Abstract
Diffuse large B-cell lymphomas (DLBCLs) can be divided into germinal-center B cell–like (GCB) and activated-B cell–like (ABC) subtypes by gene-expression profiling (GEP), with the latter showing a poorer outcome. Although this classification can be mimicked by different immunostaining algorithms, their reliability is the object of controversy. We constructed tissue microarrays with samples of 157 DLBCL patients homogeneously treated with immunochemotherapy to apply the following algorithms: Colomo (MUM1/IRF4, CD10, and BCL6 antigens), Hans (CD10, BCL6, and MUM1/IRF4), Muris (CD10 and MUM1/IRF4 plus BCL2), Choi (GCET1, MUM1/IRF4, CD10, FOXP1, and BCL6), and Tally (CD10, GCET1, MUM1/IRF4, FOXP1, and LMO2). GEP information was available in 62 cases. The proportion of misclassified cases by immunohistochemistry compared with GEP was higher when defining the GCB subset: 41%, 48%, 30%, 60%, and 40% for Colomo, Hans, Muris, Choi, and Tally, respectively. Whereas the GEP groups showed significantly different 5-year progression-free survival (76% vs 31% for GCB and activated DLBCL) and overall survival (80% vs 45%), none of the immunostaining algorithms was able to retain the prognostic impact of the groups (GCB vs non-GCB). In conclusion, stratification based on immunostaining algorithms should be used with caution in guiding therapy, even in clinical trials.
Introduction
Diffuse large B-cell lymphoma (DLBCL), although considered a single category in the World Health Organization classification, most likely includes different clinicopathologic entities difficult to separate using standard techniques.1,2 From the clinical standpoint, the introduction of immunochemotherapy in the treatment of DLBCL has dramatically improved the outcome of these patients compared with chemotherapy alone.3-8 However, a significant proportion of these patients (20%-30%) become refractory or eventually relapse.9 Therefore, the identification of factors, either biologic or clinical, that can identify patients at a higher risk is a priority. Different prognostic factors for response and survival have been described for DLBCL, but in the rituximab era, the role of these biologic prognostic factors has yet to be determined.10
DLBCLs can be divided by gene-expression profiling (GEP) studies into germinal center B cell–like (GCB) and activated B cell–like (ABC) subtypes, with the latter having a significantly poorer outcome than the GCB group.11 These molecular subtypes are associated with different outcomes, even after the introduction of immunochemotherapy.12 However, GEP techniques are not applicable to the routine clinical practice, and different approaches using immunophenotypic algorithms with small panels of biomarkers have been developed to translate the robust information from molecular studies into a routine clinical platform.13-17 Two of these algorithms were designed in the preimmunotherapy era and the other 3 were used in cohorts of patients treated with rituximab. The biomarkers used included the GCB markers CD10, BCL6, GCET1, and LMO2 and antigens related to postgerminal center (non-GCB) differentiation, such as MUM1/IRF4 and FOXP1. In addition, the categorization of DLBCL into a GCB or non-GCB subgroup was shown to be associated with clinicopathologic features of tumors and predicted patient outcome in some studies but not in others.13-23 The reasons for these contradictory results may be complex, and include difficulties in the standardization of both the staining methodologies and evaluation of the results24 and heterogeneity in the characteristics of the patients in the different studies.13-18 However, it is not clear whether any of the algorithms is superior to the others in obtaining GEP molecular information, because a comparative study using all of them has not been performed and only 3 studies have associated the immunophenotypic algorithm with the GEP molecular classification.
The aim of the present study was to compare the above-mentioned algorithms in a series of patients with DLBCL homogeneously treated with immunochemotherapy to assess their correlation with GEP data and their usefulness in predicting patient outcome.
Methods
Patients
Two-hundred eighty-seven patients were diagnosed with DLBCL from January 2002 to December 2006 in 5 institutions from the Grup per l'Estudi dels Limfomes de Catalunya i Balears (GELCAB). Patients with a recognized disease phase of follicular lymphoma or another type of indolent lymphoma with subsequent transformation into a DLBCL, as well as those with immunodeficiency-associated tumors, posttransplant lymphoproliferative disorders, and those with intravascular, central nervous system, primary effusion, or primary mediastinum lymphomas were excluded from the study. In 157 of 287 patients, the material necessary to construct a tissue microarray (TMA) and to assess the expression of the different antigens was available. These patients constituted the subjects of the present study. No significant differences were observed regarding main initial features and outcome between the 157 patients with available TMA and the remainder (data not shown). The study was approved by the Ethical Committee of the Hospital Clínic, University of Barcelona. Informed consent was obtained in accordance with the Declaration of Helsinki.
Staging measures included patient history and physical examination; blood cell counts; serum biochemistry, including lactate dehydrogenase (LDH) and β2-microglobulin (β2m) levels; chest, abdomen, and pelvis computerized tomography scans; and bone marrow biopsy. Posttherapy restaging consisted of a repetition of the previous tests and/or biopsies. Response was assessed according to conventional criteria.25
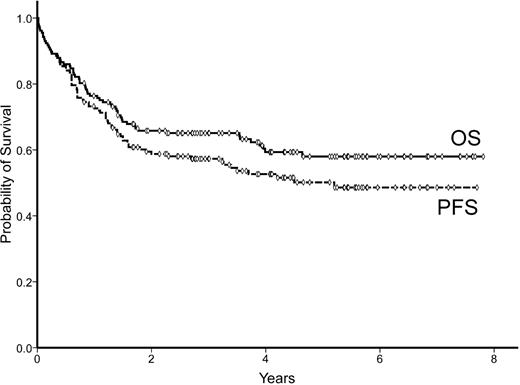
The median age of the patients was 65 years (range 17-91) and the male/female distribution was 77/80. The main characteristics of the patients are listed in Table 1. All patients received rituximab-containing chemotherapies, including regimens with anthracyclines in 133 (85%) patients. Response to treatment was as follows: 119 (76%) complete responses (CRs), 4 partial responses (2%), and 34 nonresponses (22%). The median follow-up for surviving patients was 4.3 years (range 0.8-8.6). The 5-year progression free-survival (PFS) of the series was 50% (95% CI: 42%-58%). Sixty-one patients died during the follow-up, with a 5-year overall survival (OS) of 58% (95% CI: 50%-66%). PFS and OS curves are shown in Figure 1. The main initial and evolutive variables, including the histologic parameters indicated in “Histologic review and TMA construction,” were recorded and analyzed for prognosis.
Initial and evolutive features of 157 patients with DLBCL
| Feature . | n (%) . |
|---|---|
| Age | |
| median (range) | 65 (17-91) |
| ≤ 60 y | 65 (41%) |
| Sex | |
| Female | 80 (51%) |
| Male | 77 (49%) |
| Poor performance status (ECOG-PS> 1) | 66 (42%) |
| B-symptoms | 55 (35%) |
| Extranodal involvement | 63 (40%) |
| Bone marrow involvement | 46 (29%) |
| Ann Arbor stage III-IV | 94 (60%) |
| High serum LDH | 93 (59%) |
| High serum β2-m* | 64 (47%) |
| IPI | |
| Low risk | 47 (30%) |
| Low/intermediate risk | 35 (22%) |
| High/intermediate risk | 38 (25%) |
| High risk | 37 (23%) |
| Response to therapy | |
| CR | 119 (76%) |
| Partial response | 4 (2%) |
| Nonresponse/progression | 34 (22%) |
| Feature . | n (%) . |
|---|---|
| Age | |
| median (range) | 65 (17-91) |
| ≤ 60 y | 65 (41%) |
| Sex | |
| Female | 80 (51%) |
| Male | 77 (49%) |
| Poor performance status (ECOG-PS> 1) | 66 (42%) |
| B-symptoms | 55 (35%) |
| Extranodal involvement | 63 (40%) |
| Bone marrow involvement | 46 (29%) |
| Ann Arbor stage III-IV | 94 (60%) |
| High serum LDH | 93 (59%) |
| High serum β2-m* | 64 (47%) |
| IPI | |
| Low risk | 47 (30%) |
| Low/intermediate risk | 35 (22%) |
| High/intermediate risk | 38 (25%) |
| High risk | 37 (23%) |
| Response to therapy | |
| CR | 119 (76%) |
| Partial response | 4 (2%) |
| Nonresponse/progression | 34 (22%) |
β2m levels were available in 135 cases.
ECOG-PS indicates Eastern Cooperative Oncology Group performance status.
Histologic review and TMA construction
The diagnosis of DLBCL was based on the criteria established in the World Health Organization classification.1 The diagnostic samples were reviewed by expert hematopathologists from the 5 hospitals of the GELCAB involved in the study. All immunohistochemical studies were performed after constructing TMAs, including two 1-mm representative cores of each case using a tissue arrayer (MTA I; Beecher Instruments). Standardized methods for tissue fixation (10% buffered formalin) and processing were used at all participating centers. The immunostains were performed on formalin-fixed, paraffin-embedded tissue sections using a fully automated immunostainer (Bond-Max; Vision BioSystems). All immunostains were centralized in one of the centers involved in the study and evaluated in common in a multiheaded microscope. The conditions and antibodies are shown in Table 2. CD10, BCL6 (kindly provided by Dr M. A. Piris, Centro Nacional de Investigaciones Oncológicas, Madrid, Spain), MUM1/IRF4, BCL2, FOXP1 (kindly provided by Dr A. H. Banham, John Radcliffe Hospital, Oxford, United Kingdom), GCET1 (kindly provided by Dr M. A. Piris), and LMO2 were the markers used to build the algorithms. The assessment of CD10, BCL6, MUM1/IRF4, and BCL2 was performed according to the original studies describing the algorithms and following the recent guidelines recommended for their interpretation by the Lunenburg Lymphoma Biomarker Consortium. Appropriate internal controls were necessary to evaluate the immunostains.26 FOXP1, GCET1, and LMO2 were evaluated based on previous publications.27-29 Endothelial cells were used as internal controls for FOXP1 and LMO2. For GCET1, tonsil sections were used as an external control.
Immunophenotyping study: antibodies and conditions of use
| Antigen . | Source . | Dilution . | Incubation, min . |
|---|---|---|---|
| CD20/CD79a | Dako | 1:80 | 30 |
| CD10 | Novocastra | 1:25 | 30 |
| BCL6 | Centro Nacional de Investigaciones Oncológicas | 1:25 | 60 |
| GCET1 | Centro Nacional de Investigaciones Oncológicas | 1:1 | 30 |
| HGAL | Dako | 1:500 | 60 |
| IRF8 | Santa Cruz Biotechnology Sc 13043 rabbit polyclonal | 1:100 | 60 |
| LMO2 | Santa Cruz Biotechnology Sc 65736 mouse monoclonal | 1:4000 | 60 |
| JAW1 | Santa Cruz Biotechnology Sc 11688 goat polyclonal | 1:100 | 60 |
| MUM1/IRF4 | Dako | 1:400 | 60 |
| FOXP1 | A. H. Banham | 1:80 | 30 |
| BLIMP1 | Santa Cruz Biotechnology Sc 13206 | 1:2 | 60 |
| XBP1 | Santa Cruz Biotechnology Sc 7160 | 1:400 | 120 |
| BCL2 | Dako | 1:75 | 60 |
| Antigen . | Source . | Dilution . | Incubation, min . |
|---|---|---|---|
| CD20/CD79a | Dako | 1:80 | 30 |
| CD10 | Novocastra | 1:25 | 30 |
| BCL6 | Centro Nacional de Investigaciones Oncológicas | 1:25 | 60 |
| GCET1 | Centro Nacional de Investigaciones Oncológicas | 1:1 | 30 |
| HGAL | Dako | 1:500 | 60 |
| IRF8 | Santa Cruz Biotechnology Sc 13043 rabbit polyclonal | 1:100 | 60 |
| LMO2 | Santa Cruz Biotechnology Sc 65736 mouse monoclonal | 1:4000 | 60 |
| JAW1 | Santa Cruz Biotechnology Sc 11688 goat polyclonal | 1:100 | 60 |
| MUM1/IRF4 | Dako | 1:400 | 60 |
| FOXP1 | A. H. Banham | 1:80 | 30 |
| BLIMP1 | Santa Cruz Biotechnology Sc 13206 | 1:2 | 60 |
| XBP1 | Santa Cruz Biotechnology Sc 7160 | 1:400 | 120 |
| BCL2 | Dako | 1:75 | 60 |
The diagnostic samples and immunohistochemical studies were reviewed by at least 3 expert hematopathologists blinded to the clinical details. The cores were evaluated in a semiquantitative manner, and all markers used to build the algorithms were dichotomized at a positive cutoff that depended on the algorithm used. Small biopsy samples that could not be included in a TMA and cores that dropped off of the TMAs were studied as whole-tissue sections. For individual cores with discordant results, the core with the most representative and highest number of positive cells was scored. The discrepancies between the observers were resolved by reaching consensus.
In addition to the markers included in the immunophenotypic algorithms, we also studied the new GCB markers HGAL, IRF8, and JAW1 and the non-GCB markers XBP1 and BLIMP1. These additional biomarkers were evaluated semiquantitatively and the samples were stratified into 5 groups according to the percentages of positive tumor cells: 1 (0%- ≤ 10%), 2 (10%-25%), 3 (26%-50%), 4 (51%-75%), and 5 (≥ 75%). The positive values and cutoff for these new markers was assessed based on previous studies.29-32
Immunophenotypic algorithms
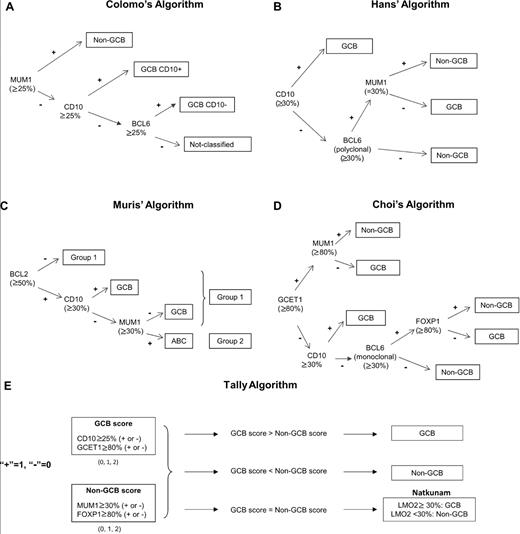
To determine the origin of GCB or non-GCB, we applied 5 different algorithms described previously using the following panel of antibodies: CD10, BCL6, MUM1/IRF4, GCET1, FOXP1, LMO2, and BCL2. The thresholds used were those described previously in the literature for each algorithm and are detailed in Figure 2. In the Colomo algorithm (Figure 2A), the non-GCB phenotype was established if MUM1/IRF4 was positive. The cases that were negative for MUM1/IRF4 and positive for CD10 were considered to be the GCB phenotype. Cases negative for MUM1/IRF4 and CD10 and positive for BCL6 were also assigned to the GCB group. Finally, cases negative for the 3 markers were considered as not classified.13 For the Hans algorithm (Figure 2B), the cases were assigned to the GCB phenotype if CD10 alone or both CD10 and BCL6 were positive. If both CD10 and BCL6 were negative, the case was considered to be of non-GCB origin.14 In the Muris algorithm (Figure 2C), cases positive for CD10 were assigned to the GCB phenotype. Cases negative for CD10 were differentiated according to MUM1/IRF4 expression into the ABC or the GCB phenotype. Thereafter, BCL2 immunostaining was used to separate 2 different prognostic groups (1 and 2).15 The 2 steps of the Muris algorithm were used to assess GCB versus ABC status and the prognostic impact of the algorithms was applied. In the Choi algorithm (Figure 2D), the cases positive for MUM1/IRF4 and/or FOXP1 or negative for CD10 and BCL6 were assigned to the non-GCB group. The cases positive for CD10, GCET1 without MUM1 expression, or positive for BCL6 without FOXP1 expression were classified as GCB.16 In the Tally algorithm (Figure 2E) described recently, the method included an equal number of GCB markers (GCET1 and CD10) and non-GCB markers (FOXP1 and MUM1/IRF4). This algorithm was constructed from the immunophenotype pair with more positive antigens. Because 2 antibodies are used for each type, the LMO2 antigen determines the phenotype (GCB or non-GCB) when the score is equal in the 2 categories.17
Five immunostaining algorithms used to assess differentiation profile (GCB vs non-GCB) in patients with DLBCL.
Five immunostaining algorithms used to assess differentiation profile (GCB vs non-GCB) in patients with DLBCL.
GEP
A subset of 62 samples with RNA extracted from fresh frozen lymph nodes was investigated using Affymetrix HG U133 plus 2.0 gene expression arrays. All gene expression array data were normalized using MAS5.0 software, and were log2 transformed. We used the Bayesian compound covariate predictor of the ABC/GCB DLBCL described previously.12,33 All samples predicted as ABC DLBCL at greater than 90% were called ABC DLBCL. The samples that showed less than 10% of probability of being called ABC DLBCL were classified as GCB DLBCL. All the other cases were considered unclassified DLBCL.
Statistical analysis
The definitions of CR, PFS, and OS were according to standard criteria.25 Categorical data were compared using the Fisher exact test and the 2-sided P value, whereas nonparametric tests were used for ordinal data. The multivariate analysis of the variables predicting response was performed with the logistic regression method.34 The actuarial survival analysis was performed according to the Kaplan-Meier method, and the curves were compared with the log-rank test.35 The multivariate analysis for survival was performed using the stepwise proportional hazards model (Cox).36
Results
Immunophenotypic profile
The immunohistochemistry expression of single antigens is detailed in Table 3. The distribution of the patients according to the differentiation phenotype (GCB vs non-GCB) after applying the different algorithms is shown in Table 4. The distribution of the cases according to the phenotype was similar for the Colomo and Hans algorithms. In the Choi and Tally algorithms, the proportion of non-GCB cases assigned was significantly higher (non-GCB vs GCB, 67% vs 33%, and 63% vs. 37%, respectively). In the Muris algorithm, the majority of the cases were allocated as the GCB phenotype (GCB vs non-GCB, 57% vs 43%). The Colomo algorithm is the only one of the 5 algorithms that considers a category of “not classified,” and 23 tumors were included in this group. Although most of these cases would be categorized in the non-GCB subtype in the other classifiers, we excluded the Colomo algorithm for the comparison between the other algorithms. Therefore, the other 4 algorithms (Hans, Muris, Choi, and Tally) were completely assessed in 135 patients, with 111 cases (82%) being allocated to the same GCB or non-GCB group.
Immunohistochemical expression of single antigens in 157 patients
| Antigen . | No. of assessable samples . | No. of positive samples (%) . |
|---|---|---|
| CD10* | 157 | 41 (26%) |
| BCL6* | 146 | 94 (64%) |
| GCET1* | 146 | 67 (46%) |
| HGAL | 139 | 61 (44%) |
| IRF8 | 147 | 42 (29%) |
| LMO2* | 143 | 60 (42%) |
| Jaw1 | 149 | 126 (85%) |
| MUM1/IRF4* | 148 | 41 (28%) |
| FOXP1* | 143 | 112 (78%) |
| BLIMP1 | 147 | 66 (45%) |
| XBP1 | 104 | 28 (27%) |
| BCL2* | 152 | 76 (50%) |
| Antigen . | No. of assessable samples . | No. of positive samples (%) . |
|---|---|---|
| CD10* | 157 | 41 (26%) |
| BCL6* | 146 | 94 (64%) |
| GCET1* | 146 | 67 (46%) |
| HGAL | 139 | 61 (44%) |
| IRF8 | 147 | 42 (29%) |
| LMO2* | 143 | 60 (42%) |
| Jaw1 | 149 | 126 (85%) |
| MUM1/IRF4* | 148 | 41 (28%) |
| FOXP1* | 143 | 112 (78%) |
| BLIMP1 | 147 | 66 (45%) |
| XBP1 | 104 | 28 (27%) |
| BCL2* | 152 | 76 (50%) |
Antigens used to build up the algorithms.
Distribution, CR rate, PFS, and OS of 157 patients with DLBCL according to immunophenotype (GCB vs non-GCB)
| Algorithm . | n (%) . | CR rate, n (%) . | 5-year PFS . | 5-year OS . |
|---|---|---|---|---|
| Colomo | ||||
| GCB | 53 (44%) | 39 (74%) | 48% | 54% |
| Non-GCB | 68 (56%) | 53 (78%) | 55% | 62% |
| Hans | ||||
| GCB | 61 (41%) | 47 (77%) | 54% | 60% |
| Non-GCB | 88 (59%) | 67 (76%) | 52% | 59% |
| Muris | ||||
| GCB | 87 (57%) | 63 (72%) | 48% | 57% |
| Non-GCB | 65 (43%) | 51 (78%) | 56% | 63% |
| Choi | ||||
| GCB | 45 (33%) | 32 (71%) | 48% | 54% |
| Non-GCB | 90 (67%) | 70 (78%) | 52% | 61% |
| Tally | ||||
| GCB | 55 (37%) | 45 (82%) | 63% | 56% |
| Non-GCB | 92 (63%) | 65 (71%) | 54% | 47% |
| Algorithm . | n (%) . | CR rate, n (%) . | 5-year PFS . | 5-year OS . |
|---|---|---|---|---|
| Colomo | ||||
| GCB | 53 (44%) | 39 (74%) | 48% | 54% |
| Non-GCB | 68 (56%) | 53 (78%) | 55% | 62% |
| Hans | ||||
| GCB | 61 (41%) | 47 (77%) | 54% | 60% |
| Non-GCB | 88 (59%) | 67 (76%) | 52% | 59% |
| Muris | ||||
| GCB | 87 (57%) | 63 (72%) | 48% | 57% |
| Non-GCB | 65 (43%) | 51 (78%) | 56% | 63% |
| Choi | ||||
| GCB | 45 (33%) | 32 (71%) | 48% | 54% |
| Non-GCB | 90 (67%) | 70 (78%) | 52% | 61% |
| Tally | ||||
| GCB | 55 (37%) | 45 (82%) | 63% | 56% |
| Non-GCB | 92 (63%) | 65 (71%) | 54% | 47% |
GEP: concordance with immunohistochemistry
GEP data were available for 62 patients: 30 cases were allocated to GCB origin (48%), 22 cases to ABC origin (36%), and 10 cases (16%) were considered unclassified. No differences in terms of initial features and outcome were observed between the patients with GEP information and those without (data not shown).
The relationship between the expression of single antigens and GEP is shown in Table 5. Only CD10 and JAW1 expression were significantly correlated with GCB origin according to GEP (P = .007 and P = .01, respectively), whereas MUM1/IRF4 expression was correlated with ABC origin (P = .02).
Correlation between GEP and different single antigens in 52 DLBCL patients with available microarray data
| Antigen . | GEP . | |||
|---|---|---|---|---|
| No. of assessable patients . | GCB, n (%) . | ABC, n (%) . | ||
| CD10* | (−) (+) | 52 | 19 (47) 11 (92) | 21 (53) 1 (8) |
| BCL6 | (−) (+) | 48 | 12 (44) 15 (71) | 15 (56) 6 (36) |
| MUM1/IRF4† | (−) (+) | 49 | 22 (68) 6 (35) | 10 (32) 11 (65) |
| IRF8 | (−) (+) | 50 | 21 (64) 7 (41) | 12 (34) 10 (59) |
| GCET1 | (−) (+) | 48 | 12 (52) 16 (64) | 11 (48) 9 (36) |
| GCET2 | (−) (+) | 50 | 12 (55) 16 (57) | 10 (45) 12 (43) |
| FOXP1 | (−) (+) | 49 | 6 (86) 21 (50) | 1 (14) 21 (50) |
| LMO2 | (−) (+) | 48 | 10 (48) 18 (67) | 11 (52) 9 (33) |
| JAW1‡ | (−) (+) | 50 | 0 (0) 29 (63) | 4 (100) 17 (37) |
| BLIMP1 | (−) (+) | 49 | 14 (54) 15 (65) | 12 (46) 8 (35) |
| XBP1 | (−) (+) | 25 | 10 (63) 7 (78) | 6 (37) 2 (22) |
| BCL2 | (−) (+) | 51 | 20 (67) 9 (43) | 10 (33) 12 (21) |
| Antigen . | GEP . | |||
|---|---|---|---|---|
| No. of assessable patients . | GCB, n (%) . | ABC, n (%) . | ||
| CD10* | (−) (+) | 52 | 19 (47) 11 (92) | 21 (53) 1 (8) |
| BCL6 | (−) (+) | 48 | 12 (44) 15 (71) | 15 (56) 6 (36) |
| MUM1/IRF4† | (−) (+) | 49 | 22 (68) 6 (35) | 10 (32) 11 (65) |
| IRF8 | (−) (+) | 50 | 21 (64) 7 (41) | 12 (34) 10 (59) |
| GCET1 | (−) (+) | 48 | 12 (52) 16 (64) | 11 (48) 9 (36) |
| GCET2 | (−) (+) | 50 | 12 (55) 16 (57) | 10 (45) 12 (43) |
| FOXP1 | (−) (+) | 49 | 6 (86) 21 (50) | 1 (14) 21 (50) |
| LMO2 | (−) (+) | 48 | 10 (48) 18 (67) | 11 (52) 9 (33) |
| JAW1‡ | (−) (+) | 50 | 0 (0) 29 (63) | 4 (100) 17 (37) |
| BLIMP1 | (−) (+) | 49 | 14 (54) 15 (65) | 12 (46) 8 (35) |
| XBP1 | (−) (+) | 25 | 10 (63) 7 (78) | 6 (37) 2 (22) |
| BCL2 | (−) (+) | 51 | 20 (67) 9 (43) | 10 (33) 12 (21) |
P = .007.
P = .01.
P = .02.
The concordance between the GEP profiles and the patterns of immunohistochemistry as assessed by the 5 algorithms is shown in Table 6. A significant correlation was observed between GEP data and the Colomo (P = .02), Hans (P = .015), Muris (P = .04), and Tally (P = .02) algorithms, whereas the correlation did not reach significance with Choi algorithm (P = .1). A higher percentage of misclassified cases was observed in the GCB than in the non-GCB subgroup. The proportion of GCB-DLBCL cases that were not correctly allocated by immunohistochemistry was 41%, 48%, 30%, 60%, and 46% for the Colomo, Hans, Muris, Choi, and Tally algorithms, respectively. Conversely, the proportion of ABC patients who were not properly assigned according to the different algorithms was 19%, 15%, 38%, 16%, and 20% for the Colomo, Hans, Muris, Choi, and Tally algorithms, respectively. The sensitivity in the GCB group was 59%, 52%, 70%, 40%, and 53% for the Colomo, Hans, Muris, Choi, and Tally algorithms, respectively; the sensitivity in the non-GCB group was 81%, 85%, 62%, 84%, and 80%, respectively.
Concordance between GEP (GCB vs ABC) and the immunohistochemistry patterns (GCB vs non-GCB) as assessed by 5 algorithms in 52 patients with available GEP information
| Algorithm . | GEP . | |
|---|---|---|
| GCB, n (%) . | ABC, n (%) . | |
| Colomo* | ||
| GCB | 13 (59%) | 3 (19%) |
| Non-GCB | 9 (41%) | 13 (81%) |
| Hans† | ||
| GCB | 15 (52%) | 3 (15%) |
| Non-GCB | 14 (48%) | 17 (85%) |
| Muris‡ | ||
| GCB | 21 (70%) | 8 (38%) |
| Non-GCB | 9 (30%) | 13 (62%) |
| Choi§ | ||
| GCB | 10 (40%) | 3 (16%) |
| Non-GCB | 15 (60%) | 16 (84%) |
| Tally¶ | ||
| GCB | 15 (54%) | 4 (20%) |
| Non-GCB | 13 (46%) | 16 (80%) |
| Algorithm . | GEP . | |
|---|---|---|
| GCB, n (%) . | ABC, n (%) . | |
| Colomo* | ||
| GCB | 13 (59%) | 3 (19%) |
| Non-GCB | 9 (41%) | 13 (81%) |
| Hans† | ||
| GCB | 15 (52%) | 3 (15%) |
| Non-GCB | 14 (48%) | 17 (85%) |
| Muris‡ | ||
| GCB | 21 (70%) | 8 (38%) |
| Non-GCB | 9 (30%) | 13 (62%) |
| Choi§ | ||
| GCB | 10 (40%) | 3 (16%) |
| Non-GCB | 15 (60%) | 16 (84%) |
| Tally¶ | ||
| GCB | 15 (54%) | 4 (20%) |
| Non-GCB | 13 (46%) | 16 (80%) |
P = .02.
P = .02.
P = .04.
P = .1.
P = .02.
Clinical significance of GEP and immunohistochemistry profiles
The CR rate of the series was 76%. CR achievement was observed more frequently in patients with absence of B-symptoms, ambulatory performance status, absence of extranodal involvement or bulky mass, normal serum β2-m, adriamycin-containing chemotherapy, and low-risk International Prognostic Index (IPI) scores. Neither single-antigen expression nor the differentiation profile as assessed by the immunohistochemistry algorithms was able to predict CR (Table 4). GEP profiles also did not predict CR.
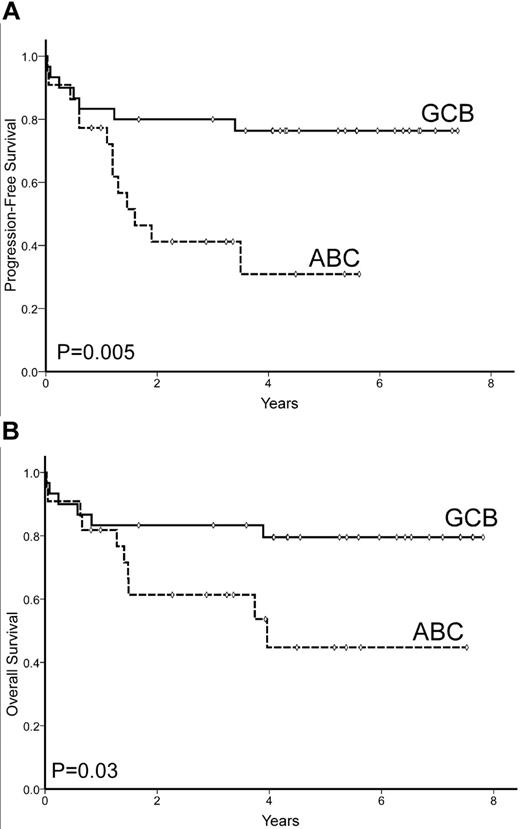
Seventy-four of 157 patients eventually progressed, including 36 of the 119 patients who reached a CR. The 5-year PFS was 50% (95% CI: 42%-58%; Figure 1). Variables with unfavorable prognostic value for PFS were: presence of B-symptoms, nonambulatory Eastern Cooperative Oncology Group performance status (ECOG-PS > 1), extranodal involvement at ≥ 1 sites, advanced Ann Arbor stage (III-IV), high serum LDH, high serum β2-m levels, and high-risk IPI score. High BCL2 protein expression also predicted poor PFS (5-year PFS 27% vs 56% for BCL2 > 75% vs ≤ 75%, respectively; P = .003). In addition, the differentiation profile (GCB vs ABC) as assessed by GEP was able to predict PFS (5-year PFS, 76% vs 31%, respectively; P = .005), as shown in Figure 3A. Conversely, the immunophenotypic profiles as assessed by the 5 different algorithms did not show significant prognostic value for PFS (Table 4).
PFS and OS of 52 patients with GEP data available according to molecular subtype (GCB vs ABC) profile.
PFS and OS of 52 patients with GEP data available according to molecular subtype (GCB vs ABC) profile.
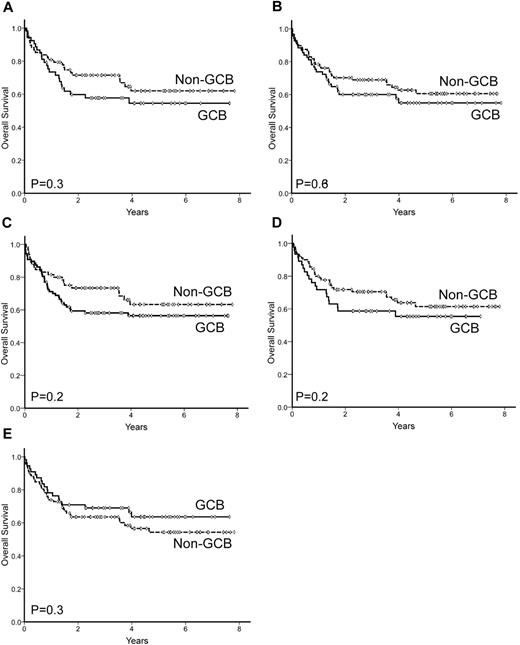
Sixty-one patients died during follow-up, with a 5-year OS of 58% (95% CI: 50%-66%; Figure 1). Unfavorable variables predicting OS were: age > 60 years, poor performance status (ECOG-PS > 1), advanced Ann Arbor stage (III-IV), high serum LDH levels, high serum β2-m level, and high-risk IPI score. Patients treated with adriamycin-containing chemotherapy also showed a significant advantage in terms of OS. Among the single antigens tested, only BCL2 overexpression (> 75%) was correlated with poor OS. Differentiation profile as assessed by GEP (GCB vs ABC) showed a significant prognostic value for OS in the subset of patients with this GEP information, with a 5-year OS of 80% vs 45%, respectively (P = .03; Figure 3B). In contrast, none of the profiles assessed by immunostaining was able to predict OS, as shown in Table 4 and Figure 4. When the 2-step Muris algorithm, which included BCL2 immunostaining, was used, the algorithm was also unable to predict OS (5-year OS 59% vs. 48% for groups 1 and 2, respectively; P = .4). The results were the same when the study was performed in 133 patients receiving strictly R-CHOP (cyclophosphamide, hydroxydaunorubicin, oncovin, and prednisone/prednisolone plus rituximab; supplemental Figures 1-2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). A multivariate analysis was performed in the 52 patients with GEP information, including differentiation profile (GCB vs ABC profile), BCL2 expression (≤ 75% vs > 75%), and IPI score. In the final model with 52 cases, differentiation profile was the most important variable in predicting OS (risk ratio 3.3; P = .04).
OS of 157 patients with DLBCL according to the differentiation profile (GCB vs non-GCB) as assessed by 5 immunohistochemistry algorithms. (A) Colomo algorithm, (B) Hans algorithm, (C) Muris algorithm, (D) Choi algorithm, and (E) Tally algorithm.
OS of 157 patients with DLBCL according to the differentiation profile (GCB vs non-GCB) as assessed by 5 immunohistochemistry algorithms. (A) Colomo algorithm, (B) Hans algorithm, (C) Muris algorithm, (D) Choi algorithm, and (E) Tally algorithm.
Discussion
Two types of DLBCL can be recognized on the basis of GEP data: GCB-DLBCL and ABC-DLBCL. These groups have a different cell origin and a distinct clinical behavior, with patients carrying an ABC-DLBCL tumor showing a poorer outcome than patients with a GCB tumor.11 This fact, originally described in the pre-rituximab era, has been recently confirmed in patients receiving immunochemotherapy.12 In addition to the prognostic interest, the molecular classification might be useful in selecting treatments that could be active only in specific subtypes of DLBCL, such as the effective use of bortezomib in ABC-DLBCLs.37-39 Therefore, GEP information is not only of academic interest, but could also be an important decision element in the management of the patients in the near future. Unfortunately, GEP assessment using microarrays is not feasible in routine clinical practice. For this reason, different attempts have been made at capturing the prognostic categorization of GEP information using a more friendly technical approach such as immunohistochemistry. This method can be performed in archival, formalin-fixed, paraffin-embedded tissues and is feasible at any pathology laboratory. Different algorithms have been created combining the expression of well-known antigens, including the ones analyzed in the present study (Colomo, Hans, Muris, Choi, and Tally; Figure 2).13-17 The real value of these algorithms as a surrogate for GEP data and to define the prognosis of patients with DLBCL is the subject of controversy and was the reason for the present study.
This study was performed in a homogeneous and representative series of patients with DLBCL diagnosed and treated with immunochemotherapy in 5 hospitals from the GELCAB. Clinical features and outcome were as expected for an unselected series of DLBCLs. TMA construction and the assessment of the different antigens were performed based on information from original descriptions.13-17 Although a significant association between GEP and immunophenotypic profiles was observed (Table 6), the positive and negative predictive values were poor for the 5 algorithms tested. In fact, 30%-50% of GCB-DLBCLs and 15%-25% of ABC-DLBCLs were incorrectly allocated by immunohistochemistry, making its use difficult in clinical practice. Furthermore, in prognostic terms, the applicability of the algorithms was poor in the current series. Although, the number of cases studied by GEP was relative low, the molecular GCB and ABC subtypes showed a clearly differentiated outcome in terms of PFS and OS. As reflected in Figure 4, none of the 5 algorithms was capable of defining groups with prognostic impact.
The current results contradict other studies supporting an excellent correlation between immunohistochemistry and GEP in terms of prognosis.14,16,17 However, some others studies agree with ours in regard to the lack of clinical significance of the immunophenotypic profiles in patients treated with immunochemotherapy.18 Several factors may account for these discrepancies, including the population studied and the methodology used. First, retrospective analysis of a heterogeneous series with different therapies may have confounded the results. However, the present study was an unbiased, population-based series, with patients homogenously treated with immunochemotherapy. On the other hand, we have included both nodal and extranodal DLBCLs. The main clinical features, including IPI scores, seem to be very similar to other publications.17 Secondly, technical shortcomings and interobserver discrepancies in the interpretations of the immunostaining slides most likely have an important role in the divergent data. The liability of immunohistochemistry has been pointed out in several studies, including the Lunenburg Lymphoma Biomarker Consortium study, which was an important step forward in standardizing the immunohistochemical studies of these lymphomas.26 In our study, we followed the Lunenburg indications to evaluate the antigens described in the consortium. However, some of the antigens used to build up the algorithms are relatively new and the evaluation criteria have not been thoroughly investigated. Therefore, we cannot rule out the possibility that differences in the evaluation of the immunostaining with the newer markers could be part of the discrepancy compared with more recently described algorithms. The low-positive values of markers considered absolutely typical of GCB (ie, GCET1 and LMO2) were positive in 64% and 67% of the cases, respectively, and for the ABC subtype (ie, FOXP1) were positive in 50% of the cases (Table 5). However, we also observed discrepant results using the more established algorithms, suggesting that the reasons for the discordance may not be merely attributable to interpretative differences. We believe that one of the most important difficulties in immunohistochemical attempts to reproduce GEP results is the limitation of capturing information obtained from complex gene-expression signatures on a large number of genes using a very small number of antigens. Therefore, to reliably reproduce GEP information and to transfer it to the clinical practice, new technologies, probably different from standard immunohistochemistry, are warranted.
In summary, in the present study, none of the 5 immunohistochemical algorithms was able to accurately predict the GEP subtype or to separate molecular groups with prognostic value. As a consequence, stratification based on immunohistochemical algorithms for guiding therapy should be viewed very cautiously, even in clinical trials.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from the Ministry of Education and Science of Spain (SAF 2008 03630-O to E.C. and FIS PI 070409 to A.L.G.) and from the Red Temática de Investigación Cooperativa en Cáncer (RTICC U.2006-RET2039-O to E.C.)
A complete list of GELCAB members appears in the online supplemental Appendix.
Authorship
Contribution: G.G.-G., T.C.-S., E.C., L.C., and A.L.-G. designed and performed research, analyzed data, and wrote the paper; F.C., J.L.M., S.S., S.M., A.V., A.M., P.J., M.P., and A.G.-H. analyzed data; E.G.-B., S.M., J.M.S., L.A., L.E., A.M.-T., and E.G. performed research; and N.V. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr A. López-Guillermo, MD, Department of Hematology, Hospital Clínic, Villarroel 170, 08036 Barcelona, Spain; e-mail: alopezg@clinic.ub.es.
References
Author notes
G.G.-G. and T.C.-S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal