Abstract
Hypoxia-inducible factor (HIF) regulates the major transcriptional cascade central to the response of all mammalian cells to alterations in oxygen tension. Expression arrays indicate that many hundreds of genes are regulated by this pathway, controlling diverse processes that in turn orchestrate both oxygen delivery and utilization. However, the extent to which HIF exerts direct versus indirect control over gene expression together with the factors dictating the range of HIF-regulated genes remains unclear. Using chromatin immunoprecipitation linked to high throughput sequencing, we identify HIF-binding sites across the genome, independently of gene architecture. Using gene set enrichment analysis, we demonstrate robust associations with the regulation of gene expression by HIF, indicating that these sites operate over long genomic intervals. Analysis of HIF-binding motifs demonstrates sequence preferences outside of the core RCGTG-binding motif but does not reveal any additional absolute sequence requirements. Across the entire genome, only a small proportion of these potential binding sites are bound by HIF, although occupancy of potential sites was enhanced approximately 20-fold at normoxic DNAse1 hypersensitivity sites (irrespective of distance from promoters), suggesting that epigenetic regulation of chromatin may have an important role in defining the response to hypoxia.
Introduction
Oxygen is essential to the survival of all eukaryotic cells, and adaptation to altered oxygen tension occurs at both the systemic level and at the cellular level through regulation of both oxygen utilization and delivery. The maintenance of adequate levels of red blood cells and hemoglobin is essential to the delivery of sufficient oxygen to the tissues and is controlled through erythropoiesis. Erythropoietin production is regulated in response to tissue oxygenation by the transcription factor hypoxia-inducible factor (HIF). Indeed, genome-wide association studies in Han and Tibetan Chinese populations have linked the EPAS1 gene locus with altered hemoglobin concentration,1,2 and mutations in HIF-2α and its regulatory proteins lead to disorders of erythropoiesis.3-5 Composed of an α/β-heterodimer, HIF is primarily controlled through oxygen-regulated modification of the HIF-α subunit, by 2-oxoglutarate-dependent dioxygenase enzymes6,7 that in turn modulate stability and transactivating activity of the HIF-α subunit.7-9 Small-molecule analogs of 2-oxoglutarate (eg, dimethyloxalylglycine [DMOG]) inhibit these enzymes, stabilizing HIF-α and activating target genes, such as erythropoietin.10,11
However, in addition to regulating erythropoietin, HIF drives a major transcriptional cascade responsible for orchestrating the cellular response to oxygen availability in many different tissues. It controls processes as diverse as growth factor production, proliferation, differentiation, apoptosis, energy metabolism, and angiogenesis, and is important in embryonic development and adaptation to altitude, as well as the pathophysiology of inflammation, cancer, and ischemic vascular disease.12-14 Expression arrays indicate that many hundreds of genes are regulated by the HIF pathway.15-20 In addition to direct gene regulation, HIF also regulates many other transcription factors indicating that HIF controls a network of responses to hypoxia. However, until recently, the extent of direct versus indirect transcriptional regulation of gene expression by the HIF cascade was unclear.21-25
Analysis of validated HIF-dependent regulatory elements at approximately 50 HIF-target genes, including the erythropoietin enhancer, has identified a core response element 5′-RCGTG-3′.26 However, occurrences of this motif across the genome are too numerous to permit prediction of HIF binding. Although attempts have been made to identify potential HIF-binding sites based on mammalian conservation and distance from the transcriptional start site (TSS),20 this approach is limited, by definition, to short range interactions, does not provide any mechanistic insight into the factors restricting HIF binding at the molecular level and requires experimental verification.
To identify direct HIF target genes, several recent studies have used chromatin immunoprecipitation coupled to tiled microarrays (ChIP-chip) to examine genome-wide binding of HIF-1α and HIF-2α isoforms to gene promoters.27-30 However, compared with newer techniques of chromatin immunoprecipitation coupled to next-generation high-throughput sequencing (ChIP-seq), these techniques have a relatively low signal-to-noise ratio and are intrinsically biased, being restricted to sequences used in the tiled array.
We describe the use of ChIP coupled to next-generation high-throughput sequencing to define HIF-binding sites across the whole genome irrespective of whether they coincide with a promoter. The work demonstrates the existence of large numbers of HIF-binding sites that are remote from known promoters. Robust associations with the regulation of distant target genes were defined by gene set enrichment analysis (GSEA), strongly suggesting that these sites promote transcriptional responses to hypoxia over very long genomic intervals. Refined analysis of the HIF-recognition motif based on more precise localization of HIF-binding regions defines sequence preferences beyond the core RCGTG motif. However, these additional primary sequence preferences do not greatly restrict the number of potential binding sites in the genome, indicating that epigenetic factors must play a major role in defining HIF binding. Correlation of HIF-binding data with genome-wide DNAse1 hypersensitivity data obtained by the ENCODE consortium in normoxic cells31,32 demonstrates that “resting-state” chromatin accessibility is a strong, but not sole, predictor of hypoxia-inducible HIF binding to an RCGTG motif.
Methods
Cell culture
MCF-7 (breast cancer) cells were grown in DMEM, 2mM l-glutamine, and 10% FBS (Sigma-Aldrich). Subconfluent cell cultures were exposed to 2mM DMOG (Frontier Scientific) or 0.5% ambient oxygen for 16 hours before harvest.
ChIP
ChIPs were performed on MCF-7 cells as previously described, using rabbit polyclonal anti-sera to HIF-1α (PM14), HIF-2α (PM9),27 HIF-1β (Novus NB-100-110), and RNA Pol2 (Santa Cruz Biotechnology SC-899). Preimmune serum or IgG were used as negative controls.
High-throughput sequencing
Libraries were prepared from immunoprecipitated chromatin using the Illumina ChIP-Seq kit; 36-bp single end sequence analysis was performed on the Illumina GAII platform. Sequences were mapped to NCBI build 36 (hg18) using ELAND software. Subsequent assessment, 2-sample peak finding, motif analysis, and mapping of RCGTG motifs were undertaken using the CisGenome software suite (www.biostat.jhsph.edu/∼hji/cisgenome).33-36 In addition, 3 additional filters were applied to define high-stringency peaks. First, the number of reads in each direction was at least one-third of the total. Second, the estimated false discovery rate was less than 0.05. Finally, loci with more than 4 binding sites were excluded as manual analysis revealed these to be regions of high background or repeat elements.
Independent confirmation of chromatin enrichment by real-time quantitative PCR
SYBR Green-based real-time quantitative PCR using a StepOne thermocycler was used. β-actin was used for normalization and fold enrichment at each site was determined using the ΔCt method. Primer sequences are given in supplemental Table 3 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Expression arrays
The microarray analysis of MCF-7 cells exposed to 1% ambient oxygen for 16 hours in the presence of either control or combined HIF-1α and HIF-2α siRNA has been previously described.15 Correlation of ChIP-seq data with microarray analysis of gene expression was performed using GSEA software Version 2 (www.broadinstitute.org/gsea/).36,37
Expression of quantitative PCR
cDNA was synthesized using the High Capacity cDNA Kit (Applied Biosystems). Expression analysis was performed using the SYBR Green method and the ΔCt method. Primer sequences are given in supplemental Table 3.
Luciferase vectors and reporter assays
HIF-binding sites were cloned into pGL3 promoter (Promega) using BglII (primers arrestin domain containing 3 [ARRDC3]: ACAGGAGTAGCGCGAGCAG, CTGCAAAGACCTTTGAGTTCC; ERBB receptor feedback inhibitor 1 [ERRFI1] CATGTGACGTCAGAAAGGCT, CTGCAAAGACCTTTGAGTTCC). U2OS cells, cotransfected with the pGL3 reporter and β-galactosidase reporter, were cultured in normoxia or 0.5% hypoxia for 16 hours. Luciferase activity (Promega Luciferase kit) was normalized to β-galactosidase activity.
Results
Definition of HIF-α-binding sites
An important challenge with large ChIP-seq datasets is to identify an appropriate threshold to define binding peaks. Using gene loci identified by previous ChIP-chip analysis27 as a “seed” dataset, we examined the effect of altering the threshold used to define ChIP-seq peaks, on the degree of overlap between the 2 datasets. This identified a threshold of approximately 20 reads per window for HIF-1α (and 25 reads per window for HIF-2α), below which the overlap with the ChIP-chip data fell to background (supplemental Figure 1A-B). A comparable threshold was defined when previously identified HIF-regulated genes15 were used as the “seed” dataset, indicating that this approach to identifying suitable thresholds for ChIP-seq analyses should be generalizable (data not shown).
To define high-stringency ChIP-seq-binding sites more rigorously, we used these thresholds, together with 3 additional filters (see “High-throughput sequencing”). This identified 400 high-stringency HIF-1α-binding sites at 356 gene loci (Table 1; supplemental Table 1) and 425 high-stringency HIF-2α-binding sites at 357 gene loci (Table 2; supplemental Table 2).
Top 30 HIF-1 binding sites
| HIF-1 binding sites . | Chromosome . | Start . | End . | Peak height, no. of reads per window . | Covered by human promoter 1.0R array . | Nearest gene locus . | RefSeq number . |
|---|---|---|---|---|---|---|---|
| 1 | 7 | 156986147 | 156987039 | 314 | No | DNAJB6 | NM_058246 |
| 2 | 8 | 134456859 | 134457261 | 180 | No | NDRG1 | NM_006096 |
| 3 | 1 | 114156539 | 114157002 | 288 | Yes | RSBN1 | NM_018364 |
| 4 | 16 | 29984123 | 29984574 | 160 | Yes | ALDOA | NM_184041 |
| 5 | 15 | 91153222 | 91153655 | 160 | No | CHD2 | NM_001042572 |
| 6 | 20 | 48720371 | 48720786 | 136 | No | C20orf175 | NM_080829 |
| 7 | 3 | 5003621 | 5003966 | 128 | No | BHLHB2 | NM_003670 |
| 8 | 14 | 61287408 | 61287886 | 126 | No | SNAPC1 | NM_003082 |
| 9 | 10 | 6283489 | 6283830 | 126 | Yes | PFKFB3 | NM_004566 |
| 10 | 19 | 39541937 | 39542408 | 123 | Yes | GPI | NM_000175 |
| 11 | 1 | 178403857 | 178404013 | 118 | Yes | QSCN6 | NM_002826 |
| 12 | 15 | 70306014 | 70306475 | 116 | No | PKM2 | NM_002654 |
| 13 | 6 | 87921667 | 87922197 | 109 | Yes | CGA | NM_000735 |
| 14 | 14 | 74791306 | 74791781 | 103 | No | FOS | NM_005252 |
| 15 | 14 | 22840340 | 22840855 | 102 | Yes | BCL2L2 | NM_004050 |
| 16 | 14 | 33477245 | 33477551 | 98 | No | EGLN3 | NM_022073 |
| 17 | 6 | 43743806 | 43744203 | 96 | No | MRPS18A | NM_018135 |
| 18 | 12 | 8014473 | 8014830 | 95 | No | SLC2A3 | NM_006931 |
| 19 | 3 | 48569558 | 48569870 | 95 | Yes | PFKFB4 | NM_004567 |
| 20 | 7 | 55216284 | 55216762 | 94 | No | EGFR | NM_201283 |
| 21 | 2 | 173128811 | 173129189 | 94 | Yes | PDK1 | NM_002610 |
| 22 | 4 | 186554370 | 186554738 | 89 | Yes | ANKRD37 | NM_181726 |
| 23 | 3 | 123585527 | 123585631 | 88 | Yes | C3orf28 | NM_014367 |
| 24 | 8 | 135913721 | 135913971 | 87 | No | ZFAT1 | NM_020863 |
| 25 | 1 | 8861349 | 8862115 | 86 | Yes | ENO1 | NM_001428 |
| 26 | 17 | 78009247 | 78009655 | 86 | Yes | NARF | NM_001038618 |
| 27 | 1 | 200320766 | 200321048 | 83 | No | GPR37L1 | NM_004767 |
| 28 | 2 | 86521270 | 86521896 | 81 | Yes | JMJD1A | NM_018433 |
| 29 | 15 | 88509909 | 88510331 | 81 | No | SEMA4B | NM_020210 |
| 30 | 4 | 120441100 | 120441561 | 87 | Yes | LOC401152 | NM_001001701 |
| HIF-1 binding sites . | Chromosome . | Start . | End . | Peak height, no. of reads per window . | Covered by human promoter 1.0R array . | Nearest gene locus . | RefSeq number . |
|---|---|---|---|---|---|---|---|
| 1 | 7 | 156986147 | 156987039 | 314 | No | DNAJB6 | NM_058246 |
| 2 | 8 | 134456859 | 134457261 | 180 | No | NDRG1 | NM_006096 |
| 3 | 1 | 114156539 | 114157002 | 288 | Yes | RSBN1 | NM_018364 |
| 4 | 16 | 29984123 | 29984574 | 160 | Yes | ALDOA | NM_184041 |
| 5 | 15 | 91153222 | 91153655 | 160 | No | CHD2 | NM_001042572 |
| 6 | 20 | 48720371 | 48720786 | 136 | No | C20orf175 | NM_080829 |
| 7 | 3 | 5003621 | 5003966 | 128 | No | BHLHB2 | NM_003670 |
| 8 | 14 | 61287408 | 61287886 | 126 | No | SNAPC1 | NM_003082 |
| 9 | 10 | 6283489 | 6283830 | 126 | Yes | PFKFB3 | NM_004566 |
| 10 | 19 | 39541937 | 39542408 | 123 | Yes | GPI | NM_000175 |
| 11 | 1 | 178403857 | 178404013 | 118 | Yes | QSCN6 | NM_002826 |
| 12 | 15 | 70306014 | 70306475 | 116 | No | PKM2 | NM_002654 |
| 13 | 6 | 87921667 | 87922197 | 109 | Yes | CGA | NM_000735 |
| 14 | 14 | 74791306 | 74791781 | 103 | No | FOS | NM_005252 |
| 15 | 14 | 22840340 | 22840855 | 102 | Yes | BCL2L2 | NM_004050 |
| 16 | 14 | 33477245 | 33477551 | 98 | No | EGLN3 | NM_022073 |
| 17 | 6 | 43743806 | 43744203 | 96 | No | MRPS18A | NM_018135 |
| 18 | 12 | 8014473 | 8014830 | 95 | No | SLC2A3 | NM_006931 |
| 19 | 3 | 48569558 | 48569870 | 95 | Yes | PFKFB4 | NM_004567 |
| 20 | 7 | 55216284 | 55216762 | 94 | No | EGFR | NM_201283 |
| 21 | 2 | 173128811 | 173129189 | 94 | Yes | PDK1 | NM_002610 |
| 22 | 4 | 186554370 | 186554738 | 89 | Yes | ANKRD37 | NM_181726 |
| 23 | 3 | 123585527 | 123585631 | 88 | Yes | C3orf28 | NM_014367 |
| 24 | 8 | 135913721 | 135913971 | 87 | No | ZFAT1 | NM_020863 |
| 25 | 1 | 8861349 | 8862115 | 86 | Yes | ENO1 | NM_001428 |
| 26 | 17 | 78009247 | 78009655 | 86 | Yes | NARF | NM_001038618 |
| 27 | 1 | 200320766 | 200321048 | 83 | No | GPR37L1 | NM_004767 |
| 28 | 2 | 86521270 | 86521896 | 81 | Yes | JMJD1A | NM_018433 |
| 29 | 15 | 88509909 | 88510331 | 81 | No | SEMA4B | NM_020210 |
| 30 | 4 | 120441100 | 120441561 | 87 | Yes | LOC401152 | NM_001001701 |
HIF indicates hypoxia-inducible factor.
Top 30 HIF-2 binding sites
| HIF-2 binding sites . | Chromosome . | Start . | End . | Peak height, no. of reads per window . | Covered by human promoter 1.0R array . | Nearest gene locus . | RefSeq number . |
|---|---|---|---|---|---|---|---|
| 1 | 7 | 156986122 | 156987013 | 268 | No | DNAJB6 | NM_058246 |
| 2 | 20 | 48720413 | 48720792 | 142 | No | C20orf175 | NM_080829 |
| 3 | 15 | 88509900 | 88510359 | 130 | No | SEMA4B | NM_020210 |
| 4 | 21 | 42669493 | 42669912 | 119 | No | TFF1 | NM_003225 |
| 5 | 8 | 134456898 | 134457355 | 117 | No | NDRG1 | NM_006096 |
| 6 | 7 | 127882809 | 127883250 | 105 | Yes | HIG2 | NM_013332 |
| 7 | 2 | 218226277 | 218226673 | 103 | No | TNS1 | NM_022648 |
| 8 | 3 | 4995548 | 4996080 | 95 | Yes | BHLHB2 | NM_003670 |
| 9 | 3 | 5003471 | 5003914 | 91 | No | BHLHB2 | NM_003670 |
| 10 | 12 | 8014461 | 8014804 | 91 | No | SLC2A3 | NM_006931 |
| 11 | 14 | 61287435 | 61287871 | 90 | No | SNAPC1 | NM_003082 |
| 12 | 7 | 156890452 | 156890887 | 89 | No | DNAJB6 | NM_058246 |
| 13 | 11 | 72583304 | 72583693 | 88 | No | P2RY2 | NM_002564 |
| 14 | 12 | 50823667 | 50824115 | 87 | No | KRT80 | NM_182507 |
| 15 | 14 | 99628951 | 99629206 | 86 | No | EVL | NM_016337 |
| 16 | 16 | 29984146 | 29984554 | 85 | Yes | ALDOA | NM_184041 |
| 17 | 1 | 200320701 | 200321105 | 86 | No | GPR37L1 | NM_004767 |
| 18 | 7 | 55216313 | 55216811 | 79 | No | EGFR | NM_201283 |
| 19 | 8 | 22511476 | 22511809 | 79 | Yes | C8orf58 | NM_001013842 |
| 20 | 2 | 1614654 | 1615038 | 77 | No | PXDN | NM_012293 |
| 21 | 7 | 41579152 | 41579497 | 77 | No | INHBA | NM_002192 |
| 22 | 4 | 120441169 | 120441511 | 75 | Yes | LOC401152 | NM_001001701 |
| 23 | 5 | 3979121 | 3979463 | 74 | No | IRX1 | NM_024337 |
| 24 | 20 | 48215760 | 48216285 | 73 | No | Kua | NM_199129 |
| 25 | 19 | 39541912 | 39542320 | 72 | Yes | GPI | NM_000175 |
| 26 | 1 | 200258697 | 200259111 | 71 | No | ELF3 | NM_004433 |
| 27 | 12 | 123888666 | 123889062 | 71 | No | SCARB1 | NM_005505 |
| 28 | 14 | 74791304 | 74791735 | 70 | No | FOS | NM_005252 |
| 29 | 12 | 32988395 | 32988693 | 70 | No | PKP2 | NM_001005242 |
| 30 | 15 | 62024988 | 62025281 | 70 | No | DAPK2 | NM_014326 |
| HIF-2 binding sites . | Chromosome . | Start . | End . | Peak height, no. of reads per window . | Covered by human promoter 1.0R array . | Nearest gene locus . | RefSeq number . |
|---|---|---|---|---|---|---|---|
| 1 | 7 | 156986122 | 156987013 | 268 | No | DNAJB6 | NM_058246 |
| 2 | 20 | 48720413 | 48720792 | 142 | No | C20orf175 | NM_080829 |
| 3 | 15 | 88509900 | 88510359 | 130 | No | SEMA4B | NM_020210 |
| 4 | 21 | 42669493 | 42669912 | 119 | No | TFF1 | NM_003225 |
| 5 | 8 | 134456898 | 134457355 | 117 | No | NDRG1 | NM_006096 |
| 6 | 7 | 127882809 | 127883250 | 105 | Yes | HIG2 | NM_013332 |
| 7 | 2 | 218226277 | 218226673 | 103 | No | TNS1 | NM_022648 |
| 8 | 3 | 4995548 | 4996080 | 95 | Yes | BHLHB2 | NM_003670 |
| 9 | 3 | 5003471 | 5003914 | 91 | No | BHLHB2 | NM_003670 |
| 10 | 12 | 8014461 | 8014804 | 91 | No | SLC2A3 | NM_006931 |
| 11 | 14 | 61287435 | 61287871 | 90 | No | SNAPC1 | NM_003082 |
| 12 | 7 | 156890452 | 156890887 | 89 | No | DNAJB6 | NM_058246 |
| 13 | 11 | 72583304 | 72583693 | 88 | No | P2RY2 | NM_002564 |
| 14 | 12 | 50823667 | 50824115 | 87 | No | KRT80 | NM_182507 |
| 15 | 14 | 99628951 | 99629206 | 86 | No | EVL | NM_016337 |
| 16 | 16 | 29984146 | 29984554 | 85 | Yes | ALDOA | NM_184041 |
| 17 | 1 | 200320701 | 200321105 | 86 | No | GPR37L1 | NM_004767 |
| 18 | 7 | 55216313 | 55216811 | 79 | No | EGFR | NM_201283 |
| 19 | 8 | 22511476 | 22511809 | 79 | Yes | C8orf58 | NM_001013842 |
| 20 | 2 | 1614654 | 1615038 | 77 | No | PXDN | NM_012293 |
| 21 | 7 | 41579152 | 41579497 | 77 | No | INHBA | NM_002192 |
| 22 | 4 | 120441169 | 120441511 | 75 | Yes | LOC401152 | NM_001001701 |
| 23 | 5 | 3979121 | 3979463 | 74 | No | IRX1 | NM_024337 |
| 24 | 20 | 48215760 | 48216285 | 73 | No | Kua | NM_199129 |
| 25 | 19 | 39541912 | 39542320 | 72 | Yes | GPI | NM_000175 |
| 26 | 1 | 200258697 | 200259111 | 71 | No | ELF3 | NM_004433 |
| 27 | 12 | 123888666 | 123889062 | 71 | No | SCARB1 | NM_005505 |
| 28 | 14 | 74791304 | 74791735 | 70 | No | FOS | NM_005252 |
| 29 | 12 | 32988395 | 32988693 | 70 | No | PKP2 | NM_001005242 |
| 30 | 15 | 62024988 | 62025281 | 70 | No | DAPK2 | NM_014326 |
HIF indicates hypoxia-inducible factor.
Quantitative PCR analysis to independently assess the veracity of a random selection of these peaks confirmed a very high specificity (18 of 20 HIF-1α sites and 9 of 12 HIF-2α sites; supplemental Figure 2A-B). Conversely, ChIP–quantitative PCR analysis of sites identified previously by ChIP-chip,27 not meeting all inclusion criteria for high-stringency definition by ChIP, yielded signals at these sites, albeit with low levels of enrichment in some cases (supplemental Figure 2C).
Overlap between HIF-α- and HIF-1β-binding sites
Although HIF-α proteins were originally defined as part of a dimer with HIF-1β that bound DNA at hypoxia response elements, interactions with other transcription factors have been defined that might result in DNA-binding without HIF-1β at other sites.38 Therefore, to determine the pan-genomic overlap of HIF-α- and HIF-1β-binding sites, we undertook a comparable ChIP-seq analysis of HIF-1β DNA-binding in MCF-7 cells that were exposed to hypoxia for 16 hours.
HIF-1β-binding sites were ranked according to peak height, and the number that overlapped with high-stringency HIF-1α-binding sites was plotted against this rank. This revealed the existence of 356 overlapping HIF-1β peaks, after which overlap fell to a background level. Similar analysis of HIF-2α-binding sites revealed 301 overlapping HIF-1β peaks. The very high level of concordance between HIF-α and HIF-1β binding is consistent with very accurate definition of heterodimeric binding complexes by ChIP-seq and suggests that under these conditions the large majority of HIF-α binding is complexed with HIF-1β and vice-versa. To test this, we again plotted the degree of overlap against rank (supplemental Figure 3B,D). This revealed even higher levels of association for the higher ranked peaks; for instance, when the top 200 HIF-1α and HIF-2α sites were considered, overlap with HIF-1β was 95% and 94.5% respectively. Conversely the top 200 HIF-1β sites manifest 95% overlap with HIF-α sites (supplemental Figure 3E-F). Taken together, these findings are consistent with the large majority of HIF-α DNA-binding being in complex with HIF-1β and vice-versa. Furthermore, very strong concordance between HIF-binding sites, in cells treated with either DMOG or hypoxia, suggests that at the level of DNA-binding these stimuli have very similar effects.
The association of apparently nonoverlapping sites with lower-ranked sites suggested that nonoverlap might largely reflect false assignment in one or the other dataset. We therefore retested a selection of apparently exclusive HIF-α-binding sites by ChIP–quantitative PCR and found enrichment at only 25%, suggesting that these sites are indeed mainly composed of false positive signal (data not shown). Consequently, we based all subsequent analyses on HIF-α sites at which HIF-1β was also present.
Distribution of HIF-binding and HIF-regulated gene loci
Distribution in the vicinity of the TSS.
The ability to identify HIF-binding sites, independently of the coverage afforded by tiled promoter microarrays, permits true genome-wide analysis of HIF-α DNA binding, thus enabling a complete description of the distribution of HIF-binding sites. Of the 356 high-stringency HIF-1-binding sites (HIF-1α and HIF-1β) identified by ChIP-seq, 170 (48%) were novel sites, distant from promoter regions (as defined by the Human Promoter 1.0R microarray), whereas 212 (70%) of 301 HIF-2-binding sites (HIF-2α and HIF-1β) lay outside these regions. These differing proportions probably account for the greater number of novel HIF-2-binding sites identified by ChIP-seq and suggest differing distributions of the 2 isoforms with respect to the promoter.
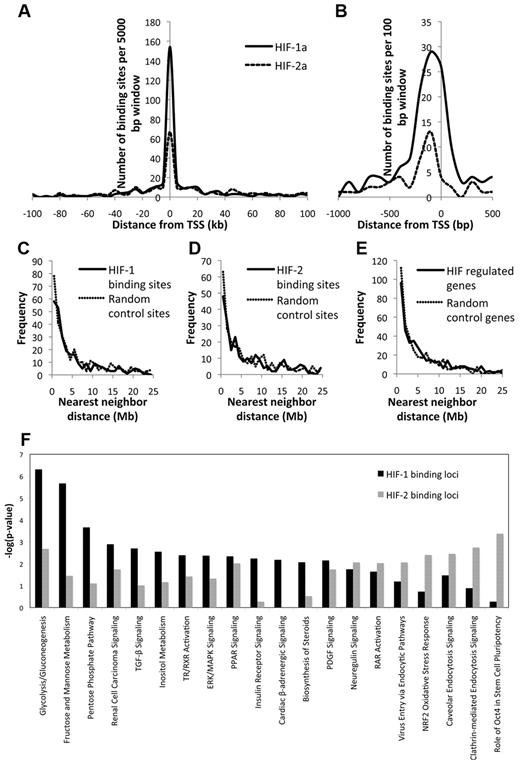
To test this, we examined the frequency distribution of HIF-1 and HIF-2-binding sites about the nearest TSS. Using 5-kb windows, we observed clustering of both HIF-1 and HIF-2-binding sites close to the TSS, with a low frequency of binding extending to much greater distances. However, HIF-1 was more likely to bind close to the TSS, with approximately 40% of HIF-1-binding sites lying within 2.5 kb of the TSS, as opposed to approximately 20% of HIF-2-binding sites (Figure 1A). To exclude the possibility that an inappropriately low threshold might distort the binding distribution of the HIF-2 sites (by including false positives), we repeated the analysis using only the 100 top binding sites for each HIF isoform. This gave almost identical distributions to the complete sets of binding sites, confirming the more widely distributed pattern of HIF-2 binding (data not shown). More detailed analysis of the binding patterns using 100-bp windows indicated that, for either HIF isoform, the most frequent position of the binding site laid approximately 100 bp 5′ to the TSS, within the core promoter (Figure 1B). However, HIF-binding sites were also identified in intergenic, 5′-untranslated region, 3′-untranslated region, and intronic and exonic regions.
Distribution of HIF-binding sites. The position of HIF-1 and HIF-2-binding sites with respect to the nearest TSS was plotted as a frequency distribution using (A) 5-kb windows and (B) 100-bp windows. Frequency distribution of the nearest-neighbor distance between (C) HIF-1-binding gene loci, (D) HIF-2-binding gene loci, and (E) HIF-regulated gene loci. In each case, the frequency distribution of nearest-neighbor distances for an equal number of randomly selected control gene loci is shown for comparison. (F) The Ingenuity Systems Inc (IPA Version 8.8-3204) software tool was used to examine the HIF-1 and HIF-2-binding loci for functional pathways. Canonical pathways statistically over-represented among HIF-binding loci are shown together with the respective −log10 (P value) for each isoform.
Distribution of HIF-binding sites. The position of HIF-1 and HIF-2-binding sites with respect to the nearest TSS was plotted as a frequency distribution using (A) 5-kb windows and (B) 100-bp windows. Frequency distribution of the nearest-neighbor distance between (C) HIF-1-binding gene loci, (D) HIF-2-binding gene loci, and (E) HIF-regulated gene loci. In each case, the frequency distribution of nearest-neighbor distances for an equal number of randomly selected control gene loci is shown for comparison. (F) The Ingenuity Systems Inc (IPA Version 8.8-3204) software tool was used to examine the HIF-1 and HIF-2-binding loci for functional pathways. Canonical pathways statistically over-represented among HIF-binding loci are shown together with the respective −log10 (P value) for each isoform.
Distribution across the genome.
The full genome-wide coverage afforded by ChIP-seq also permits analysis of clustering of HIF-α-binding sites across the genome. Analyzing nearest-neighbor distance, for HIF-1- and HIF-2-binding loci and HIF-regulated genes, revealed non-Poisson distributions. However, the spread was comparable with the distribution of identical numbers of randomly selected genes, indicating that HIF-1 and HIF-2 target genes are randomly distributed among known transcripts (Figure 1C-E).
Distribution across biologic pathways.
HIF-binding loci were then examined using the Ingenuity Pathways Analysis software tool (Version IPA 8.8-3204). Twenty canonical pathways were statistically over-represented among HIF-binding loci with several demonstrating bias toward either HIF-1 or HIF-2 binding (Figure 1F). Network analysis identified 3 major networks, the largest containing 59 focus molecules (supplemental Figure 4) with major nodes centered on ERRB2, TGFB1, TP53, MYC/MAX, HNF1A, SP1, and NOTCH1 indicating interaction with these pathways.
HIF-binding and gene regulation
Because HIF-binding sites were identified in all regions of the genome, including many at great distances from gene promoters, we next examined relationships between the position of HIF-binding sites and effects on gene expression. The high-stringency HIF-binding loci were examined for enrichment among HIF-regulated genes using GSEA.36,37 HIF-regulated genes were identified and ranked using expression array data from MCF-7 cells.15 As expected, loci binding HIF-1 and HIF-2 strongly clustered among the genes that were most highly up-regulated by HIF. In contrast, no enrichment of HIF-binding loci was observed among the genes down-regulated by HIF (supplemental Figure 5).
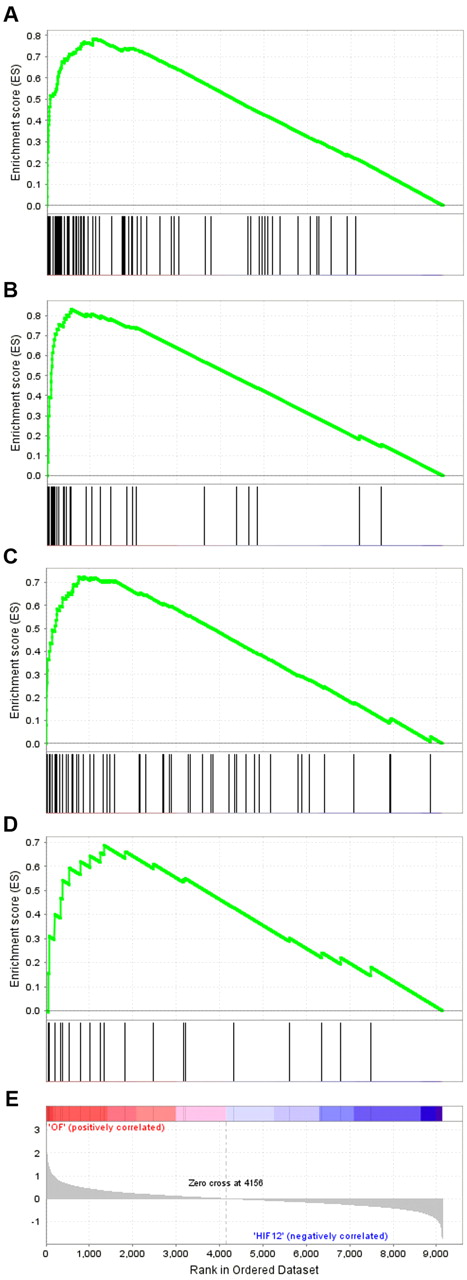
To examine the relationship between promoter-distant HIF-binding and transcriptional regulation, we stratified HIF-binding gene loci according to the distance between the TSS and the nearest HIF-binding site and ran further tests for association with up-regulation by HIF, using GSEA. These analyses revealed that HIF-1-binding loci were significantly associated with up-regulation of the nearest transcript by HIF, irrespective of the distance between the HIF-1-binding site and the TSS (Figure 2). The association was robust even for loci where HIF-1 binding was greater than 100 kb from the nearest TSS (enrichment score [ES] = 0.69, normalized enrichment score = 1.50, nominal P = .0, false discovery rate q-value = 0.09, Family-wise error rate P = .0), indicating that HIF-1 DNA-binding can exert effects on gene regulation over distances of greater than 100 kb. Comparable results were seen for HIF-2 (supplemental Figure 6).
GSEA stratified by distance between-binding site and TSS. Expression array data from MCF-7 cells, incubated for 16 hours in 1% ambient oxygen and treated either with control siRNA or siRNAs directed against both HIF-α subunits,15 were normalized, filtered for present/absent calls, and ranked according to the fold regulation by HIF-α. High-stringency HIF-1-binding gene loci were stratified according to the distance to the nearest TSS into 4 subgroups. GSEA was performed for each subgroup of genes (www.broadinstitute.org/gsea).37,39 The cumulative ESs, together with the position of the HIF-binding gene loci among the genes ranked by fold regulation, are plotted for (A) gene loci with the closest HIF-1 binding within 1 kb of the TSS, (B) between 1 kb and 10 kb of the TSS, (C) between 10 kb and 100 kb of the TSS, and (D) greater than 100 kb from the TSS. (E) The log2 fold regulation by HIF-α is illustrated.
GSEA stratified by distance between-binding site and TSS. Expression array data from MCF-7 cells, incubated for 16 hours in 1% ambient oxygen and treated either with control siRNA or siRNAs directed against both HIF-α subunits,15 were normalized, filtered for present/absent calls, and ranked according to the fold regulation by HIF-α. High-stringency HIF-1-binding gene loci were stratified according to the distance to the nearest TSS into 4 subgroups. GSEA was performed for each subgroup of genes (www.broadinstitute.org/gsea).37,39 The cumulative ESs, together with the position of the HIF-binding gene loci among the genes ranked by fold regulation, are plotted for (A) gene loci with the closest HIF-1 binding within 1 kb of the TSS, (B) between 1 kb and 10 kb of the TSS, (C) between 10 kb and 100 kb of the TSS, and (D) greater than 100 kb from the TSS. (E) The log2 fold regulation by HIF-α is illustrated.
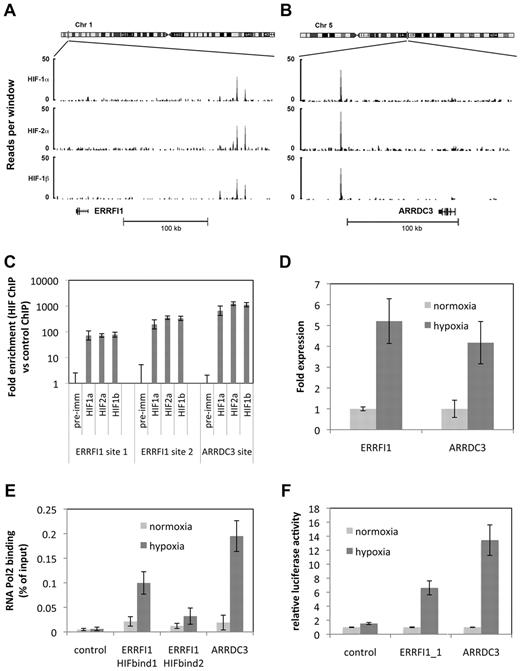
To pursue this further, we performed additional analyses on 2 of the most distant HIF-binding sites. HIF DNA binding at both the ARRDC3 and ERRFI1 loci is located more than 100 kb distant from the promoter, although in each case these are the nearest documented genes (Figure 3A-B), and both have been reported to be regulated by HIF.15 Two major peaks were present at the ERRFI1 locus and one at the ARRDC3 locus. Each peak was confirmed in ChIP–quantitative PCR analysis with high levels of enrichment (Figure 3C). Furthermore, analysis of mRNA levels by quantitative PCR confirmed hypoxia-dependent regulation of these transcripts (Figure 3D). ChIP–quantitative PCR analysis of these sites using antibody directed against RNA Pol2 demonstrated interaction with RNA Pol2 after 16 hours at 0.5% hypoxia (Figure 3E), indicating hypoxia-inducible interaction between these binding sites and the general transcriptional machinery. Finally, fusion of DNA sequences spanning these binding sites to a heterologous luciferase reporter gene conferred hypoxic regulation on luciferase activity, confirming the ability of these sequences to mediate hypoxia-inducible gene expression (Figure 3F).
HIF binding regulates gene expression over long genomic intervals. Genomic binding of HIF-1α, HIF-2α, and HIF-1β subunits at the (A) ERRFI1 and (B) ARRDC3 gene loci. The number of reads per 100-bp window and the location of each gene are shown. (C) ChIP–quantitative PCR confirmation of the ChIP-seq defined binding peaks. (D) Expression-quantitative PCR confirmation of gene regulation in response to 16 hours at 0.5% hypoxia. (E) ChIP–quantitative PCR demonstrating RNA Pol2 recruitment to the ERRFI1 and ARRDC3 HIF-binding sites in normoxia and after 16 hours at 0.5% hypoxia. (F) Luciferase reporter assay showing activity of a heterologous luciferase reporter gene fused to sequences spanning each HIF-binding site in normoxia and after 16 hours at 0.5% hypoxia.
HIF binding regulates gene expression over long genomic intervals. Genomic binding of HIF-1α, HIF-2α, and HIF-1β subunits at the (A) ERRFI1 and (B) ARRDC3 gene loci. The number of reads per 100-bp window and the location of each gene are shown. (C) ChIP–quantitative PCR confirmation of the ChIP-seq defined binding peaks. (D) Expression-quantitative PCR confirmation of gene regulation in response to 16 hours at 0.5% hypoxia. (E) ChIP–quantitative PCR demonstrating RNA Pol2 recruitment to the ERRFI1 and ARRDC3 HIF-binding sites in normoxia and after 16 hours at 0.5% hypoxia. (F) Luciferase reporter assay showing activity of a heterologous luciferase reporter gene fused to sequences spanning each HIF-binding site in normoxia and after 16 hours at 0.5% hypoxia.
At HIF-binding sites where the nearest gene was not regulated, we next examined for association between HIF binding and regulation of the second-nearest transcript. These next nearest HIF-binding gene loci also showed significant association with transcripts regulated by HIF (ES = 0.45, normalized enrichment score = 1.45, nominal P = .0, false discovery rate q-value = 0.09, Family-wise error rate P = .0), indicating that HIF commonly “skips” the nearest gene to regulate another more distant promoter (data not shown).
HIF-binding loci were then grouped according to whether the HIF-1 binding was in the intron, exon, 5′-untranslated region, upstream from the TSS or downstream from the transcriptional end site. Similar GSEA analysis showed significant association with up-regulated genes for all subsets (supplemental Figure 7).
Finally, the amplitude of transcriptional up-regulation by HIF bore no relationship to the distance of the nearest HIF-binding site from the promoter (supplemental Figure 8). Taken together, these results indicate that, at a genome-wide level, HIF-binding loci appear to be functionally similar (ie, they exert qualitatively and quantitatively similar effects on hypoxia-inducible gene expression over large genomic intervals irrespective of position with reference to the regulated gene promoter).
Analysis of transcription factor binding site motifs at HIF-binding sites
Based on analysis of approximately 50 HIF-regulated genes, the minimal cis-regulatory element required for HIF-dependent transactivation has been defined as the RCGTG motif.26 Our unbiased and more precise pan-genomic identification of more than 400 HIF-binding sites allows us to make a more definitive statement regarding sequence preferences surrounding this core motif using 2 independent approaches. First, in an unbiased analysis, HIF-binding sites were examined for common novel motifs using the Gibbs Motif Sampler module of CisGenome,35 which were in turn tested for enrichment by determining the relative number of occurrences in HIF-binding regions compared with control regions.34 As expected, the most enriched motif identified in this way for both HIF-1- and HIF-2-binding regions contained the core RCGTG motif with a preference for A over G at the R position (Figure 4A-B). However, these motifs also exhibited extended sequence preferences both 5′ and 3′ to this core motif. In both cases, preference was seen for T immediately 5′ to the RCGTG and for C immediately 3′ to this motif. The motif identified among HIF-1-binding regions also identified a preference for C or G 2-bp 5′ to the R position. In addition, this approach also identified additional motifs that occur more often in HIF-binding regions than control regions (Figure 4A-B), including an AP-1-like motif.
Characterization of the HIF-binding motif. (A-B) HIF-1- and HIF-2-binding regions were examined for commonly occurring novel motifs using the Gibbs Motif Sampler module of CisGenome. Enrichment of motifs identified in this way was then examined by determining the relative occurrence of each motif in HIF-binding regions compared with a control set of regions (r1), of phylogenetically conserved motifs within phylogenetically conserved portions of HIF-binding regions compared with control regions (r2), and of phylogenetically conserved motifs within all portions of the HIF-1 binding and control regions (r3). The sequence logos40 for motifs with all 3 enrichment ratios greater than 2 are shown. The height of each letter is proportionate to its frequency, with the most frequent at the top. The height of each stack is then adjusted to signify the information content of the sequences at that position (measured in bits). (C) HIF-1 and (D) HIF-2-binding sites containing a single RCGTG motif were analyzed for sequence conservation beyond this core motif. Sequences were extended for 100 bp upstream and downstream of the RCGTG motif and aligned. At each position, the observed base frequency was compared with the expected using a χ2 test. The relative occurrence of each nucleotide at each position is indicated as a percentage of the total by the letter height. Positions showing nonrandom base frequency (P < .01 after Bonferroni correction) are highlighted in gray.
Characterization of the HIF-binding motif. (A-B) HIF-1- and HIF-2-binding regions were examined for commonly occurring novel motifs using the Gibbs Motif Sampler module of CisGenome. Enrichment of motifs identified in this way was then examined by determining the relative occurrence of each motif in HIF-binding regions compared with a control set of regions (r1), of phylogenetically conserved motifs within phylogenetically conserved portions of HIF-binding regions compared with control regions (r2), and of phylogenetically conserved motifs within all portions of the HIF-1 binding and control regions (r3). The sequence logos40 for motifs with all 3 enrichment ratios greater than 2 are shown. The height of each letter is proportionate to its frequency, with the most frequent at the top. The height of each stack is then adjusted to signify the information content of the sequences at that position (measured in bits). (C) HIF-1 and (D) HIF-2-binding sites containing a single RCGTG motif were analyzed for sequence conservation beyond this core motif. Sequences were extended for 100 bp upstream and downstream of the RCGTG motif and aligned. At each position, the observed base frequency was compared with the expected using a χ2 test. The relative occurrence of each nucleotide at each position is indicated as a percentage of the total by the letter height. Positions showing nonrandom base frequency (P < .01 after Bonferroni correction) are highlighted in gray.
Although the unbiased approach will determine novel motifs, it is poor at distinguishing the relative abundance of bases in 2 related motifs, for instance, sequences binding HIF-1 versus HIF-2. Therefore, in a second approach, predicated on the RCGTG motif, sequences flanking each unique HIF-binding RCGTG motif were compared (Figure 4C-D). Both HIF-1- and HIF-2-binding regions are relatively GC rich, reflecting their concentration within promoter regions. For both isoforms, an extended hypoxia response element consensus motif is identified, which is remarkably similar for each. Indeed, no significant difference in base frequency was observed at any position flanking the RCGTG motifs either for the 2 isoforms or for promoter proximal versus distant sites. At position 0 (the position of R), A is preferred over G. Preference is also seen for T at position −1. Importantly, C is strikingly under-represented at position −1, implying that the CACGTG E-box motif is inhibited from binding HIF. This exclusion may result from competition by other proteins that recognize the E-box and reduce their availability to bind HIF, including the HIF-1β homodimer.41 Outside the core RCGTG motif, preference is seen for a CAC motif between positions +13 and +15. Furthermore, the frequency of CAC motifs at other positions downstream of the RCGTG motif was not increased above background (supplemental Figure 9). Mutation of the CAC motif between +13 and +15 has previously been shown to affect functional activity of the erythropoietin 3′ enhancer.42,43
Correlation of HIF binding with DNAse1 hypersensitivity
Within the reference genome, there are 1 114 925 RCGTGs, of which only 606 (0.05%) were identified in HIF-1- and 462 (0.04%) in HIF-2-binding regions. Even allowing for false-negative results arising from the high-stringency criteria used to define these binding sites, the large majority of RCGTG motifs do not bind HIF. Apart from excluding some E-box motifs, sequence analysis does not provide any definite rules to define potential binding sites further. We therefore wished to determine the extent to which epigenetic mechanisms, in particular chromatin accessibility, as defined by DNAse1 hypersensitivity, might contribute to the definition of HIF-binding sites. We therefore correlated our HIF-binding results with DNAse1 hypersensitivity peaks from the ENCODE consortium dataset 1 for Hypersensitivity by Digital DNAse1 in MCF-7 cells (wgEncodeUwDnaseSeqPeaksRep1Mcf7).31,32
Overall, 271 (76%) of HIF-1-binding sites and 246 (82%) of HIF-2-binding sites overlapped with DNAse1-hypersensitive peaks and HIF-binding regions demonstrated a much higher concentration of hypersensitive peaks than the immediate flanking regions (supplemental Figure 10). When all of the 1 114 925 RCGTG motifs, genome-wide, are considered DNAse1-hypersensitive RCGTG motifs are 19 times as likely to bind HIF-1 and 22 times as likely to bind HIF-2 as insensitive motifs. This suggests that DNAse1 hypersensitivity in normoxic cells would be an important predictor of HIF binding to its consensus recognition site.
To test this and to explore the extent to which coincidence between DNAse1 hypersensitivity and HIF binding was observed away from promoter regions (where DNAse1 hypersensitivity sites may be predicted to be enriched in any case), we ranked the HIF-binding loci defined by ChIP-seq according to distance upstream/downstream of the nearest TSS. We then plotted this rank against the distance from the center of the HIF-binding region (defined experimentally by ChIP-seq) of either all RCGTG motifs or just the RCGTG motifs that coincided with DNAse1 hypersensitivity sites (Figure 5A-B). An expectation of these analyses is that correctly identified HIF-binding RCGTG motifs will align at the center of the region defined by ChIP-seq, whereas RCGTG motifs that are not binding HIF will be randomly distributed. Figure 5A shows the plot for all RCGTGs and reveals the expected alignment against a background of “noise” from the non–HIF-binding RCGTGs. When the data are restricted to RCGTG motifs that fall within DNAse1-hypersensitive sites, then a much-improved “signal-to-noise” ratio is observed (Figure 5B) consistent with the predictive power of DNAse1 hypersensitivity. Importantly, this is observed whatever the rank distance of the HIF-binding sites from the nearest TSS, indicating that normoxic DNAse1 hypersensitivity is a strong determinant of HIF-binding, even at sites far remote from promoters, and is consistent with the functional importance of these remote sites in gene regulation.
Normoxic DNAse1 hypersensitivity predicts hypoxic binding of HIF. High-stringency HIF-1-binding regions were ranked according to distance upstream/downstream of the nearest TSS (vertical axis). (A) For each HIF-1-binding region, the position of each RCGTG motif, relative to the center of the HIF-binding region, was plotted (horizontal axis). (B) The same analysis was repeated for the subset of RCGTG motifs that coincided with a normoxic DNAse1 hypersensitivity peak (ENCODE consortium dataset 1). (C) Genomic binding of HIF-1α, HIF-2α, and HIF-1β subunits at the egl nine homolog 3 gene locus. The number of reads per 100-bp window and the location of the gene together with the positions of the RCGTG motifs, and DNAse1-hypersensitive RCGTG (DNAse1-hypersensitive site RCGTG) motifs are shown for comparison.
Normoxic DNAse1 hypersensitivity predicts hypoxic binding of HIF. High-stringency HIF-1-binding regions were ranked according to distance upstream/downstream of the nearest TSS (vertical axis). (A) For each HIF-1-binding region, the position of each RCGTG motif, relative to the center of the HIF-binding region, was plotted (horizontal axis). (B) The same analysis was repeated for the subset of RCGTG motifs that coincided with a normoxic DNAse1 hypersensitivity peak (ENCODE consortium dataset 1). (C) Genomic binding of HIF-1α, HIF-2α, and HIF-1β subunits at the egl nine homolog 3 gene locus. The number of reads per 100-bp window and the location of the gene together with the positions of the RCGTG motifs, and DNAse1-hypersensitive RCGTG (DNAse1-hypersensitive site RCGTG) motifs are shown for comparison.
The data therefore suggest that genome-wide analyses of DNAse1 hypersensitivity may be used to assist in the identification of functionally important RCGTG motifs at specific loci. For example, only one of the many RCGTG motifs at the egl nine homolog 3 locus coincides with a normoxic DNAse1 hypersensitivity peak. This previously described hypoxia response element44,45 is the only one that binds HIF in all 3 datasets (Figure 5C).
Thus, accessibility as indicated by normoxic DNAse1 hypersensitivity is a major factor in predicting HIF binding to an RCGTG motif. However, despite this, the proportion of hypersensitive RCGTG motifs that bind HIF remains low (1.1% of hypersensitive RCGTGs bind HIF-1 and 0.9% bind HIF-2), indicating that other epigenetic mechanisms also probably contribute to the definition of a HIF-binding site.
Discussion
We describe the first use of ChIP coupled to next-generation high-throughput sequencing to define HIF-α- and HIF-1β-binding sites across the whole genome. This technique enables true genome-wide coverage and more precise localization of HIF-binding sites than has previously been possible. The use of MCF-7 cells has allowed comparison with previously reported genome-wide studies of hypoxia-inducible gene expression,15 and high-resolution mapping of DNAse1 hypersensitivity sites in the same cell line, and has provided the most complete pan-genomic analysis of the transcriptional response to hypoxia to date.
The high degree of concordance between HIF-α binding in cells exposed to DMOG, and hypoxic HIF-1β binding indicates that both stimuli produce comparable genome-wide patterns of HIF DNA-binding. Furthermore, the high degree of concordance observed between HIF-α and HIF-1β binding (supplemental Figure 3) implies that either HIF-α can only bind DNA as a heterodimer with HIF-1β or that if HIF-α is able to bind DNA with an alternative partner (eg, as part of an Sp-1-binding complex38 ) that it does so at the same sites as the αβ-heterodimer, with much lower affinity/occupancy, or at too few sites to be identified in a genome-wide analysis. Conversely, it also indicates that, in these cells under these conditions, the large majority of HIF-1β is bound as a HIF-α/β heterodimer.
Using highly stringent criteria, more than 500 HIF-binding sites were identified across the genome. Many of these sites were outside of regions covered by promoter arrays and were remote from core promoters. Indeed, HIF-binding sites were observed many hundreds of kilobases distant from any documented promoter. Similar enrichment was observed for both the core HIF-binding RCGTG motif (Figure 4) and for DNAse1-hypersensitive sites (supplemental Figure 10) among these promoter-distant binding sites, compared with promoter-proximal sites, whereas CpG islands were enriched only at promoter proximal sites (supplemental Figure 10). Together with independent experimental validation by quantitative PCR, these results indicate that the identification of promoter-distant sites was accurate and probably functionally relevant.
Nearest neighbor analysis of HIF-binding and HIF-regulated genes15 revealed a distribution of distances comparable with randomly chosen control loci. This lack of clustering of both HIF-binding and particularly HIF-regulated gene loci provides evidence against widespread operon-like regulation of genes or genomic clustering of HIF transcriptional targets. However, we cannot exclude the possibility that distant chromatin elements cluster in 3-dimensional space to form hypoxia-inducible “transcription factories.”
Ingenuity pathways analysis (Figure 1F) demonstrated that many canonical pathways are targeted by HIF with various bias toward either HIF-1 or HIF-2 binding. Consistent with previously published work, pathways involved in carbohydrate metabolism are principally targeted by HIF-1, whereas genes with a role in Oct4-regulated stem cell pluripotency predominantly bind HIF-2.46 Interestingly, pathway-specific association of particular HIF isoforms was observed even though siRNA studies have suggested that in MCF7 cells HIF-2 appears to have rather little transcriptional activity,15 suggesting that isoform-specific patterns of HIF binding might extend across different cells and is consistent with post–DNA-binding mechanisms limiting transcriptional activity.17,45
The ability to interrogate pan-genomic patterns of DNA binding for HIF, which is universally expressed in human cells, raises a question as to what extent patterns of binding are cell-type specific. Previous studies of HIF-binding to promoter regions have revealed in the region of 60% concordance between U87 and HepG2 cells.28 To gain early insights into this question at a pan-genomic level, we compared sites identified in the current work with an unpublished analysis in 786-0 renal carcinoma cells (J.S., D.R.M., unpublished observations, March 2011). This revealed similar concordance both within promoters and at promoter-distant sites (overlap being observed at 42% and 36%, respectively).
Correlation of HIF-binding data with functional studies of the HIF transcriptional response using GSEA revealed the existence of robust statistical associations with patterns of hypoxia-inducible gene expression even over genomic distances in excess of 100 kb. Indeed, these studies indicated that for HIF-1 binding, the effects on hypoxia-inducible gene expression were qualitatively and quantitatively similar, irrespective of the position of HIF-1 binding at the gene locus, irrespective of distance at least up to 100 kb, and irrespective of the presence of an intervening gene that was not responsive to the HIF pathway. This is compatible with a model in which most or all of the promoter-distant HIF-binding sites are hypoxia-inducible enhancers that connect with transcriptional complexes at the promoters of target genes by DNA looping over potentially very large distances and thus exert similar effects on the transcriptional activity of these complexes irrespective of physical distance along the chromosome. Further work will be required to define physical interactions at individual loci and the temporal sequence of events governing HIF binding and the formation of transcriptional complexes after induction of HIF by hypoxia. Interestingly, in pan-genomic studies of RNA Pol2 binding in hypoxic cells, we have commonly observed a hypoxia-inducible association of RNA Pol2 with these loci, but no evidence of previously unannotated transcripts, consistent with their physical association with distant promoters (J.S., D.R.M., unpublished observations, March 2011).
The current ChIP-seq analysis confirms and extends observations on the architecture of the hypoxia-inducible response that have been derived from ChIP-chip analyses using tiled promoter arrays.27-30 In particular, even when promoter-distant sites are included in the analyses, we could find no evidence of an association with the loci of transcripts that were negatively regulated by HIF. This confirms that down-regulation of transcript abundance by the HIF pathways is very largely, or even completely, indirect with respect to the binding of HIF to DNA.
The improved accuracy and unbiased ascertainment of HIF-binding across the genome also permitted significant refinements to be made to the HIF consensus. These studies revealed preferences at a number of sites outside the RCGTG consensus, including the relative exclusion of the E-box motif CACGTG, suggesting that competitive binding by E-box proteins may exclude HIF from such sites even in hypoxic cells.
Notably, it was not possible to distinguish HIF-1- from HIF-2-binding motif preferences, even for residues outside the RCGTG consensus, although HIF-2-binding sites were differently distributed from HIF-1-binding sites, being substantially more prevalent at sites remote from promoters. This would suggest that such motifs, if they exist, are either too pleiotropic or too distant to detect in motif analysis of HIF-binding regions or that nonsequence defined, “epigenetic” chromatin markers contribute to the differential binding of the 2 isoforms.
Compared with some other transcription factors that have been analyzed at the pan-genomic level, the number of high-stringency HIF-binding sites defined across the genome was relatively modest.47 This contrasts with relatively promiscuous primary sequence preferences for HIF binding, again suggesting that epigenetic processes must determine the binding of HIF to DNA and may be particularly important in constraining the transcriptional response to hypoxia.
Because HIF-α proteins are reduced by proteolysis to very low levels in normoxia, our finding that the majority of HIF-binding sites coincided with normoxic DNAse1 hypersensitivity sites was somewhat surprising and argues against widespread opening of chromatin conformation by HIF binding or hypoxia itself. This finding is, however, consistent with observations that the HIF-binding site at the erythropoietin locus corresponded to a normoxic DNAse1-hypersensitive region,48 that chromatin accessibility can predetermine glucocorticoid receptor-binding patterns,49 and that the pattern of HIF-binding at promoters correlates with markers of normoxic transcriptional activity at these loci.28
However, many DNAse-hypersensitive motifs both close to and distant from actively transcribed genes did not bind HIF. This suggests that additional epigenetic factors determining the distribution of these enhancer sites in different cellular contexts probably have major influence on the transcriptional response to hypoxia. Given the importance of HIF to oxygen-regulated physiology and to the pathophysiology of disease, including cancer and anemia, identification of the epigenetic factors regulating these and other processes that constrain the binding of HIF to discrete sites will be important in understanding the pathophysiology and therapeutic tractability of this highly pleiotropic pathway. Our results set a framework for such analyses.
Datasets are available at the following Web sites: ChIP-seq data: http://www.ncbi.nlm.gov/geo/query/acc.cgi.?acc=GSE28352; Expression array data, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3188; and ENCODE Digital DNAse1 hypersensitivity data, http://genome.ucsc.edu/cgi-bin/hgTables.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Wellcome Trust, Cancer Research United Kingdom, and the Higher Education Funding Council for England.
Wellcome Trust
Authorship
Contribution: D.R.M., J.S., C.W.P., and P.J.R. designed the research; J.S. and D.R.M. performed the experiments; D.R.M., S.O., and J.R. analyzed and interpreted the data and performed bioinformatic analyses; and D.R.M., J.S., C.W.P., and P.J.R. wrote the manuscript.
Conflict-of-interest disclosure: P.J.R. and C.W.P. are scientific cofounders of ReOx Ltd. The remaining authors declare no competing financial interests.
Correspondence: David R. Mole, The Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, OX3 7BN, United Kingdom; e-mail: drmole@well.ox.ac.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal