Abstract
In mammalian nuclei, a select number of tissue-specific gene loci exhibit broadly distributed patterns of histone modifications, such as histone hyperacetylation, that are normally associated with active gene promoters. Previously, we characterized such hyperacetylated domains within mammalian β-globin gene loci, and determined that within the murine locus, neither the β-globin locus control region nor the gene promoters were required for domain formation. Here, we identify a developmentally specific erythroid enhancer, hypersensitive site-embryonic 1 (HS-E1), located within the embryonic β-globin domain in mouse, which is homologous to a region located downstream of the human embryonic ϵ-globin gene. This sequence exhibits nuclease hypersensitivity in primitive erythroid cells and acts as an enhancer in gain-of-function assays. Deletion of HS-E1 from the endogenous murine β-globin locus results in significant decrease in the expression of the embryonic β-globin genes and loss of the domain-wide pattern of histone hyperacetylation. The data suggest that HS-E1 is an enhancer that is uniquely required for β-like globin expression in primitive erythroid cells, and that it defines a novel class of enhancer that works in part by domain-wide modulation of chromatin structure.
Introduction
Expression of eukaryotic genes requires the modulation of chromatin structure by the transcriptional machinery. One well-studied and important component of such modulation involves the covalent modification of histones, for example by acetylation, methylation, or phosphorylation. In particular, histone acetylation is associated with transcriptional activation, and in fact hyperacetylation of promoter-proximal nucleosomes appears to be a universal feature of actively transcribed genes.1,2 Acetylation and other modifications can stabilize or destabilize the interaction of transcription factors with DNA and thus can have profound consequences on gene regulation. In this sense, the localization of such modifications to active gene promoters is logical from the standpoints of both mechanism (histone modifying activities are recruited to gene promoters by other factors) and function (modification of promoter-proximal nucleosomes stabilizes the interaction of additional factors and can disrupt local chromatin structure to increase accessibility of promoter sequences).
However, at some gene loci, patterns of histone modifications associated with gene activation are not confined to gene promoters, but instead extend over broader regions of genomic DNA; we have termed these “hyperacetylated domains.”3 Examples of gene loci that exhibit such domains include a transgenic human growth hormone locus in mouse, the interferon γ locus in T-lymphocytes, the IgH and HLA-DRA loci in B lymphocytes, and the mammalian α-globin and β-globin loci in erythroid cells.3,4 Unlike promoter-proximal histone modification patterns, neither the mechanism by which a domain is formed nor the function it serves in gene regulation is immediately clear.
The β-globin gene cluster has served as a model system for the study of gene regulation for several decades. High-level, erythroid-specific β-globin gene expression requires sequences located 5′ of the genes and termed the locus control region (LCR).5 Expression from within the cluster is developmentally regulated. Initially during embryogenesis, immature primitive erythroblasts derived from the yolk sac enter the bloodstream as nucleated cells and differentiate as a semisynchronous cohort. Subsequently, definitive erythrocytes, having matured and enucleated within the fetal liver or postnatal marrow, enter the circulation. Corresponding to these changes in lineage, mammals undergo a switch in globin-gene expression on the transition from primitive to definitive erythropoiesis, and some mammals, including humans, have fetal-specific β-globin genes as well.6-8
Corresponding with this developmental-specific program of transcription within the β-globin gene cluster, large blocks of sequence encompassing the expressed genes are associated with histones that exhibit hyperacetylation and dimethylation of lysine 4 of histone H3. Thus, in primitive erythroid cells, the embryonic-specific ϵy-globin and βh1-globin genes are embedded within a single hyperacetylated domain that is 15.7 kb in extent, but, in definitive erythroid cells where these genes are normally silent, the domain is absent.9,10 Previously, we determined that the formation of this embryonic-specific hyperacetylated domain occurs independently of the LCR and the gene promoters, suggesting that the cis-acting DNA sequence determinants of domain formation reside elsewhere.11
In this study we report the discovery of a novel, evolutionarily conserved enhancer element located within the embryonic β-globin hyperacetylated domain, termed hypersensitive site-embryonic 1 (HS-E1), which is specifically active in primitive erythroid cells. We find that upon deletion of this element from the endogenous murine β-globin locus, transcription of the embryonic ϵy-globin and βh1-globin genes, but not of the other β-globin genes, is disrupted. In addition, this decrease in gene expression is accompanied by loss of the hyperacetylated domain, although not of the domain-wide histone methylation pattern. Our results define a novel, conserved stage-specific enhancer within mammalian β-globin gene loci, provide a direct link between hyperacetylated domain formation and β-globin gene activation, and reveal an unexpected complexity to the spectrum of histone modifications that are associated with the active β-globin domain.
Methods
Cell culture
Cell lines were maintained at 37°C in a CO2-humidified atmosphere. K562 cells were cultured in Iscove modified Dulbecco medium with 10% fetal bovine serum (FBS), 1% Glutamax (Invitrogen), and 1% penicillin/streptomycin (pen/strep). HeLa and MEL cells were cultured in Dulbecco modified Eagle medium containing 10% FBS, 1% Glutamax, and 1% pen/strep.
Collection of peripheral blood and fetal liver
Mice were mated overnight and the vaginal plug verified the next morning, indicating E0.3. At E8.5, E12.5, and E14.5 (E15.5 for DNase I–hypersensitive site analysis), pregnant mice were killed by cervical dislocation for collection of embryonic blood and fetal liver.7,10 Individual embryos/fetuses were dissected and used for ChIP, mRNA expression, and genotyping. Animal experiments were approved by the University of Rochester Committee on Animal Resources.
DNase I–hypersensitive site assay
Nuclei were harvested as previously described.9 Briefly, cells were harvested, washed once with phosphate-buffered saline (PBS), resuspended in a hypertonic buffer, and dounce-homogenized. Nuclei were treated with various concentrations of DNase I (Worthington) and digestion verified by agarose gel electrophoresis. Equal quantities of DNase I–treated nuclei were digested for 1 hour with the restriction enzyme EcoRV (New England Biolabs) to fragment the chromatin further. qPCR primers amplified ∼ 250- to 300-bp fragments in regions previously determined to be DNase I–hypersensitive sites (HSs; HS2 of the LCR, the β1-globin promoter in mouse, and the ϵ-globin promoter in human), HS-E1, and an intergenic sequence as a negative control. Primer sequences are available on request. PCR was performed using iQ SYBR Green supermix (Bio-Rad) and detected using the MyiQ Single Color Real Time PCR detection system and iCycler (Bio-Rad). Loss of fluorescent detection with increasing DNase I concentrations was interpreted as hypersensitivity.
Luciferase assay
Firefly luciferase constructs were cloned using the pGL3-Basic vector as a backbone. The ϵy-globin and β1-globin promoters were cloned upstream, and HS2 and HS-E1 were cloned downstream of the luciferase gene. These inserts were derived from the murine β-globin locus (D allele) with the following coordinates (+1 set as the transcription start site for ϵy): (1) ϵyPr, −635-+54; (2) β1Pr, +28 385-+29 052; (3) HS2, −10 414-−9259; and (4) HS-E1, +7481-+8317. MEL745a cells were transfected with firefly luciferase constructs and a Renilla luciferase vector as a transfection control using Lipofectamine LTX (Invitrogen) according to manufacturer instructions, and the cells were harvested after 48 hours. Luminescence was detected using the Dual Luciferase Kit (Promega) and a Lumat LB 9507 luminometer (Berthold Tech).
Colony assay
Colony assays were performed as previously described,9 with modifications. Vectors consisted of the human γ-globin gene promoter driving neomycin resistance (neo) expression.9 HS2 and HS-E1 were cloned downstream of the neo cassette. Vectors were transfected into K562 cells using the reagents described above. Cells were placed under G418 selection (Cellgro; 900 μg/mL) 48 hours after transfection and plated at 10 000 cells/well in 24-well plates. Colonies were counted after 10 to 14 days. All colony counts were normalized to the average number of colonies obtained with the promoter-only construct.
Targeting vector
The targeting vector for HS-E1 deletion (pHS-E1-Neo) consisted of a 3.78-kb SmaI fragment located 3′ of the ϵy-globin gene, including the sequence for βh0-globin, as the 5′ homology arm, and a loxP site–flanked selectable marker (phosphoglycerate kinase I gene promoter driving the neomycin resistance gene). The 3′ homology arm consisted of a 2.66-kb NheI fragment located 5′ of the βh1-globin gene. The parent vector, pGEM-DTA, consists of the negative selection marker DT-A in a pGEM-3 backbone.12 Homologous recombination resulted in the replacement with the selectable marker of sequences located from coordinates +7500 to +8231 (ie HS-E1), with the transcription start site for the ϵy-globin gene (the first A in the sequence GTACGTACTTGCTTCTG) designated as coordinate +1. The generation of HS-E1 substitution and deletion mice was carried out as previously described.12 The selectable marker used to replace the deleted region after homologous recombination was removed by mating mice harboring the substitution with CMV-Cre transgenic mice.13
Embryo/fetus genotype analysis
Embryos and fetuses were digested overnight at 55°C using direct PCR (tail; Viagen) containing 0.5 mg/mL proteinase K (Promega) and the following morning transferred to 85°C for 50 minutes. The undigested material was removed by centrifugation at 15 700g for 1 minute. Two microliters of the supernatant was amplified using HotStarTaq (QIAGEN) for genotyping using primers that either spanned the deletion or were located within it, to distinguish wild-type from mutants. In addition, primers were designed to amplify neomycin to verify excision of the selectable marker by cre-mediated recombination.
mRNA expression analysis
RNA was isolated using TRIzol (Invitrogen) according to manufacturer instructions. One microgram of RNA was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad). Reverse transcription was also performed without enzyme as a negative control. Primers amplified ϵy-globin, βh0-globin, βh1-globin, β1-globin, and β2-globin cDNAs and ribosomal 18S as a control. Levels of globin cDNA measured by quanitative RT-PCR were converted to the corresponding percentage of 18S expression in the graphs. Primer sequences were published previously10 and are available on request. cDNA was amplified using iQ SYBR Green supermix (Bio-Rad) and detected using the MyiQ Single Color Real Time PCR detection system and iCycler (Bio-Rad).
ChIP assay
The chromatin immunoprecipitation (ChIP) assay was performed as previously described11 with some alterations. Formaldehyde–cross-inked cells were either flash-frozen in liquid N2 pending genotyping analysis or immediately lysed, sonicated, subjected to immunoprecipitation (IP), and DNA-isolated. Assays were performed on individual embryos, resulting in low cell numbers, so the following alterations were made: sonications were performed using a Misonix Sonicator 3000 with a 1/16-inch microtip at an output of 18-22 W for 6 bursts of 15 seconds each. IP was performed using ∼ 2 μg of antibody preconjugated to protein A or G Dynabeads (Invitrogen) for ∼ 2 hours. Antibody/chromatin/bead complexes were washed and the DNA isolated using Chelex-100 (Bio-Rad) described previously.14 Antibodies used for IP were (1) normal rabbit IgG (Millipore) as a control, (2) antihistone H3 acetylated at K9 and/or K16 (Millipore), (3) antihistone H3 dimethylated at K4 (Millipore and Abcam), (4) antihistone H3 dimethylated at K79 (Abcam), and (5) anti-RNA Pol II (Santa Cruz Biotechnology). Analysis was performed using qPCR as above. Primers were designed to amplify regions within the murine β-globin locus, and sequences within loci that are inactive in erythroid cells as controls, including amylase, protamine, and T-cell receptor β-chain. All primers used were described previously.10 Enrichments were calculated by using ΔCt to determine the ratio between amplification from IP and input DNA for a given test sequence, then dividing this by the average of the same ratio for the inactive controls. Error bars represent standard error of the mean (SEM) from a minimum of 3 independent ChIP experiments.
Statistical analysis
One-way analysis of variance (ANOVA) incorporated the number of data points (n), mean, and standard deviation to determine significance (P value).
Results
A developmental stage–specific hypersensitive site within the embryonic β-globin domain
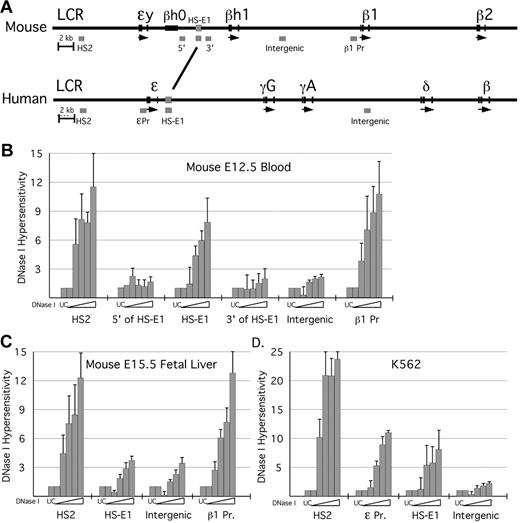
Based in part on the phenotype of the LCR deletion, we hypothesize that sequences required for formation of hyperacetylated domains within the β-globin locus will reside within them or at their boundaries. Within the domain that encompasses the murine ϵy- and βh1-globin genes in primitive erythroid cells, we have shown that the gene promoters are not required for domain formation.11 Outside of the promoters and genes themselves, we have identified a region of approximately 700 bp located between the ϵy-globin and βh1-globin genes that has significant sequence homology (53.1% identity) with a region located immediately downstream of the human ϵ-globin gene (Figure 1A; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This is the only intergenic region within the embryonic domain that exhibits such sequence conservation.
DNase I HS analysis in erythroid cells. (A) Schematic of the mouse and human β-globin loci, to scale. Positions of PCR amplimers used in the DNaseI assay are indicated as gray bars beneath each locus. The region of homology between mouse and human to which HS-E1 maps is marked by the line between the 2 loci (see also supplemental Figure 1). HS analysis was performed for the murine β-globin locus using primary erythroid cells harvested from (B) E12.5 peripheral blood and (C) E15.5 fetal liver. HS analysis was also performed for the human β-globin locus using the K562 cell line (D). Analysis was performed using qPCR and the loss in fluorescent signal indicated a decrease in amplification product. This value was interpreted as an increase in hypersensitivity, and the inverse of this value was represented heregraphically.
DNase I HS analysis in erythroid cells. (A) Schematic of the mouse and human β-globin loci, to scale. Positions of PCR amplimers used in the DNaseI assay are indicated as gray bars beneath each locus. The region of homology between mouse and human to which HS-E1 maps is marked by the line between the 2 loci (see also supplemental Figure 1). HS analysis was performed for the murine β-globin locus using primary erythroid cells harvested from (B) E12.5 peripheral blood and (C) E15.5 fetal liver. HS analysis was also performed for the human β-globin locus using the K562 cell line (D). Analysis was performed using qPCR and the loss in fluorescent signal indicated a decrease in amplification product. This value was interpreted as an increase in hypersensitivity, and the inverse of this value was represented heregraphically.
We performed DNase I–hypersensitive site analysis on mouse primary cells derived from E12.5 peripheral blood (Figure 1B) and E15.5 fetal liver (Figure 1C). DNase I hypersensitivity is commonly thought to result from localized disruption of chromatin structure and/or from DNA bending because of the binding of transcription factors. We have modified a PCR-based assay similar to those used in previous studies15,16 to measure DNase I sensitivity over specific sequences—5′HS2 of the LCR, the conserved intergenic sequence, other intergenic sequences that neighbor it, a sequence located between the βh1-globin and β1-globin genes, and the β1-globin gene promoter—using cell populations as small as 5-10 × 106 cells. We find that in both E12.5 peripheral blood (composed of more than 90% nucleated primitive erythroid cells)7 and E15.5 fetal liver, 5′HS2 and the β1-globin promoter are hypersensitive to DNase I digestion, whereas the conserved intergenic sequence is hypersensitive to digestion solely in the primitive erythroid lineage. We therefore term this sequence hypersensitive site-embryonic 1 (HS-E1).
To examine nuclease sensitivity at the corresponding sequences within the human β-globin locus, we have performed the DNase I assay using the human erythroleukemia cell line K562. In K562 cells, which express the embryonic ϵ-globin gene, we find DNase I hypersensitivity at 5′HS2 of the LCR, the ϵ-globin promoter, and the human equivalent of HS-E1, but not at a distinct, nonconserved intergenic sequence (Figure 1D). In addition, data from a genome-wide DNase-seq assay performed in K562 cells indicate that a DNaseI HS localizes to this region in the human locus.17
HS-E1 functions as an enhancer
To determine whether HS-E1 acts as an enhancer, we constructed reporter plasmids in which either the ϵy-globin or the β1-globin promoter was used to drive expression of a firefly luciferase gene, and then placed HS-E1 or 5′HS2 of the LCR downstream of the gene (Figure 2A). We find that linking HS-E1 to either expression construct results in an increase in luciferase expression of almost 5-fold when transiently transfected into MEL cells (Figure 2A). This is essentially identical to the increase mediated by 5′HS2 of the LCR in this assay. Thus, HS-E1 functions as an enhancer in erythroid cells.
Assays for enhancer function of HS-E1. (A) Transient reporter assay. Reporter constructs used in the transient assay are illustrated at the left. The bar graph shows relative levels of luciferase activity measured using the indicated constructs. (B) Colony assay. The selectable marker constructs used in the colony assay are illustrated to the left. The bar graph shows colony numbers obtained using the indicated constructs, with the average colony number for pγneo normalized to 1.0. Each bar in the graph represents the average of 12 replicates of the experiment. All error bars represent standard error of the mean.
Assays for enhancer function of HS-E1. (A) Transient reporter assay. Reporter constructs used in the transient assay are illustrated at the left. The bar graph shows relative levels of luciferase activity measured using the indicated constructs. (B) Colony assay. The selectable marker constructs used in the colony assay are illustrated to the left. The bar graph shows colony numbers obtained using the indicated constructs, with the average colony number for pγneo normalized to 1.0. Each bar in the graph represents the average of 12 replicates of the experiment. All error bars represent standard error of the mean.
We have also performed stable transfection (colony) assays in human K562 cells to examine the activity of HS-E1 within a chromosomal context. This assay measures the ability of a randomly integrated transgene construct to achieve levels of expression of a selectable marker sufficient to confer drug resistance. For example, linking 5′HS2 to the expression cassette results in an approximately 4-fold increase in the number of colonies obtained, reflecting a similar increase in the number of integration sites rendered permissive for expression (Figure 2B). HS-E1 mediates a smaller but still statistically significant increase in colony formation, consistent with the enhancer function demonstrated by the transient transfection assay.
HS-E1 is required for normal embryonic β-globin gene expression
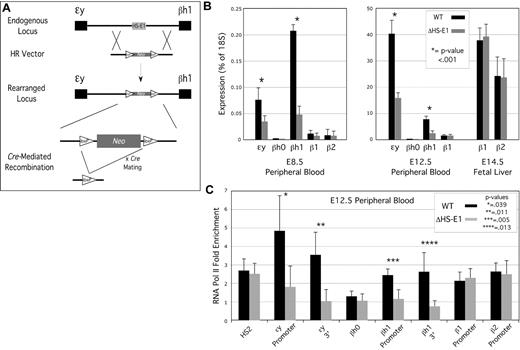
To evaluate the function of HS-E1 within the context of the native β-globin locus, we deleted it from the mouse genome using the strategy outlined in Figure 3A. The conserved region was replaced with a selectable marker by homologous recombination in embryonic stem (ES) cells, which were then used to derive a mouse line harboring the replacement. The selectable marker, flanked by lox recombinase sites, was then removed by crossing this mouse line with a Cre-expressing line. Mice heterozygous for the deletion were mated and embryos taken at embryonic day 8.5 (E8.5), E12.5, or E14.5 to harvest peripheral blood (from E8.5 or E12.5, representing the primitive erythroid lineage) or fetal liver (from E14.5, representing the definitive erythroid lineage). Genotyping, mRNA isolation, and ChIP analysis were performed using cells derived from individual embryos. Timed matings were performed using mice heterozygous for the deletion, so comparisons between homozygous mutant and homozygous wild-type embryos could be made between littermates.
Effect of deletion of HS-E1 from the endogenous β-globin locus on gene expression and RNA Pol II association. (A) Homologous recombination strategy for the deletion of HS-E1. (B) β-globin mRNA expression analysis for E8.5 and E12.5 peripheral blood, and for E14.5 fetal liver, derived from wild-type embryos and embryos homozygous for the deletion of HS-E1. All values are derived from averages of at least 6 individual embryos. (C) RNA Pol II ChIP analysis in E12.5 peripheral blood. Enrichment for RNA polymerase II association with the indicated sequences is calculated using inactive control loci as background controls (ie, 1.0).
Effect of deletion of HS-E1 from the endogenous β-globin locus on gene expression and RNA Pol II association. (A) Homologous recombination strategy for the deletion of HS-E1. (B) β-globin mRNA expression analysis for E8.5 and E12.5 peripheral blood, and for E14.5 fetal liver, derived from wild-type embryos and embryos homozygous for the deletion of HS-E1. All values are derived from averages of at least 6 individual embryos. (C) RNA Pol II ChIP analysis in E12.5 peripheral blood. Enrichment for RNA polymerase II association with the indicated sequences is calculated using inactive control loci as background controls (ie, 1.0).
Notably, mice homozygous for the deletion of HS-E1 are viable, and we observed no decrease in the proportion of progeny that were heterozygous or homozygous for the mutation. Cytospin preparations of primitive blood from wild-type versus mutant embryos revealed no obvious morphologic differences, except that in general primitive erythrocytes from mutant embryos appeared to be somewhat smaller (supplemental Figure 2).
Deletion of HS-E1 resulted in significant decreases in embryonic ϵy-globin and βh1-globin mRNA levels in primitive erythroid cells at either E8.5 or E12.5 (Figure 3B), with expression of both genes reduced to 20%-40% of that observed in wild-type littermates. The deletion had no effect on the βh0-globin, or adult β1-globin or β2-globin genes, which are active at low levels in these cells. It has been shown previously that β-globin gene expression in primitive erythrocytes undergoes a process termed maturational switching, in which levels of βh1-globin mRNA are relatively high at early stages (such as E8.5), whereas later in embryonic development, more mature primitive erythrocytes express predominantly ϵy-globin.10 The effects of the HS-E1 deletion are roughly equivalent in magnitude at E8.5 and E12.5 (Figure 3B), and so this sequence does not appear to have a significant effect on this switching phenomenon.
We observed no significant effects on expression of the adult β1-globin or β2-globin genes in definitive erythroid cells derived from E14.5 fetal liver (Figure 3B), consistent with an activity for HS-E1 that is restricted to the primitive erythroid lineage and to the embryonic β-globin genes.
We also performed ChIP analysis in E12.5 peripheral blood to assess the effect of the HS-E1 deletion on RNA Pol II occupancy. We observe a significant decrease in RNA Pol II detectable by ChIP at the ϵy-globin and βh1-globin genes (Figure 3C). The magnitudes of the decreases observed at the promoters and the 3′ ends (third exons) of the genes are similar, which suggests that the loss of RNA pol II association reflects a loss of RNA pol II recruitment to the promoters, as opposed to a subsequent step such as transcriptional elongation. We observe no significant difference in RNA Pol II enrichments at 5′ HS2 of the LCR or the promoters of the βh0-globin, β1-globin, or β2-globin promoters in E12.5 peripheral blood.
Deletion of HS-E1 results in a loss of domain-wide hyperacetylation
We originally identified HS-E1 as the lone conserved intergenic sequence within the embryonic β-globin hyperacetylated domain, and thus as a logical candidate for a cis-acting DNA sequence required for domain formation. We therefore performed ChIP analysis of individual embryos harboring the deletion of HS-E1 to determine the effect of this deletion on the hyperacetylated domain. We have previously defined the embryonic domain as extending from the ϵy-globin gene promoter to a region between 15.7-16.7 kb 3′ of this. This definition stems from two observations: (1) the LCR deletion affects histone acetylation levels determined using probes located from 3.7 to 1.2 kb 5′ of the ϵy-globin promoter, but did not affect any sequences further 3′11 ; and (2) a sharp transition from high levels of enrichment to background levels of histone acetylation normally occurs between probes located at 15.7 and 16.7 kb 3′ of the ϵy-globin promoter.
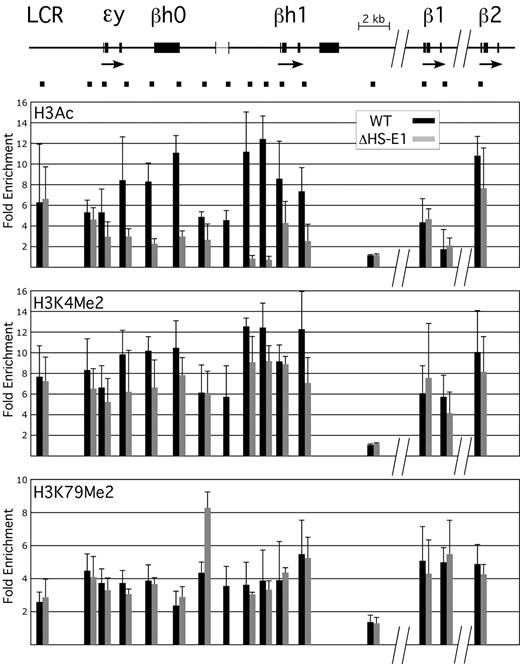
In embryos harboring the HS-E1 deletion, we find a significant decrease in histone H3 acetylation (H3Ac) extending from the ϵy-globin promoter to the 3′ end of the βh1-globin gene in E12.5 peripheral blood (Figure 4A); this is the entirety of the embryonic hyperacetylated domain. Levels of H3Ac enrichment compared with control inactive gene loci decrease to 2-fold or lower throughout the domain, which is similar to the pattern observed across this region in definitive erythroid cells, where these genes are silent.9-11 Thus, HS-E1 is required for normal domain formation across this region in primitive erythroid cells.
Effect of deletion of HS-E1 on domain-wide histone modifications. A schematic of the murine β-globin locus is shown at the top, to scale but with the indicated discontinuities. The bracketed region between the ϵy-globin and βh1-globin genes shows the location of the HS-E1 deletion. Black bars beneath the locus correspond to the positions of amplimers used for ChIP analysis, and the corresponding ChIP data are shown directly below each amplimer in the bar graphs. Values in the bar graphs represent enrichments observed for the indicated histone modifications relative to inactive control loci, as measured in E12.5 primitive erythroid cells from wild-type mice (black) or from mice homozygous for the deletion of HS-E1 (gray). Note that one amplimer lies within the deleted region.
Effect of deletion of HS-E1 on domain-wide histone modifications. A schematic of the murine β-globin locus is shown at the top, to scale but with the indicated discontinuities. The bracketed region between the ϵy-globin and βh1-globin genes shows the location of the HS-E1 deletion. Black bars beneath the locus correspond to the positions of amplimers used for ChIP analysis, and the corresponding ChIP data are shown directly below each amplimer in the bar graphs. Values in the bar graphs represent enrichments observed for the indicated histone modifications relative to inactive control loci, as measured in E12.5 primitive erythroid cells from wild-type mice (black) or from mice homozygous for the deletion of HS-E1 (gray). Note that one amplimer lies within the deleted region.
Surprisingly, however, we observe a more modest decrease in another modification associated with the hyperacetylated domain, dimethylation of histone H3 lysine 4 (H3K4Me2) (Figure 4B). Although the overall pattern of H3K4Me2 is affected domain-wide, the magnitude of the decrease varies from 0%-40%, depending on the probe examined. We also find that another modification associated with the domain, dimethylation of histone H3 lysine 79 (H3K79Me2), is affected minimally, if at all, with the exception of a single region located immediately adjacent to the deletion (Figure 4C), which actually appears to become more highly enriched. The results indicate that although HS-E1 is required for the establishment of H3Ac across the embryonic domain, it is not the only contributing factor to the pattern of histone methylation within this region.
Discussion
In this study, we have defined a novel, stage-specific enhancer element, termed HS-E1, within the mammalian β-globin locus. This enhancer is located within the hyperacetylated domain that encompasses the embryonic ϵy-globin and βh1-globin genes in primitive erythroid cells, and is required for normal domain formation, although not for domain-wide patterns of histone methylation. HS-E1 is also required for normal levels of embryonic β-globin gene expression, suggesting a link between elevated domain-wide levels of histone acetylation and gene activation.
HS-E1 is marked by DNaseI hypersensitivity in primitive but not definitive erythroid cells, and the effects on gene expression of its deletion from the endogenous murine β-globin locus are limited to the primitive erythroid lineage, clearly defining this element as a developmental stage-specific enhancer. Notably, however, HS-E1 exhibits enhancer activity in luciferase reporter assays using MEL cells, which express solely adult β-globin genes and are generally used as a model for definitive erythropoiesis. This suggests that within the endogenous locus HS-E1 is inactivated in definitive erythroid cells by a mechanism that supercedes its enhancer activity and is absent in the artificial constructs used in the reporter assay in MEL cells. In support of this possibility, we have previously shown that deletion of the ϵy-globin and βh1-globin gene promoters resulted in the formation of the embryonic hyperacetylated domain in definitive erythroid cells.11 Thus, in definitive erythropoiesis the silencing effects of the gene promoters appear to predominate over the enhancer function of HS-E1. However, all of these effects are strictly limited to the embryonic region of the β-globin locus, because deletion of HS-E1 has no effect on expression of the adult β-globin genes, nor do the promoters appear to exert any effect on the adult genes or the β-globin LCR.
Enhancer activity of HS-E1 is demonstrated not only by the transient reporter assay, but also by the decrease in ϵy-globin and βh1-globin gene expression on deletion of HS-E1 from the endogenous locus. This decrease in gene expression is accompanied by a domain-wide loss of histone H3 acetylation as well, suggesting that the mechanism by which HS-E1 enhances β-globin gene expression involves domain-wide modulation of chromatin structure. Within the context of a native locus, the only precedent for this association comes from studies of transgenic human growth hormone loci in mice. In these studies, deletion of binding sites for the transcription factor Pit-1, within the pituitary-specific enhancer element located 15 kb 5′ of the gene promoter, resulted in complete loss of growth hormone gene expression and of a hyperacetylated domain that normally extends for 30 kb throughout the locus.18
Thus, HSI from the human growth hormone locus and HS-E1 from the mouse and human β-globin loci appear to constitute members of a novel class of enhancers whose function is associated with the formation of broad domains of histone hyperacetylation. We have previously presented evidence against models of domain formation in which multiple sequences, or all sequences, within the domain serve to recruit histone acetyltransferases independently (mass action), or in which domain-wide histone acetylation is a consequence of transcriptional elongation throughout the region by RNA polymerase II.11 The current evidence suggests that enhancers like HS-E1 serve to nucleate the formation of a histone acetyltransferase–containing complex which then spreads bidirectionally along the chromatin fiber in a manner analogous to that involved in the best available models for heterochromatin formation. Activation is then mediated by the domain-wide alteration of chromatin structure itself, or is accomplished by interactions of the spreading complex with other factors when it reaches a gene promoter; the latter model has been described previously and is termed linking19,20 or oozing.21
Deletion of HS-E1 has relatively small effects on histone methylation within the embryonic domain. We have previously defined the hyperacetylated domain as a region within which levels of histone hyperacetylation and H3K4 dimethylation are significantly elevated,3,11 and in this study we add H3K79 dimethylation to this definition as well. However, the phenotype of the HS-E1 deletion demonstrates that although these modifications comap throughout the hyperacetylated domain, they originate via different mechanisms and have different cis-acting sequence determinants. Importantly, the evidence we have presented against a role for transcriptional elongation in histone hyperacetylation within the domain does not rule out a role for elongation in domain-wide histone methylation.11 The partial decrease in histone methylation levels seen at some regions within the embryonic hyperacetylated domain on deletion of HS-E1 is, in fact, consistent with the significant but partial loss of ϵy-globin and βh1-globin gene expression. In mice harboring deletions of the ϵy-globin and βh1-globin gene promoters, however, histone methylation was not affected in primitive erythroid cells, and re-establishment of the hyperacetylated domain in definitive erythroid cells, which included H3K4Me2, was not associated with any measurable genic transcription. Thus, the domain-wide histone methylation pattern arises either from transcriptional elongation that is a consequence of nongenic or intergenic transcription originating from promoters that have not yet been defined within the locus, or alternatively from an entirely distinct mechanism.
In a previous study, we analyzed the effects on domain formation of deletion of the ϵy-globin and/or βh1-globin gene promoters from the endogenous mouse β-globin locus. Interestingly, deletion of both promoters resulted in the formation of the hyperacetylated domain over the embryonic β-globin genes in definitive erythroid cells, where these genes are normally silent and the domain normally absent.11 Notably, deletion of the ϵy-globin promoter alone resulted in formation of only a portion of this domain, extending from the ϵy-globin gene to the approximate location of HS-E1. In addition, the βh1-globin gene, but not ϵy-globin, is expressed at low levels in the initial wave of definitive erythropoiesis in early fetal liver, and this activity is associated with histone hyperacetylation over the βh1-globin but not the ϵy-globin gene.22 The available evidence, therefore, suggests that the hyperacetylated domain encompassing the embryonic β-globin genes in mouse is not monolithic, but composed of 2 regions that can be independently regulated. However, both require the presence of HS-E1.
The phenotype resulting from deletion of HS-E1 from the endogenous murine β-globin locus provides a direct link between domain-wide histone modification patterns and gene activation. Moreover, the characterization of this sequence as an evolutionarily conserved, stage-specific enhancer expands the repertoire of mechanisms involved in normal regulation of endogenous mammalian β-globin loci beyond the well-studied contributions of the gene promoters and the LCR. It remains to be seen if additional elements with similar activities are also involved in the control of gene expression within the β-globin locus at later developmental stages, and in a broader context it will be of interest to characterize the full range of gene loci that are influenced by the activity of distal regulatory elements that modulate chromatin structure on the domain scale.
The online version of this article includes a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank James Palis for critical reading of the paper and the Dartmouth Transgenic and Genetic Construct Shared Resource for assistance in generating mutant ES cells and mice.
This work was supported by National Institutes of Health grant R01 DK070687, a Burroughs Wellcome Development Award, and a Beckman Young Investigators Award (M.B.), and National Institutes of Health grant T32 GM068411 (G.F. and B.C.-R.).
National Institutes of Health
Authorship
Contribution: G.F. designed and performed research, analyzed data, and wrote the paper; B.C.-R. performed research and analyzed data; C.d.V., M.G., K.E.M., and P.D.K. performed research; J.F. and S.F. contributed vital new reagents; and M.B. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Michael Bulger, Department of Pediatrics, Center for Pediatric Biomedical Research, University of Rochester Medical Center, Box 703, 601 Elmwood Ave, Rochester, NY 14642; e-mail: Michael_bulger@urmc.rochester.edu.