carrying variant translocations among 153 newly diagnosed CP cases (6.5%). The results of that study suggested that the variants might confer an unfavorable clinical outcome, because only 2 patients achieved an optimal response to tyrosine kinase inhibitor (imatinib and nilotinib) treatment according to European LeukemiaNet recommendations.33 The investigators assumed that the involvement of additional chromosomes in the BCR/ABL rearrangement could adversely affect outcome when a selective tyrosine kinase inhibitor such as imatinib was used, because these genetic changes might be markers of genomic instability and can be considered as clonal evolution.38
In the present study, we report a large series of 559 early CP CML patients treated with IM as frontline therapy within 3 clinical trials of the GIMEMA Working Party on CML. Variant translocations occurred in 30 patients (5%), a frequency consistent with previous reports. We were not able to determine a cluster of breakpoints on specific chromosomes, because 15 different chromosomes were involved as the third or fourth chromosomes in the variant translocations.
In our series of early CP patients, the presenting features were not different between the groups with and without variant translocations (Table 1). The proportion of patients treated with high-dose IM was higher in the variant translocations group, but the difference was not statistically significant. Although the number of patients with variant translocations was relatively small, in our experience, the presence of variant translocations does not influence the response to IM therapy. The rates of CCgR and MMolR (Table 3), the stability of CCgR, and the outcome (Figures 2 and 3) of 30 patients with variant translocations were all similar to those of patients with classic translocations. According to the International Randomized Study of Interferon and STI571 (IRIS) trial,39 progression to the advanced phase and events are more frequent within the first 4 years of IM treatment. In our study, neither group has reached the median time to progression, but it might be reasonable to predict that significant differences will not appear with further observations.
The availability of double-fusion FISH provides a useful tool with which to investigate the mechanism of the genesis of the variant translocations. In most cases, one chromosome additional to chromosomes 9 and 22 is involved by 2 different mechanisms: 1-step or 2-step. The 1-step mechanism is characterized by a simultaneous translocation of 3 chromosomes in a 3-break event, and the 2-step by a classic t(9;22) translocation, followed by a second translocation between chromosome 9 and the third involved chromosome in a 4-break event. Other cases may be characterized by additional mechanisms in which multiple simultaneous rearrangements occur in association with insertions and/or deletions. The role of these different mechanisms has been described previously, and no effect on response to IM therapy was shown.8 In the present study, the 1-step mechanism occurred in 18 of 24 (75%) evaluable cases, as shown by FISH signal characterization, whereas we observed a minority of cases with a 2-step mechanism (4 of 24; 16.7%) or with a complex mechanism (2 of 24; 8.3%). In the previous studies, the 1-step mechanism was also more frequent than the 2-step mechanism.4,6,40 We did not notice any strong differences in terms of response rate or survival related to the different mechanisms of rearrangement or the number of involved chromosomes; however, we only found 6 cases with 2-step or more complex mechanisms and only two 4-way translocation cases.
Deletions of a sizable portion of the derivative chromosome 9 have been described in 10%-15% of CML patients. These deletions have been found to occur more frequently in patients with variant Ph translocations.4,7,8,11,41 However, a recent study42 asserted that deletions of der(9) do not influence the response or outcome of CML in early CP patients treated with IM. In the present study, 6 of 30 patients (20% vs 10% without variant translocations) showed deletions of der(9) by FISH, suggesting a trend toward a higher number of deletions in the group with variant translocations. It has been hypothesized that each recombination event has a probability of erroneous repair leading to genomic loss next to the breakpoint, potentially involving a tumor suppressor.4,8,41 Therefore, a variant translocation with more recombination events has a greater chance of genomic loss.4 In addition, some studies10 have reported that each recombination at 9q34 should have a finite probability leading to deletion around the region, and it could be postulated that many of those cases with a der(9) deletion had a 4-break translocation and therefore were more likely to have a deletion than a 3-break translocation. So et al40 has hypothesized that a 4-break mechanism operates at the initial stages of leukemogenesis, favoring a 9q deletion at the time of translocation. However, in the present study, in 3 of 4 cases with deletion on der(9), we suggested a 2-step or a multistep mechanisms. However, the presence of additional cytogenic abnormalities at diagnosis in only one patient did not allow us to investigate its prognostic role in cases with variant translocations.
In conclusion, this large series of early CP CML patients treated with IM as a frontline therapy confirms that the clinical characteristics and outcome of patients with variant Ph translocations are similar to those with classic Ph translocations. Therefore, our data suggest that patients with variant translocations do not constitute a “warning” category in the imatinib era.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Mrs Katia Vecchi for her valuable assistance.
This study was supported by Associazione Italiana contro le Leucemie, i linfomi e i mielomi Bologna (BolognaAIL), Progetti di Ricerca di Interesse Nazionale (PRIN) 2005, and PRIN 2007 (University of Bologna); by the Associazione Italiana per la Ricerca sul Cancro (AIRC); by the Fondazione del Monte di Bologna e Ravenna; and by the European LeukemiaNet.
Authorship
Contribution: G. Marzocchi, F.C., and N.T. analyzed the data and wrote the manuscript; G. Marzocchi, S.L., C.B., M. Stacchini, U.G., L.V., G.D., A.M., L.B., G. Giudici, A.M.C., and F.G. performed molecular cytogenetic analysis; M.A. coordinated molecular analysis; F.C., G. Gugliotta, G. Specchia, M. Sassarego, A.S., and F. Palandri enrolled the study patients and collected clinical data; F. Pane, G. Saglio, G. Martinelli, G.R., and M.B. designed and supervised the trials; and G.R., M.B., and N.T. supervised the study and gave final approval for submission.
Conflict-of-interest disclosure: F. Pane receives research support from Novartis, is a consultant for Novartis and Bristol-Myers Squibb, and serves on the Novartis speakers' bureau. G. Saglio is a consultant for Novartis and Bristol-Myers Squibb and serves on the speakers' bureaus of Novartis and Bristol-Myers Squibb. G. Martinelli serves on the speakers' bureaus of Novartis, Bristol-Myers Squibb, and Pfizer. M. Baccarani receives research support from Novartis, Bristol-Myers Squibb, and Pfizer; is a consultant for Novartis, Bristol-Myers Squibb, and Pfizer; and serves on the Novartis speaker's bureau. G. Rosti is a consultant with Novartis and serves on the speakers' bureaus of Novartis and Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
A complete list of participants from the GIMEMA Working Party on CML can be found in the supplemental Appendix.
Correspondence: Nicoletta Testoni, Department of Hematology and Oncologic Sciences Seragnoli Hospital, University of Bologna, Via Massarenti 9, 40138 Bologna, Italy; e-mail: nicoletta.testoni@unibo.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal