Abstract
Although deregulated expression of specific microRNAs (miRNAs) has been described in solid cancers and leukemias, little evidence of miRNA deregulation has been reported in ALK-positive (ALK+) anaplastic large cell lymphomas (ALCL). These tumors overexpress the major antiapoptotic protein myeloid cell leukemia 1 (MCL-1), a situation that could compensate for the lack of BCL-2. We report that ALK+ ALCL cell lines and biopsy specimens (n = 20) express a low level of miR-29a and that this down-modulation requires an active NPM-ALK kinase. Murine models (transgenic mice and mouse embryonic fibroblast [MEF] cells), which allow conditional NPM-ALK fusion protein expression, showed an increase of miR-29a expression in the absence of NPM-ALK. Concordant results were observed after the abolition of NPM-ALK kinase activity (siALK or PF-2341066) in NPM-ALK+ ALCL cell lines. In addition, we showed that low expression of miR-29a, probably through methylation repression, plays an important regulatory role in MCL-1 overexpression that could promote tumor cell survival by inhibiting apoptosis. Enforced miR-29a expression was found to modulate apoptosis through inhibition of MCL-1 expression in ALCL cell lines and in a xenografted model, with a concomitant tumor growth reduction. Thus, synthetic miR-29a represents a potential new tool to affect tumorigenesis in these lymphomas.
Introduction
Anaplastic lymphoma kinase–positive (ALK+) anaplastic large cell lymphoma (ALCL) is now recognized as a distinct entity in the World Health Organization (WHO) classification of hematopoietic tumors1,2 and is characterized by the expression of an oncogenic fusion protein involving the ALK tyrosine kinase receptor.3,4 Most of the activated pathways downstream to this fusion protein with a constitutive tyrosine kinase activity have been characterized and play major roles in lymphomagenesis in ALK+ ALCL, controlling key cellular processes such as proliferation, survival, and cell migration (for review see Chiarle et al5 ). Another characteristic of ALK+ ALCLs is the lack or low expression of the antiapoptotic proteins BCL-2 and BCL-XL, suggesting a possible explanation for their relatively good prognosis. However, these tumors overexpressed MCL-1 (myeloid cell leukemia-1), an oncogene of particular interest, belonging to BCL-2 family of apoptosis-regulating proteins,6 and also involved in programming differentiation7 and promoting cell viability.8 MCL-1 expression could compensate for the lack of immunohistochemically detectable BCL-2 and BCL-XL expression in ALK+ ALCL.9-11 Some studies have suggested that the Jak-STAT and PI3K pathways activated in ALK+ tumors could be involved in up-regulating MCL-1 but other pathways at the posttranscriptional level, such as microRNAs, might also contribute to its high expression in ALK+ ALCL cases (see reviews by Akgul12 and Michels et al13 ).
Micro-RNAs (miRNAs) are small noncoding RNAs that regulate target gene expression posttranscriptionally through base pairing within the 3′-UTR regions of the target messenger RNAs and inducing their degradation, translational inhibition, or both of the encoded proteins.14,15 MiRNAs play key regulator roles in fundamental biologic processes including cell differentiation, apoptosis, cell proliferation, organ development, and hematopoiesis (see review by Kluiver et al16 ). MicroRNA-29 (MiR-29) family members have been shown to be down-regulated in several hematopoietic neoplasms, including chronic lymphocytic leukemia with poor prognosis,17 acute myeloid leukemia,18 and mantle cell lymphoma,19 as well as solid cancers such as lung cancer,20 hepatocellular carcinoma,21 and invasive breast cancer.22 More particularly, miR-29a, miR-29b, or both directly target the antiapoptotic protein MCL-1 in cholangiocarcinoma,23 hepatocellular carcinoma,21 and acute myeloid leukemia (AML).18 However, to date, only one study has addressed the expression and role of miRNAs in ALCL.24 We decided to focus our attention on miR-29 family members because they have recently been shown to regulate MCL-1 oncogene.
Methods
Murine models
Generation of conditional transgenic mice and tetracycline/doxycycline (Tet)-off mouse embryonic fibroblast (MEF) cells (Clontech) for expression of NPM-ALK oncogene in our laboratory was described by Giuriato et al.25,26 The tetracycline regulatory system was used to control the transgene transcription; the addition of doxycycline (an analog of tetracycline) allowed silencing of NPM-ALK expression (off condition), and doxycycline removal permitted NPM-ALK conditional expression. Lymph nodes from on and off mice were used for RNA extractions.
Human cell lines and tumoral and normal samples
Of the 5 ALCL cell lines studied, 4 were t(2;5)(p23;q35)+ (Karpas 299,27 SU-DHL-1,28 COST,29 and PIO [own laboratory unpublished cell line]), and one NPM-ALK− (FE-PD).30 Peripheral blood mononuclear cells (PBMCs) from 3 healthy donors used as normal lymphoid cells controls were kindly provided by Dr J. J. Fournié (UMR U1037).
Tumor samples from 20 ALK+ ALCL, 12 ALK− ALCL, and 3 reactive lymph nodes (RLN) were retrieved from our tumor tissue bank. The study was carried out in accordance with the Université de Toulouse institutional review board–approved protocols, and the procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2000. The diagnosis of ALCL was based on morphologic and immunophenotypic criteria as described in the last WHO classification.1,2 The percentage of malignant cells was assessed by ALK1 or CD30 staining and was greater than 50% for all selected cases. Antibody binding was detected with Dako REAL Detection System (Code K5001).
Cell treatments
ALK inhibitor treatment.
Racemic PF-2341066 [3-[1-(2,6-dichloro-3-fluoro-phenyl)-ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)-pyridin-2-ylamine] was synthesized according to the method described in the patent international application WO 2006/021881. ALCL cell lines were treated for 16 hours with 100nM PF-2341066.
Demethylation treatment.
5-aza-2′-deoxycytidine (5-aza-DC; Sigma-Aldrich) was dissolved in culture media at 10μM and frozen at −20°C until use. Cells were seeded at a density of 2 × 105 cells/mL and the drug was added at a final concentration of 3μM. Cells were harvested at day 3 for analysis.
Bisulfite modification, PCR, and sequencing
The methylation status of the CpG sites upstream from miR-29 cluster was determined by bisulfite sequencing. Genomic DNA was extracted, denatured, and modified using EZ DNA Methylation-Gold kit (Zymo Research) and then PCR-amplified using the following primers: Fwd-CpG-miR-29 5′-TGTGATTTATTTGGTTTTTGAGTTA-3′ and Rev-CpG-miR-29 5′-AAAAAAACATACCTAAAACTCCCTT-3′. PCR products were purified and cloned into the TOPO cloning system (Invitrogen). 7 clones were sequenced with T7 primer for each cell line.
Array–comparative genomic hybridization analysis
Tumoral genomic DNA was extracted from ALCL tumor samples, and 250 ng was labeled and cohybridized with normal unrelated reference DNA on the 4 × 180K Human Genome CGH Microarray (Agilent Technologies). After washing, the arrays were scanned and data were analyzed using Feature Extraction and Genomic Workbench 5.0 software.
RNAs extraction
Total RNA extraction was performed from 40 frozen sections of tumor biopsies or from 10 × 106 cells, using Trizol Reagent total RNA isolation reagent (Invitrogen) after the manufacturer's protocol. The concentration was quantified by NanoDrop Spectrophotometer (NanoDrop Technologies).
Northern blotting
Total RNA (10 μg) was separated on 15% denaturing polyacrylamide gel, electrotransferred to nylon membrane, UV-crosslinked, and prehybridized in hybridization buffer (5× SSC [saline-sodium citrate], 1% SDS, 1× Denhardt, and 150 μg/mL tRNA). Oligonucleotides complementary to mature miRNAs were end-labeled with 32P-γ-ATP by T4 polynucleotide kinase (New England Biolabs) and hybridized on membrane overnight mostly at 50°C. The blot was washed 2 × 10′ at room temperature in 2× SSC, 0.1% SDS, and then 2 × 15′ at the same stringency (0.1× SSC, 0.1% SDS). Northern blots were processed for autoradiography or phosphorimager. Experiments were repeated in triplicate.
Reverse-transcription–quantitative PCR
RT-PCR quantification for mature miRNAs.
cDNA was synthesized from total RNA using gene-specific primers for miR-29a, miR-29b, or miR-29c, according to the TaqMan MicroRNA assay protocol (PE Applied Biosystems). Real-time PCR for each individual miRNA was performed using an Applied Biosystems 7300 Sequence detection system. Normalization of target miRNAs was done with the small noncoding nucleolar RNA RNU24, which has the least variability across our cell lines, tumor samples, and control samples (average Ct = 25.41, SD = .78). Data were presented as relative quantity (RQ) of target miRNA, normalized to RNU24 and calculated by the 2−ΔCt method.
RT-qPCR quantification for Mcl-1 mRNA.
cDNA was synthesized from 1 μg of total RNA using Superscript III reverse transcriptase (Invitrogen) with random primers. Quantitative PCR was performed on the cDNA diluted at 1:10 on an Applied Biosystems 7300 Sequence detection system using SYBR Green PCR reagents. Mcl-1 primers were as follows: Forward 5′-AGCTGCATCGAACCATTAGC-3′, Reverse 5′-TCCTGATGCCACCTTCTAGG-3′. Normalization of target mRNAs was done with an endogenous standard, S14 (forward 5′-ATCAAACTCCGGGCCACAGGA-3′, reverse 5′-CTGCTGTCAGAGGGGATGGGG-3′). Data were presented as relative quantity (RQ) of target miRNA, normalized to S14 and calculated by the 2−ΔCt method.
miRNA and siRNA transfections
ALCL cells were maintained in exponential growth phase in IMDM supplemented with 15% FCS, l-glutamine (2mM), sodium pyruvate (1mM), and penicillin/streptomycin at 37°C in an atmosphere of 5% CO2. MiRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol and 1 of the following: synthetic premiRNA-29a, miR negative control 1 (Ambion), or LNA anti–miR-29a (Sigma-Aldrich) or with a double-stranded RNA siALK (5′-GGGCGAGCUACUAUAGAAA55–3′) at a final concentration of 150nM. Cells were washed twice in PBS 72 hours after transfections. RNA and protein extractions were performed for RT-qPCR or Western blotting subsequent analysis.
Western blotting
The cell pellet was resuspended in passive lysis buffer (Promega) containing protease inhibitor cocktail (Roche), phenylmethylsulfonyl fluoride (PMSF) 1mM, and phosphatase inhibitors (NaF 50mM, Na3VO4 1mM). Protein lysates were separated using 10% SDS-acrylamide gels, and transferred onto Protran nitrocellulose membranes (Whatman). For immunodetection, membranes were incubated with antibodies directed against MCL-1 at 1/1000 (S-19, Santa Cruz Biotechnology), ALKc at 1/500 (Dr B. Falini, Perugia, Italy), phospho-ALK at 1/1000 (#3341, Cell Signaling Technology), STAT-3 and P-STAT3 at 1/1000 (#9132 & #9131, Cell Signaling Technology), DNMT1 and DNMT3b at 1/700 (#5119 & #2161, Cell Signaling Technology) or β-actin at 1/10 000 (A5441, Sigma-Aldrich). Signals from horseradish-peroxidase (HRP)-coupled secondary antibodies were generated by enhanced chemiluminescence solution (ECL; Amersham Biosciences) and recorded with films. Quantification was performed using GeneTools software from Syngene. Experiments were repeated in triplicate.
Apoptosis assays
Karpas-299 and SU-DHL-1 cells were transfected with 150nM of one of the following: miR negative control, miR-29a, LNA anti–miR-29a, and miR-29a: LNA anti–miR-29a (1:1 ratio) following the protocol described. After 72 hours, cells were treated with 1μM doxorubicin (Sigma-Aldrich) for 6 hours to induce apoptosis. Flow cytometric analysis (FACS scan) of apoptotic cells was performed after annexinV/propidium iodide (PI) staining (Annexin-V-Fluos; Roche). Data were analyzed using FlowJo 7 software (TreeStar). Caspase 3/7 activities were determined using the Caspase-Glo 3/7 assay according to manufacturer's instructions (Promega) and luminescence was monitored with a Mithras plate-reader (LB940).
Tumorigenicity assay
Experiments on NOD/SCID mice were performed according to the institutional ethical guidelines of Inserm. MiR-29a or miR control-transfected Karpas-299 cells (10 × 106) were resuspended in 100 μL of PBS and injected subcutaneously into either side of the flank of the same female at 6 weeks of age. Tumor growth in the 7 mice included was examined every 2 days. Tumor volume was monitored by measuring the length (L), width (W), and thickness (T) of the tumor with calipers and was calculated with the formula L × W × T × 0.52. When control tumors reached a volume greater than 1000 mm3, mice were killed and tumors weighed. Tumors were analyzed for apoptotic cells by Caspase3 immunostaining (#AF835, 1/500, R&D Systems). Antibody binding was detected using streptaridin-biotin peroxidase complex (ABC) method and the Dako StreptABComplex/HRP Duet (mouse) kit (Dakocytomation). The determination and counting of immunopositive cells in all the specimens were performed by one pathologist (L.L.) under masked conditions, and the procedure repeated 3 times. The number of positive cells was counted for a total of 10 neighboring high-power view fields (×400), and the average number of positive cells was calculated. In parallel, MCL-1 expression was monitored by Western blotting, as described in “Western blotting.”
Results
MiR-29a is less expressed in ALK+ ALCL samples than in ALK− ones
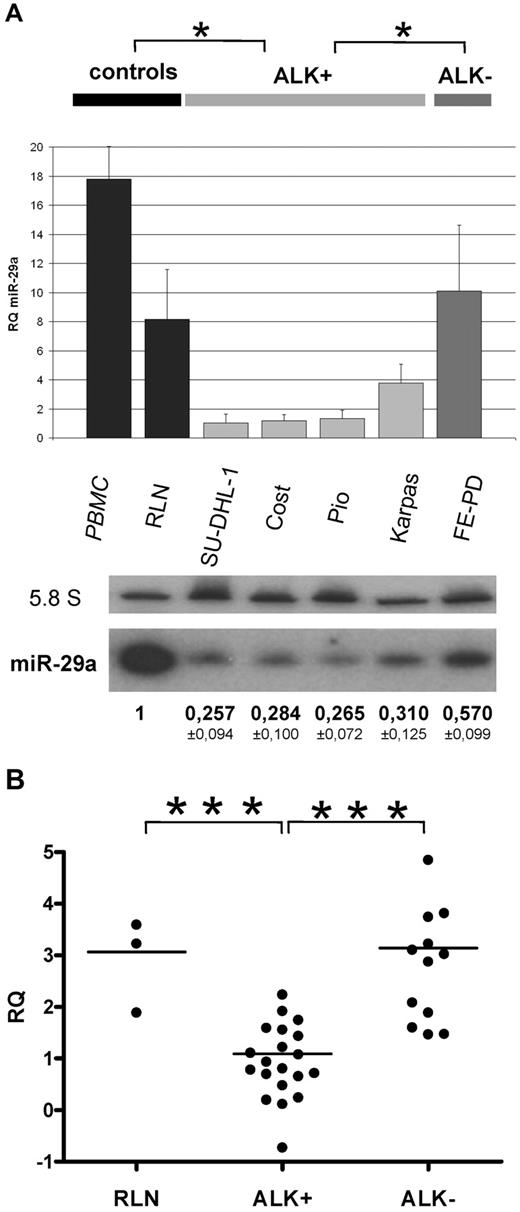
Because MCL-1 protein expression can be targeted by miR-29 family members in other malignant diseases,17-23 we focused our analysis on miR-29 family expression in ALCL cell lines and tumors using RT-qPCR and Northern blotting. As shown in Figure 1A, miR-29a down-expression ranged from 2.5-fold to 10-fold in NPM-ALK+ cell lines compared with the NPM-ALK− line, with concordant results by Northern blotting (approximately 2-fold change). Compared with peripheral blood mononuclear cells and reactive lymph nodes (RLNs) used as normal controls, miR-29a is also clearly less abundant in NPM-ALK+ ALCL cell lines, using both detection methods (RT-qPCR: P < .05; Northern blotting: P < .001, t test). By contrast, miR-29b and miR-29c were found by RT-qPCR to be weakly expressed, without any significant variation between NPM-ALK+ and NPM-ALK− cell lines (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
MiR-29a is low expressed in ALK+ ALCL. (A) MiR-29a expression in ALK+ (SU-DHL-1, COST, PIO, Karpas-299) and ALK+ ALCL cells (FE-PD) was analyzed by RT-qPCR and Northern blotting. RNA from reactive lymph node (RLN) and peripheral blood mononuclear cells (PBMC) serve as normal controls. The histogram shows the relative expression (RQ) of miR-29a after normalization with RNU24. Bars represent standard deviations (SD). P for ALK+ vs controls and ALK+ vs ALK− (*P < .05). 5.8S RNA was used as the loading control in the Northern blotting. P for ALK+ vs controls (P < .001) and ALK+ vs ALK− (P < .05). (B) RT-qPCR of miR-29a in 20 ALK+ and 12 ALK− patient tumors. Three RLNs were used as normal controls. RQ was plotted on a logarithmic scale. P for ALK+ vs RLN and ALK+ vs ALK− (***P < .001).
MiR-29a is low expressed in ALK+ ALCL. (A) MiR-29a expression in ALK+ (SU-DHL-1, COST, PIO, Karpas-299) and ALK+ ALCL cells (FE-PD) was analyzed by RT-qPCR and Northern blotting. RNA from reactive lymph node (RLN) and peripheral blood mononuclear cells (PBMC) serve as normal controls. The histogram shows the relative expression (RQ) of miR-29a after normalization with RNU24. Bars represent standard deviations (SD). P for ALK+ vs controls and ALK+ vs ALK− (*P < .05). 5.8S RNA was used as the loading control in the Northern blotting. P for ALK+ vs controls (P < .001) and ALK+ vs ALK− (P < .05). (B) RT-qPCR of miR-29a in 20 ALK+ and 12 ALK− patient tumors. Three RLNs were used as normal controls. RQ was plotted on a logarithmic scale. P for ALK+ vs RLN and ALK+ vs ALK− (***P < .001).
Twenty ALK+ and 12 ALK− tumor samples, consisting of > 50% of tumor cells, were analyzed by RT-qPCR to confirm the significant low level of miR-29a in ALK+ cells. As illustrated in Figure 1B, RT-qPCR showed that miR-29a was clearly lower expressed in ALK+ tumors than in ALK− ALCL (4-fold change) and in RLNs.
MiR-29a down-regulation depends on the expression of an active ALK fusion protein
Next, we asked whether the differential expression of miR29a observed between ALK+ and ALK− samples was correlated to NPM-ALK expression. To address this issue, we used several models.
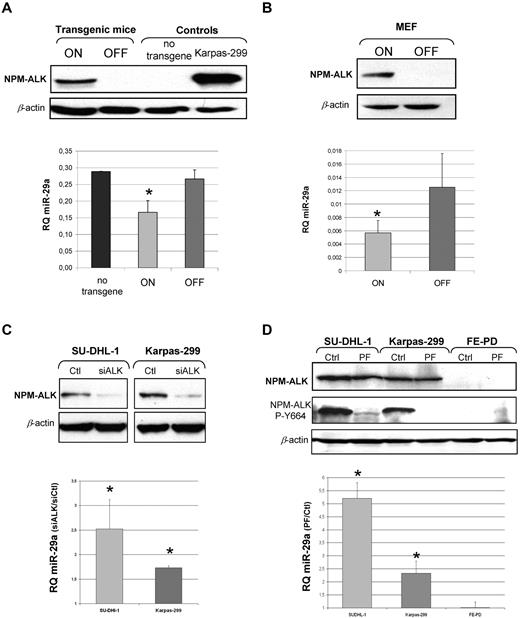
The 2 murine models developed in our laboratory, transgenic NPM-ALK mice26 and a murine cellular model25 (Tet-off MEF cells), both allow conditional NPM-ALK fusion protein expression under the control of doxycycline (Tet-off system). As expected, transgenic mice lymph nodes and Tet-off MEF cells showed ALK fusion protein expression after doxycycline removal (on condition) (Figure 2A-B). In the absence of the ALK fusion protein (off condition), miR-29a level was increased 1.6-fold in transgenic mice (Figure 2A) and 2.2-fold in MEF cells by RT-qPCR (Figure 2B).
MiR-29a expression is dependent on NPM-ALK. Using murine conditional models NPM-ALK fusion protein expression was analyzed by Western blotting (A) in lymph nodes of conditional NPM-ALK transgenic mice and (B) in MEF NPM-ALK cells in on and off conditions. β-actin was used as a loading control. In both models, miR-29a was detected by RT-qPCR in on and off conditions (histograms). Data are representative of 3 independent experiments. P for on vs off (*P < .05). (C) Human ALCL ALK+ cell lines (SU-DHL-1 and Karpas-299) were transiently transfected with an anti-ALK siRNA (siALK) or a control siRNA (Ctl) and inhibition of NPM-ALK protein expression was assessed by Western blotting. (D) ALCL cells were treated with an NPM-ALK inhibitor (PF-2341066) and its autophosphorylation status was detected with an antibody directed against phosphorylated-tyrosine 664 (NPM-ALK P-Y664). β-actin was used as a loading control. Expression levels of miR-29a, by RT-qPCR in ALCL cell lines after transfection with siALK (C) or treatment with PF-2341066 (D) are presented as the ratio of RQ (siALK/Ctl) and RQ (PF/Ctl) respectively. P for siALK vs Ctl and PF vs Ctl (*P < .05).
MiR-29a expression is dependent on NPM-ALK. Using murine conditional models NPM-ALK fusion protein expression was analyzed by Western blotting (A) in lymph nodes of conditional NPM-ALK transgenic mice and (B) in MEF NPM-ALK cells in on and off conditions. β-actin was used as a loading control. In both models, miR-29a was detected by RT-qPCR in on and off conditions (histograms). Data are representative of 3 independent experiments. P for on vs off (*P < .05). (C) Human ALCL ALK+ cell lines (SU-DHL-1 and Karpas-299) were transiently transfected with an anti-ALK siRNA (siALK) or a control siRNA (Ctl) and inhibition of NPM-ALK protein expression was assessed by Western blotting. (D) ALCL cells were treated with an NPM-ALK inhibitor (PF-2341066) and its autophosphorylation status was detected with an antibody directed against phosphorylated-tyrosine 664 (NPM-ALK P-Y664). β-actin was used as a loading control. Expression levels of miR-29a, by RT-qPCR in ALCL cell lines after transfection with siALK (C) or treatment with PF-2341066 (D) are presented as the ratio of RQ (siALK/Ctl) and RQ (PF/Ctl) respectively. P for siALK vs Ctl and PF vs Ctl (*P < .05).
We also performed siRNA anti-ALK (siALK) experiments in 2 human ALK+ cell lines (SU-DHL-1 and Karpas-299). After checking the knockdown of NPM-ALK by siALK (Figure 2C), we monitored miR-29a level by RT-qPCR. Results were in agreement with those obtained in mouse models, with an increase of 2.5-fold and 1.7-fold after NPM-ALK inhibition, in SU-DHL-1 and Karpas-299 cell lines, respectively (Figure 2C). Furthermore, we showed that miR-29a expression was dependent on the active NPM-ALK protein. NPM-ALK+ cell lines and NPM-ALK− cell lines were treated for 16 hours with 100nM concentration of the kinase inhibitor, PF-2341066, an efficient inhibitor of NPM-ALK activity in vitro and in vivo.31 It is well known that PF-2341066 is an ATP-competitive and selective small-molecule inhibitor of NPM-ALK and c-Met under our experimental conditions (100nM). However, we checked by Western blotting that the level of c-Met phosphorylation was very low in basal conditions and was not affected by PF-2341066 treatment (data not shown). NPM-ALK kinase activity was inhibited with loss of NPM-ALK autophosphorylation on the tyrosine-664 residue (Figure 2D). Following this inhibition, miR-29a was increased by factors of 5.2 and 2.4 in the SU-DHL-1 and Karpas-299 cell lines, respectively (Figure 2D). No change in miR-29a expression was observed in NPM-ALK− cells. Moreover, this effect on miR-29a was specific, because miR-29b, miR-29c, and the unrelated miR-222 expression remained unaffected by PF-2341066 treatment (data not shown). Altogether, these data confirmed an NPM-ALK–dependent repression of miR-29a expression.
DNA methylation in NPM-ALK–expressing cells contributes partially to silencing of miR-29a
To identify the mechanisms involved in miR-29a low expression in ALK-expressing cells, we searched for DNA structural or epigenetic modifications.
MiR-29a is expressed from only one locus, located on chromosome 7 (Chr 7q32.3), inside common fragile site FRA7H, and coding for miR-29a and miR-29b1. In the 20 ALK+ ALCL cases tested, high-resolution array-based comparative genomic hybridization analysis (CGH arrays) did not show any deletion of this locus (data not shown).
We further investigated whether miR-29a weaker expression in NPM-ALK+ cells could be explained by epigenetic mechanisms, given that miRNAs genes can be regulated by DNA methylation.32-35 First, NPM-ALK+ and NPM-ALK− cell lines were treated with a demethylating agent (5-aza-2′-deoxycytidine [5-aza-DC]) for 3 days and miR-29a level was subsequently determined by RT-qPCR. A significantly higher level of miR-29a was detected in Karpas-299 and SU-DHL-1 treated cells (8-fold and 3.6-fold increase, respectively, Figure 3A), whereas there was no significant change in the FE-PD cell line. The level of miR-29b, which is also expressed from another locus (Chr 1q32.2), was not affected (not shown). These results led us to look for the presence of CpG sites upstream from miR-29a coding gene using bioinformatic software (Emboss CpGPlot from EMBL-EBI: http://www.ebi.ac.uk/Tools/emboss/cpgplot/). One CpG-enriched site was predicted in this region and contains 5 CpG dinucleotides in 72 bp (Figure 3B). Bisulfite sequencing on nontreated or 5-aza-DC–treated cells was performed to identify methylation status of these CpG sites. As shown in Figure 3B, the 5 CpG were hypermethylated in NPM-ALK+ Karpas-299 and SU-DHL-1 cells (97%-100%), compared with the NPM-ALK− ones (66%). Methylation was reverted for the majority of the CpG sites after demethylation treatment, with a greater efficiency in Karpas-299. To demonstrate the link between the methylation status of these CpG sites and NPM-ALK expression, we also sequenced this region in Karpas-299 and SU-DHL-1 cells transfected with an anti-ALK siRNA (siALK). Following NPM-ALK knockdown (Figure 2C), we observed a partial demethylation of these CpG sites, ranging from 26% to 37% (Figure 3B), not observed with the siRNA control (data not shown). Concomitant increase of miR-29a expression was observed with an increase of 2.5-fold and 1.7-fold, in SU-DHL-1 and Karpas 299 cell lines, respectively (Figure 2C).
Epigenetic regulation of miR-29a. (A) MiR-29a expression levels by RT-qPCR with or without 5-aza-2′-deoxycytidine (5-aza-DC) treatment in Karpas-299, SU-DHL-1 (ALK+) and FE-PD (ALK−) cells are shown. Data represent the mean of triplicates with standard deviations (SD) and *P < .05. (B) Schematic representation of miR-29 locus on chromosome 7, coding for miR-29b1 and miR-29a (black boxes). The prediction of a CpG-enriched site with Emboss CpGPlot software (http://www.ebi.ac.uk/Tools/emboss/cpgplot/index.html) is represented as a dashed box and contains 5 CpG sites. Bisulphite sequencing of these 5 CpG sites in cells treated with 5-aza-DC or transfected with siALK, and control ALCL cells. Each circle indicates a CpG dinucleotide (●: methylated CpG; ○: unmethylated CpG).
Epigenetic regulation of miR-29a. (A) MiR-29a expression levels by RT-qPCR with or without 5-aza-2′-deoxycytidine (5-aza-DC) treatment in Karpas-299, SU-DHL-1 (ALK+) and FE-PD (ALK−) cells are shown. Data represent the mean of triplicates with standard deviations (SD) and *P < .05. (B) Schematic representation of miR-29 locus on chromosome 7, coding for miR-29b1 and miR-29a (black boxes). The prediction of a CpG-enriched site with Emboss CpGPlot software (http://www.ebi.ac.uk/Tools/emboss/cpgplot/index.html) is represented as a dashed box and contains 5 CpG sites. Bisulphite sequencing of these 5 CpG sites in cells treated with 5-aza-DC or transfected with siALK, and control ALCL cells. Each circle indicates a CpG dinucleotide (●: methylated CpG; ○: unmethylated CpG).
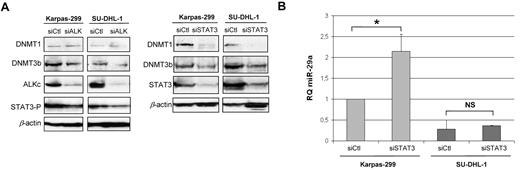
As STAT3 protein is able to induce methylation of some genes, through transcriptional activation of DNA methyltransferases (DNMTs), we have determined the level of DNMT1 and 3b in ALK and STAT3 knockdown conditions (Figure 4A). In siALK conditions, we observed, as expected, a decrease of STAT3 activation, and in parallel a decrease of DNMT3b protein, although DNMT1 expression was poorly affected. In conditions of strong inhibition of STAT3 in Karpas-299 (compared with its inhibition via siALK), both DNMT3b and DNMT1 proteins were decreased, with concomitant increase of miR-29a expression (Figure 4B). In SU-DHL-1, STAT3 inhibition by siRNA was less effective, with a weaker down-regulation of DNMT3b but a complete abolition of DNMT1 expression. However, in this cell line, the miR-29a re-expression was not significant, suggesting that DNMT3b is much more involved in miR-29a methylation than is DNMT1. These findings suggest that miR-29a expression was partly repressed by methylation in NPM-ALK-expressing cells, via STAT3 activation.
DNMT and STAT3 role in miR-29a low-expression. (A) DNMT1 and DNMT3b endogenous expression in ALK+ ALCL cell lines were detected by Western blotting following NPM-ALK or STAT3 knockdown by siRNA. (B) RQ miR-29a after inhibition of STAT3 in Karpas-299 and SU-DHL-1 cells. P for siCtl vs siSTAT3 in Karpas-299 (*P < .05). NS: not significant.
DNMT and STAT3 role in miR-29a low-expression. (A) DNMT1 and DNMT3b endogenous expression in ALK+ ALCL cell lines were detected by Western blotting following NPM-ALK or STAT3 knockdown by siRNA. (B) RQ miR-29a after inhibition of STAT3 in Karpas-299 and SU-DHL-1 cells. P for siCtl vs siSTAT3 in Karpas-299 (*P < .05). NS: not significant.
Mcl-1 is a target of miR-29a in NPM-ALK+ ALCL cells
We investigated whether miR-29a was able to modulate MCL-1 protein level in ALK+ ALCL tumors and cell lines, because Mcl-1 has been proved to be targeted by miR-29 in cholangiocarcinoma cell line,23 hepatocellular carcinoma,21 and AML.18 First, we checked the basal level of Mcl-1 mRNA by RT-qPCR in 20 cases of ALK+ tumors and 12 ALK− samples. Figure 5A shows a significantly high level of Mcl-1 mRNA in the ALK+ cases compared with the ALK− tumors and RLN (P < .05, t test). In cell lines, RT-qPCR assays also demonstrated an up-regulation of Mcl-1 mRNA in NPM-ALK+ cell lines compared with the FE-PD cell line, with a strikingly high level present in Karpas-299 compared with SU-DHL-1 and COST cell lines (Figure 5B). Similar results were obtained in Western blotting analysis for MCL-1 protein expression (Figure 5C). The higher mean relative value of Mcl-1 mRNA in ALK+ cell lines compared with ALK+ ALCL biopsies could be explained by the percentage of tumor cells expressing MCL-1 in cell lines compared with biopsies.
MiR-29a targets MCL-1 in NPM-ALK+ ALCL cells. (A) mRNA levels of Mcl-1 in ALK+ (19 cases) and ALK− (12 cases) ALCL tumors by RT-qPCR. Three RLNs serve as controls. RQ of Mcl-1 was calculated using the S14 as standard control. P for ALK+ vs RLN and ALK+ vs ALK− (*P < .05). MCL-1 endogenous expression in ALCL cell lines is shown in panel B by RT-qPCR with RQ plotted on a logarithmic scale and in panel C by Western blotting with the ratio of MCL-1/βactin. (D) MCL-1 protein levels were monitored 72 hours after Karpas-299, SU-DHL-1, and COST transfection with 1 or 2 (1:1 ratio) of the following: miR negative control, miR-29a, or LNA-anti–miR-29a. The long isoform of MCL-1 (40 kDa) was mainly detected. β-actin was used as a loading control to determine the relative expression of MCL-1 (long MCL-1 at 40 kDa and short MCL-1* at 28 kDa forms added) as the ratio of MCL-1/β actin. Data are representative of 3 independent experiments.
MiR-29a targets MCL-1 in NPM-ALK+ ALCL cells. (A) mRNA levels of Mcl-1 in ALK+ (19 cases) and ALK− (12 cases) ALCL tumors by RT-qPCR. Three RLNs serve as controls. RQ of Mcl-1 was calculated using the S14 as standard control. P for ALK+ vs RLN and ALK+ vs ALK− (*P < .05). MCL-1 endogenous expression in ALCL cell lines is shown in panel B by RT-qPCR with RQ plotted on a logarithmic scale and in panel C by Western blotting with the ratio of MCL-1/βactin. (D) MCL-1 protein levels were monitored 72 hours after Karpas-299, SU-DHL-1, and COST transfection with 1 or 2 (1:1 ratio) of the following: miR negative control, miR-29a, or LNA-anti–miR-29a. The long isoform of MCL-1 (40 kDa) was mainly detected. β-actin was used as a loading control to determine the relative expression of MCL-1 (long MCL-1 at 40 kDa and short MCL-1* at 28 kDa forms added) as the ratio of MCL-1/β actin. Data are representative of 3 independent experiments.
To ascertain that MCL-1 can be targeted by miR-29a in ALK+ cells, we performed a gain-of-function experiment of miR-29a in Karpas-299, SU-DHL-1, and COST cells. MiR-29a or a miR negative control was transiently transfected and endogenous MCL-1 expression was detected by Western blotting (Figure 5D). The long isoform of MCL-1 (40 kDa) was mainly detected. Densitometric measurement of the bands showed that MCL-1 protein level was reduced by 56% to 77% in miR-29a–expressing NPM-ALK+ cells, compared with the control. Cotransfection with an LNA anti–miR-29a double-strand oligonucleotide abolished the miR-29a inhibitory effect on MCL-1 protein level. No change was observed in Mcl-1 expression at the mRNA level between the different transfection conditions (data not shown). Moreover, we did not observe any effect of miR-29a overexpression on NPM-ALK (Figure 5D) and STAT3 protein expression (supplemental Figure 2).
MiR-29a sensitizes NPM-ALK+ ALCL cells to apoptosis through MCL-1 targeting
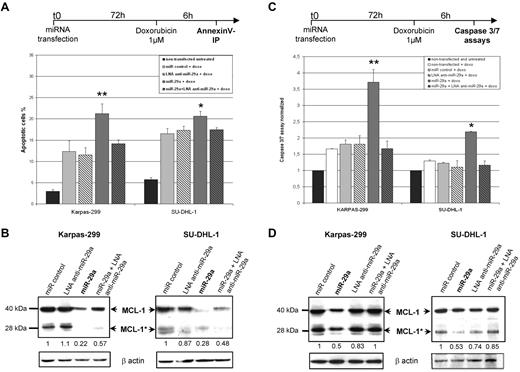
As the antiapoptotic protein MCL-1 is targeted by miR-29a in our model, we examined the effect of miR-29a overexpression on apoptosis in ALK+ cells. Karpas-299 and SU-DHL-1 cells were transiently transfected with miR-29a, miR negative control, or locked nucleic acid (LNA) anti–miR-29a double-strand oligonucleotide. Cells were then induced to undergo apoptosis with doxorubicin, a drug commonly used in the treatment of patients with ALCL. After annexin-V/PI assays, miR-29a–transfected Karpas-299 cells presented an apoptotic cell rate of 21.2% compared with 12.3% with the miR-negative control (Figure 6A). Transfection of miR-29a together with LNA anti–miR-29a partly reverted miR-29a sensitization to apoptosis. We confirmed these observations by Caspase 3/7 activities measurements, showing an apoptosis enhancement with miR-29a between 2-fold and 3.6-fold compared with controls (Figure 6C). In parallel, we checked MCL-1 proteins that showed a caspase-cleaved pattern after apoptosis induction, as previously described36 : in addition to the long form of 40 kDa (MCL-1), a shorter protein of 28 kDa (MCL-1*) appeared in all transfected and doxorubicin-treated cells. Both MCL-1 isoforms are reduced by at least 2-fold in miR-29a transfected cells with respect to controls (Figure 6B and 6D). Altogether, these data demonstrated that miR-29a overexpression sensitizes NPM-ALK+ ALCL cells to Doxorubicin-induced apoptosis through MCL-1 targeting.
MiR-29a overexpression enhances doxorubicin-induced apoptosis in NPM-ALK+ ALCL cells. Karpas-299 and SU-DHL-1 NPM-ALK+ cells were transiently transfected with miR negative control, miR-29a, or LNA anti–miR-29a oligonucleotides for 72 hours. Doxorubicin (1μM) was then used for 6 hours to induce apoptosis. AnnexinV/PI staining (A) and Caspase 3/7 assays (C) were used to assess apoptosis. Histograms showed the percentage of apoptotic cells and the normalized Caspase3/7 activities, respectively (*P < .05 and **P < .01). (B,D) Western blotting for MCL-1 in miRNA-transfected and doxorubicin-treated cells demonstrated the presence of long (MCL-1 at 40 kDa) and short caspase-cleaved (MCL-1* at 28 kDa) MCL-1 proteins. Densitometry measurements of both forms were added and normalized with β-actin.
MiR-29a overexpression enhances doxorubicin-induced apoptosis in NPM-ALK+ ALCL cells. Karpas-299 and SU-DHL-1 NPM-ALK+ cells were transiently transfected with miR negative control, miR-29a, or LNA anti–miR-29a oligonucleotides for 72 hours. Doxorubicin (1μM) was then used for 6 hours to induce apoptosis. AnnexinV/PI staining (A) and Caspase 3/7 assays (C) were used to assess apoptosis. Histograms showed the percentage of apoptotic cells and the normalized Caspase3/7 activities, respectively (*P < .05 and **P < .01). (B,D) Western blotting for MCL-1 in miRNA-transfected and doxorubicin-treated cells demonstrated the presence of long (MCL-1 at 40 kDa) and short caspase-cleaved (MCL-1* at 28 kDa) MCL-1 proteins. Densitometry measurements of both forms were added and normalized with β-actin.
Effects of miR-29a overexpression in a xenograft model
An in vivo model was subsequently used to evaluate miR-29a tumor suppressor effect. Karpas-299 cells were transfected with miR-29a or miR-control and inoculated in right and left flanks, respectively, of immunocompromised NOD/SCID mice (n = 7). MiR-29a–transfected cells revealed a delayed tumor formation over time, and tumors weighed significantly less at the end of the experiment (0.7 g for miR-29a–transfected tumors compared with 1.2 g for miR-control transfected tumors; Figure 7A-B). We analyzed apoptosis using Caspase-3 immunohistochemistry (Figure 7C). MiR-29a–transfected tumors displayed a significant increase of apoptotic cells with respect to the control (P < .01), that correlated with MCL-1 protein down-expression (Figure 7 D). These results support a tumor suppression effect of miR-29a in vivo, via a decreased MCL-1 expression.
Effect of miR-29a on tumor growth. NOD-SCID mice were xenografted with Karpas-299 cells transfected with miR-29a or miR-control (miR-Ctl). (A) Curves of tumor growth showing tumor volumes (in cubic millimeters) at the indicated days and (B) histograms of tumor weight (grams) averages at the end of the experiment. P for miR-29a vs control (*P < .05). (C) Representative images of immunostaining with Caspase-3 antibody for miR-Ctl and miR-29a tumors (Leica Digital Module R[DMR] microscope equipped with DFC300FX camera and 200×/0.85 NA objective lens, original magnification ×400). Image processing was performed using IM50 software from Leica. Table shows the average of Caspase-3–positive cells for all 7 mice, counted 3 times each, with SD and P value. (D) Representative Western blotting for MCL-1 in miR-Ctl and miR-29a–expressing tumors. Normalized mean value and SD of densitometry measurements are shown (n = 7).
Effect of miR-29a on tumor growth. NOD-SCID mice were xenografted with Karpas-299 cells transfected with miR-29a or miR-control (miR-Ctl). (A) Curves of tumor growth showing tumor volumes (in cubic millimeters) at the indicated days and (B) histograms of tumor weight (grams) averages at the end of the experiment. P for miR-29a vs control (*P < .05). (C) Representative images of immunostaining with Caspase-3 antibody for miR-Ctl and miR-29a tumors (Leica Digital Module R[DMR] microscope equipped with DFC300FX camera and 200×/0.85 NA objective lens, original magnification ×400). Image processing was performed using IM50 software from Leica. Table shows the average of Caspase-3–positive cells for all 7 mice, counted 3 times each, with SD and P value. (D) Representative Western blotting for MCL-1 in miR-Ctl and miR-29a–expressing tumors. Normalized mean value and SD of densitometry measurements are shown (n = 7).
Discussion
Our study showed that ALK+ ALCL cell lines, as well as biopsy specimens, presented a low expression of miR-29a compared with ALK− cell lines. Furthermore, we demonstrated that NPM-ALK kinase activity is critical for miR-29a down-regulation in ALK+ cells, partly through methylation regulation. This down-regulation contributes in part to the expression of MCL-1, the main antiapoptotic BCL-2 family member present in ALK+ ALCL, and thus to tumor cell survival.
Expression levels of antiapoptotic BCL-2 family members such as BCL-XL, BCL-2, and MCL-16 have already been assessed in ALCL.9-11 These studies underlined the absence of BCL-2 expression and low or undetectable expression of BCL-XL in ALK+ ALCL compared with ALK− tumors. MCL-1, in absence of BCL-2, could promote tumor cell survival by inhibiting cell death.10,11 Rust et al found no difference in MCL-1 staining intensity or percentage of positive tumor cells between ALK+ and ALK− cases.11 By contrast, Rassidakis et al reported that MCL-1 was detected in only 61.5% of ALK− ALCL with a lower number of positive cells.10 Our results showing significantly more abundant Mcl-1 mRNA in ALK+ compared with ALK− ALCL are in agreement with this latter study.
The mechanisms of MCL-1 expression in ALCL tumors are not completely understood. In a recent study, Amin et al demonstrated the STAT3-mediated induction of Mcl-1 transcription in 2 ALK+ ALCL cell lines,37 but the phosphatidylinositol 3-kinase (PI3K) pathway has also been demonstrated in other models.38,39 Moreover, Mott et al were the first to demonstrate in a cholangiocarcinoma cell model that MCL-1 could also be regulated at the posttranscriptional level, notably by miR-29b.23 The results of our study are in line with those reported by Mott et al,23 and more recently by Garzon et al18 and Xiong et al,21 because we found that miR-29a, another member of the miR-29 family, contributes to the up-regulation of MCL-1 in ALK+ ALCL.
Using RT-qPCR, we observed a significant lower expression of miR-29a in ALK+ compared with ALK− ALCL cell lines, whereas miR-29b and miR-29c were poorly detected and not differentially expressed. Although miR-29a and miR-29b are partially cotran-scribed (miR-29b being also transcribed from another locus with miR-29c), the discrepancy between miR-29a and miR-29b expression has already been observed by Hwang et al, who suggest a specific posttranscriptional mechanism preventing the accumulation of miR-29b but not that of miR-29a.40 We confirmed the low expression of miR-29a in a series of ALK+ ALCL tumor samples, compared with ALK− ones or normal controls (ie, PBMCs and reactive lymph nodes, the normal counterpart of ALCL being yet unknown).
Moreover, using 2 different murine models, we demonstrated that miR-29a lower expression was dependent on the expression of an active NPM-ALK kinase. Indeed, both transgenic mice and MEF cells, which allow conditional NPM-ALK fusion protein expression,25,26 showed a clear increase of miR-29a expression in the absence of the fusion protein. Comparable results were observed when NPM-ALK expression or its phosphorylation was abolished in human NPM-ALK+ ALCL cells, using an anti-ALK siRNA and the ALK kinase inhibitor PF-2341066, respectively. This molecule is known to inhibit both ALK and c-Met kinase activity. However, the unaffected level of c-Met phosphorylation in our PF-2341066–treated cell lines excluded a bystander effect on miR-29a expression. Altogether these observations showed that miR-29a weaker expression is strongly correlated with the expression of a constitutively active NPM-ALK fusion protein.
Next, we attempted to elucidate the mechanisms underlying miR-29a down-regulation. Previous studies have demonstrated that inactivation of miR-15a and miR-16-1 in chronic lymphocytic leukemia and prostate carcinoma can be regulated by genetic changes41 (eg, translocations or deletions) or epigenetic modifications such as hypermethylation of regulatory elements.32-35 The lack of DNA copy number alterations in the genomic loci containing miR-29 gene (chr7q32.3), as demonstrated by our CGH-array analysis, made it unlikely that miR-29a inactivation was related to genetic changes. We postulated that miR-29 gene could be silenced by epigenetic mechanisms as demonstrated for an increasing number of micro-RNAs in other various types of cancers (review by Valeri et al42 ). Because methylation can be reversed by the use of epigenetics drugs, we treated 2 NPM-ALK+ cell lines with 5-aza-DC. This demethylating agent induced an increase in miR-29a levels in NPM-ALK+ treated cells, whereas no significant change was observed in NPM-ALK− cell line. The promoter of the miR-29 cluster on chromosome 7, encoding miR-29a, has not yet been identified. However, it contains a CpG-enriched site in its upstream 1kb region, as predicted by bioinformatics. These 5 CpG sites were identified as heavily methylated in NPM-ALK+ cell lines and this methylation was partially reverted by treatment with 5-aza-DC and, to a lesser extent, after NPM-ALK knockdown by siRNA. These latter observations suggested that NPM-ALK is involved in the hypermethylation of these sites, contributing to miR-29a repression. This genomic region, upstream of miR-29 cluster, probably contains regulatory elements for miR-29 expression that could be targeted by NPM-ALK effectors. For instance, STAT3 protein is able to induce methylation of the ZAP70 gene, in ALK+ ALCL, through transcriptional activation of DNMT1 (DNA methyltransferase 1).43 To determine whether the effect on methylation of the miR29 cluster is mediated by STAT3 activation downstream to NPM-ALK, we invalidated STAT3 with an siRNA. We observed that DNMT3b and DNMT1 proteins were decreased, with concomitant increase of miR-29a expression. These results suggest that methylation of the miR29 cluster could be partly regulated by STAT3 in NPM-ALK–expressing cells. Moreover, DNMT3, targeted by miR-29a (data not shown), could amplify this mechanism in a positive feedback loop.44
MiR-29 family members have been shown to be down-regulated in several hematopoietic neoplasms, including chronic lymphocytic leukemia with poor prognosis,17 acute myeloid leukemia,18 and mantle cell lymphoma,19 as well as solid cancers such as lung cancer,20 hepatocellular carcinoma,21 and invasive breast cancer.22 More particularly, miR-29a, miR-29b, or both directly targets the antiapoptotic protein MCL-1 in cholangiocarcinoma,23 hepatocellular carcinoma,21 and AML.18 Results from the current study showed that miR-29a is also able to target MCL-1 in NPM-ALK+ ALCL cell lines. Indeed, endogenous MCL-1 protein expression was strikingly decreased in NPM-ALK+ cells transfected with miR-29a. It is known that miR-29a acts directly at the Mcl-1 3′UTR.21 The fact that cotransfection with a LNA anti–miR-29a in NPM-ALK+ cells abolishes the miR-29a inhibitory effect on MCL-1 protein level suggests a direct role of this miRNA on MCL-1 expression. However, Mcl-1 mRNA expression was not affected after miR-29a transfection. This could explain why this messenger was not found as down-regulated, in our gene array study, after miR-29a transfection of ALK+ ALCL cell lines (see supplemental data and supplemental Table 1). Nevertheless, although MCL-1 is transcriptionally induced in particular by STAT3,37 miR-29a low expression could contribute to support high MCL-1 expression.
Given that MCL-1 is an antiapoptotic protein, we assessed the induction of apoptosis by exogenous miR-29a in the Karpas 299 and SU-DHL-1 cell lines and in a xenograft model. In ALCL cell lines, miR-29a overexpression followed by doxorubicin treatment (commonly used in ALCL chemotherapy) resulted in an increased rate of apoptosis compared with the control. These results are in agreement with those reported by Garzon et al in AML18 and by Xiong et al in hepatocellular carcinoma.21 Moreover, we can assume that the impact of miR-29a on apoptosis is mediated by translation inhibition of the MCL-1 long form, because this isoform is mainly detected in miR-29a–transfected cells, before doxorubicin treatment. Furthermore, the specific effect of miR-29a was confirmed using an anti–miR-29a to restore MCL-1 protein expression. The tumor suppressor effect of miR-29a via a decreased MCL-1 expression was also demonstrated in vivo in mice xenografted with miR-29a–transfected Karpas-299 cells. These observations therefore support a role of miR-29a low expression in apoptosis blockade through MCL-1 high expression in ALK+ cells.
In this study, we demonstrated that, in addition to its transcriptional regulation,37,38 MCL-1 is also regulated at the posttranscriptional level by miR-29a in ALK+ ALCL, thus suggesting a survival advantage in silencing miR-29a. For the first time, we showed that miR-29a low expression is regulated by NPM-ALK oncogene, partly through epigenetic mechanism. Thus, miR-29a re-expression could modulate apoptosis by inhibiting MCL-1 expression in ALK+ ALCL, and might be a new tool to affect tumorigenesis in these lymphomas.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Sophie Peries, Celine Lopez, Jeannine Boyes, and Marianne Foisseau for technical support and help. We are grateful to Prof Pierre Brousset and Dr Karen Pulford for critically reviewing the manuscript.
This work was supported by grants from the Association pour la Recherche contre le Cancer (ARC), INCa (projet PAIR Lymphomes), Projet Hospitalier de Recherche Clinique (INCa, France), and Région Midi-Pyrénées (France).
Authorship
Contribution: C.D., M.H.-R., J.B., N.J., and E.D. performed functional experiments; C.D., M.-H.R., E.E., F.M., J.C., and L.L. designed experiments; C.D., M-.H.R., and L.L. analyzed data; A.K. provided PF-2341066 inhibitor; J.S. performed and analyzed CGH arrays experiments; and C.D., G.D., and L.L. wrote the manuscript. All the authors critically reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laurence Lamant, CRCT, Inserm UMR1037, Université Paul Sabatier, 31024 Toulouse, Cedex 3, France; e-mail: laurence.lamant@inserm.fr.



![Figure 7. Effect of miR-29a on tumor growth. NOD-SCID mice were xenografted with Karpas-299 cells transfected with miR-29a or miR-control (miR-Ctl). (A) Curves of tumor growth showing tumor volumes (in cubic millimeters) at the indicated days and (B) histograms of tumor weight (grams) averages at the end of the experiment. P for miR-29a vs control (*P < .05). (C) Representative images of immunostaining with Caspase-3 antibody for miR-Ctl and miR-29a tumors (Leica Digital Module R[DMR] microscope equipped with DFC300FX camera and 200×/0.85 NA objective lens, original magnification ×400). Image processing was performed using IM50 software from Leica. Table shows the average of Caspase-3–positive cells for all 7 mice, counted 3 times each, with SD and P value. (D) Representative Western blotting for MCL-1 in miR-Ctl and miR-29a–expressing tumors. Normalized mean value and SD of densitometry measurements are shown (n = 7).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/24/10.1182_blood-2010-09-301994/4/m_zh89991172150007.jpeg?Expires=1767759583&Signature=eMpheEYST0rkZrs-7Nc0m4-BMBAheFKOwyPHivqID689iPRmXgZlAx24CZ8~WwfPhxOF3197QWFxUqZzAd8VA9I0-fFKKS9oN7MzNRkKpT7UfMkcUCGLmzPrzzSk3~TDKT3sEIat23LoroZ3rXvt7f5nmEaj1SGX1cvQ3v1HoQ9q5lCw-KRk7Pkkmmg~UOfttIs-IvLZ9c6LIcl2KfoZgLKZ~~jCI74LZJ3z86mgH1S1ex-YQ4A0S1JrPRYLFT6co1mbOlKYzfkxy1nusmYGMbFvgA4tVdi3B67NahevvWx~~0YBmYh~2AyORDsBawmGdGc5G~Ec6V6BhFs8KJsjEg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal