Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate target mRNAs by binding to their 3′ untranslated regions. There is growing evidence that microRNA-155 (miR155) modulates gene expression in various cell types of the immune system and is a prominent player in the regulation of innate and adaptive immune responses. To define the role of miR155 in dendritic cells (DCs) we performed a detailed analysis of its expression and function in human and mouse DCs. A strong increase in miR155 expression was found to be a general and evolutionarily conserved feature associated with the activation of DCs by diverse maturation stimuli in all DC subtypes tested. Analysis of miR155-deficient DCs demonstrated that miR155 induction is required for efficient DC maturation and is critical for the ability of DCs to promote antigen-specific T-cell activation. Expression-profiling studies performed with miR155−/− DCs and DCs overexpressing miR155, combined with functional assays, revealed that the mRNA encoding the transcription factor c-Fos is a direct target of miR155. Finally, all of the phenotypic and functional defects exhibited by miR155−/− DCs could be reproduced by deregulated c-Fos expression. These results indicate that silencing of c-Fos expression by miR155 is a conserved process that is required for DC maturation and function.
Introduction
MicroRNAs (miRNAs) are small, single-stranded, noncoding RNAs that regulate mRNAs by binding to their 3′ untranslated (3′UTR) regions.1,2 More than 9000 miRNAs have been identified in more than 100 species. Most miRNA genes are transcribed by RNA polymerase II into primary miRNA transcripts that are processed in the nucleus by a complex containing the RNase III endonuclease Drosha.1 The resulting precursor miRNAs are transported to the cytoplasm, where the mature miRNAs are excised by a complex containing the endonuclease Dicer.1 Mature miRNAs are incorporated into the RNA-induced silencing complex, which binds to the 3′UTRs of target mRNAs, inducing their degradation and/or repressing their translation. Posttranscriptional regulation of gene expression by miRNAs is critical for a wide range of physiologic and pathologic processes, including cell proliferation, apoptosis, differentiation, morphogenesis, development, and oncogenesis.1-4
Several miRNAs play pivotal roles in the immune system.5-7 MicroRNA-155 (miR155) has emerged as a particularly prominent player in innate and adaptive immune responses.5,7 miR155 is derived from an exon of the B-cell integration cluster (BIC) gene, which was identified as a common integration site of avian leucosis virus in chicken B-cell lymphomas.8,9 BIC is a non-protein-coding gene for which the only known function is the production of miR155. Subsequent studies revealed that miR155 expression is deregulated in diverse cancers.10,11 The molecular mechanisms underlying the oncogenic role of miR155 remain unclear.
miR155 expression is induced during the activation of T cells, B cells, monocytes, macrophages, and dendritic cells (DCs), suggesting that it plays multiple roles in the immune system.5 In agreement with this, the immune system of miR155-deficient mice is compromised by defects in several cell types.12,13 Activated T cells from miR155−/− mice exhibit a bias toward Th2 differentiation and express elevated levels of IL4, IL5, and IL10. This was attributed to the fact that miR155 targets the mRNA coding for c-Maf, a transcription factor implicated in IL-4 expression and Th2 differentiation.12 The B-cell compartment in miR155−/− mice exhibits defects in germinal center development and in the generation of efficient antibody responses. miR155 is critical for affinity maturation because the generation of plasma cells produces high-affinity isotype-switched antibodies and the development of memory B cells.12-14 The B-cell defects in miR155−/− mice result at least in part from miR155 repressing the expression of the transcription factor PU.114 and activation-induced cytidine deaminase.15,16 Lastly, bone marrow–derived DCs (BM-DCs) from miR155−/− mice are impaired in their ability to activate T cells.12
We recently reported that the induction of miR155 expression in human monocyte–derived DCs (Mo-DCs) exposed to the TLR4 ligand lipopolysaccharide (LPS) leads to modulation of the IL1 signal transduction pathway.17 Another study found that miR155 induces down-regulation of DC-specific intercellular adhesion molecule-3 grabbing nonintegrin in human Mo-DCs by inhibiting the expression of PU.1.18 Neither study elucidated the T cell–activation defect exhibited by miR155-deficient BM-DCs.
To define the role of miR155 in DCs, we analyzed its expression and function in human and mouse DCs exposed to various stimuli. Activation of miR155 expression was found to be a general evolutionarily conserved process that was correlated with maturation induced by diverse stimuli in all DC subtypes tested. Microarray experiments revealed that silencing of c-Fos expression is a key function of miR155 in DCs. Finally, functional experiments performed with miR155-deficient DCs and DCs in which c-Fos expression was deregulated demonstrated that the repression of c-Fos by miR155 is required for DC maturation and the ability of DCs to promote antigen-specific T-cell activation.
Methods
Mice
Cells
The mouse DC2114 cell line21 was cultured in IMDM supplemented with 10% FCS, 0.05mM β-mercaptoethanol, 1000 units/mL of penicillin, and 1000 μg/mL of streptomycin. 293T cells were cultured in DMEM supplemented with 10% FCS, 1000 units/mL of penicillin, and 1000 μg/mL of streptomycin. Cells were cultured under 5% CO2 in a humidified incubator.
Human Mo-DCs were prepared as described previously.22 Bone marrow–derived plasmacytoid DCs (BM-pDCs) were derived from tibia and femur bone marrow suspensions from 8- to 10-week-old mice, as described previously.23 BM-DC differentiation was performed by incubation of 1 × 106 bone marrow cells per milliliter in DMEM medium supplemented with 10% FCS and 5% of a supernatant from a hybridoma-producing GM-CSF. CD11c+ BM-DCs, CD11c+B220+ BM-pDCs, and splenic CD11c+CD8α+ and CD11c+CD8α− DCs were purified by sorting with a FACSVantage SE (Becton Dickinson). DC maturation was induced with 25 ng/mL of LPS (Alexis), 0.05 mg/mL of poly (I:C) (Amersham Biosciences), 0.2nM CpG oligodeoxynucleotide 1826 (TriLink BioTechnologies), 10 μg/mL of peptidoglycan (PGN; Sigma), 500 ng/mL of Pam3CysSerLys4 (PAM3CSK4; InvivoGen), 200 ng/mL of flagellin (InvivoGen), 100 ng/mL of TNFα, 100 ng/mL of fibroblast-stimulating lipopeptide-1 (FSL-1), 10 μg/mL of muramyl dipeptide (MDP; Calbiochem), 3 μg/mL of imiquimod (InvivoGen), or CpG plus anti-CD40 antibodies (rat FGK45 hybridoma). Splenic CD4+ T cells were purified from 2D2 or OTII mice using a CD4+ T cell–isolation kit (Miltenyi Biotec).
Lentiviral transductions
A fragment of the BIC gene encoding miR155 was amplified by PCR from mouse genomic DNA using the primers 5′-GTGCTGCAAACCAGGAAG-3′ and 5′-CCTTACAAAGAGTTGTTCATC-3′. This BIC fragment was cloned into the pDONR221 vector using the Gateway BP Clonase Enzyme Mix (Invitrogen). This vector was recombined with pDONRP4-P1R into the 2K7 green fluorescent protein (GFP) lentiviral vector24 using the Gateway LR Plus Clonase Enzyme Mix (Invitrogen) to generate a vector expressing the BIC precursor under control of the EF1α promoter. This vector also expresses GFP to permit the evaluation of transduction efficiencies and the purification of transduced cells. The mutated BIC expression vector was generated by mutating the miR155 sequence in the BIC expression vector. c-Fos cDNA was amplified using the primers 5′-ATGACGTTTAAACGCCACCATGATGTTCTCGGGTTTC-3′ and 5′-ATGACGTTTAAACTCACAGGGCCAGCAGCGT-3′. This c-Fos cDNA was cloned into the lentiviral pWPI vector. Transduction of mouse DC2114 cells was performed as described previously.25
Microarray experiments
Microarray experiments and miRNA target site analyses were performed as detailed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Microarray data reported in our study have been deposited in the ArrayExpress database under accession numbers E-MTAB-497 (Figure 2B array) and E-MTAB-498 (Figure 2C array).
Quantitative RT-PCR
RNA was extracted with TRIzol. Human and mouse miR155 cDNAs were generated using specific primers and MultiScribe Reverse Transcriptase, and real-time PCR was performed using hsa-miR155 and Mmu-miR155 TaqMan MicroRNA Assays (Applied Biosystems). Mouse and human mRNAs were quantified by real-time RT-PCR using the iCycler iQ Real-Time PCR Detection System (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). Expression levels were normalized using β-actin mRNA, TATA-binding protein mRNA, or 18S rRNA. Results were quantified using a standard curve generated with serial dilutions of input cDNA. Primers are listed in supplemental Table 1.
Luciferase reporter assays
The complete 3′UTRs of human (762 bp) and mouse (800 bp) c-Fos mRNAs were amplified by PCR and inserted downstream of the Renilla luciferase gene in the dual luciferase reporter plasmid psiCHECK-2 (Promega). The QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene) was used to mutate the putative miR155-binding sites. Luciferase reporter assays were performed as described previously.17
Flow cytometry
Flow cytometry was performed with a FACSCalibur (Becton Dickinson) and analyzed with WinMDI 2.8 software. Staining was performed in the presence of saturating concentrations of 2.4G2 anti-FcγRII/III monoclonal antibodies. Antibodies are listed in supplemental Table 2.
Western blotting
Protein extracts were fractionated by SDS-PAGE and Western blotting was performed with the antibodies listed in supplemental Table 2.
T-cell stimulation
LPS-treated BM-DCs or CpG-treated DC2114 cells were loaded with 20 μg/mL of myelin oligodendrocyte glycoprotein (MOG35-55) peptide, 1 μg/mL of OVA peptide, or 1 mg/mL of OVA protein, and cocultured with 105 2D2 or OTII T cells. Control cultures contained equal numbers of unloaded DCs. T-cell activation was assessed after 18 (BM-DC cocultures) or 6 (DC2114 cocultures) hours. CD69+ cells were quantified by flow cytometry. Secretion of IL2 was measured by ELISA according to the manufacturer's instructions (eBioscience). T-cell proliferation was assessed by [3H]-thymidine incorporation.
Immunofluorescence microscopy
Cells were seeded on glass coverslips, cultured for 24 hours in the absence or presence of LPS (BM-DCs) or CpG (DC2114 cells), and fixed for 10 minutes at room temperature with 1% paraformaldehyde in PBS. BM-DCs were stained using the antibodies indicated in supplemental Table 2. Nuclei were stained with DAPI. DC2114 cells were visualized on the basis of endogenous GFP expression. Cells were observed in a Zeiss Axiocam microscope using Axiovision LE software.
Statistical analysis
P values were calculated using the Student t test with 2-tailed distribution and 2-sample unequal variance parameters.
Results
Induction of miR155 expression in human Mo-DCs
miRNA expression profiles were compared between immature Mo-DCs and Mo-DCs activated with LPS using a human miRNA microarray and a multispecies miRNA microarray (supplemental Figure 1A-B). Maturation was verified by examining the up-regulation of MHC class II (MHCII), CD83, CD86, and CD40 expression (supplemental Figure 1C). The miRNA that was up-regulated the most strongly and reproducibly was miR155.
Real-time RT-PCR experiments confirmed that miR155 expression was increased strongly in mature Mo-DCs relative to monocytes and immature Mo-DCs (supplemental Figure 2A). Maturation was controlled by assessing the induction of IL6 and IL12-p40 mRNA expression and the reduced expression of the mRNA coding for the MHCII transactivator CIITA.22 Time-course experiments indicated that miR155 accumulation was induced rapidly and increased progressively, reaching maximal levels after 24-48 hours of stimulation with LPS (supplemental Figure 2B). Quantification of its corresponding precursor miRNA indicated that miR155 accumulation resulted from a rapid and strong transcriptional activation of the BIC gene (supplemental Figure 2B). In addition, time-course experiments indicated that miR155 and BIC expression were induced by poly (I:C) with kinetics similar to those observed for stimulation with LPS (supplemental Figure 2B).
miR155 and BIC transcripts were next quantified in Mo-DCs exposed to the TLR2 ligand PGN, the TLR3 ligand poly (I:C), the TR5 ligand flagellin, and TNFα (supplemental Figure 3A). Maturation was again controlled by examining IL12-p40, IL6, and CIITA mRNA expression. miR155 and BIC expression were induced by all 4 stimuli, although the increase was weaker than that observed for exposure to LPS. BIC expression was also induced in Mo-DCs by IFNα (supplemental Figure 3B). These results indicate that ligands that trigger DC maturation induce an increase in miR155 expression.
Maturation-induced miR155 expression is conserved in mouse DCs
To determine whether the induction of miR155 expression during DC activation is a conserved process, we extended our analysis to mouse DCs. miR155 expression was first studied in a DC cell line (DC2114) derived from a transgenic mouse–expressing SV40 T-antigen under control of the CD11c promoter.21 DC2114 cells correspond to CD8α+ DCs and reproduce faithfully most of the key features of their in vivo counterparts: they can capture, process, and present antigens to CD4+ T cells; cross-present antigens to CD8+ T cells; be activated by classic maturation stimuli; and produce a pattern of chemokines and proinflammatory cytokines typical of primary DCs. A potent maturation stimulus for DC2114 cells is the TLR9 ligand CpG in combination with anti-CD40 (αCD40) antibodies. Maturation induced by CpG + αCD40 was assessed by examining cell-surface MHCII (I-Ab), CD80, CD86, and CD40 expression (supplemental Figure 4A). We performed miRNA expression-profiling experiments to identify miRNAs that undergo changes in expression in DC2114 cells stimulated with CpG + αCD40, and miR155 was found to be up-regulated strongly and reproducibly (supplemental Figure 4B).
Real-time RT-PCR experiments confirmed that miR155 expression increased dramatically in CpG + αCD40–stimulated DC2114 cells (supplemental Figure 4C). This induction was correlated with efficient maturation, as assessed by quantifying IL6, IL12-p40, and CIITA mRNA expression (supplemental Figure 4C). Time-course experiments indicated that miR155 accumulation in DC2114 cells was induced rapidly and increased progressively, reaching maximal levels after 12-24 hours of stimulation with CpG + αCD40 (supplemental Figure 4D). Induction of miR155 expression resulted from a rapid and strong activation of the BIC gene (supplemental Figure 4D). We also studied miR155 and BIC expression in DC2114 cells subjected to other signals (supplemental Figure 4E). Stimuli that promoted efficient maturation (PGN and poly (I:C) induced a strong increase in miR155 and BIC expression. Conversely, neither miR155 nor BIC expression was induced by stimuli that failed to promote efficient maturation (PAM3CSK4, LPS, and flagellin).
We next studied the activation of miR155 and BIC expression in primary mouse DCs. miR155 expression was induced strongly upon the maturation of conventional BM-DCs, BM-pDCs, and splenic CD8α− and CD8α+ DCs (supplemental Figure 5A). Furthermore, BIC expression was induced in BM-DCs by all maturation stimuli tested, including LPS, poly (I:C), PGN, PAM3CSK4, flagellin, FSL-1, imiquimod, CpG, MDP, and IFNα (supplemental Figure 5B-C).
miR155 is required for DC maturation and function
Numbers of conventional DCs (CD11c+CD8α+, CD11c+CD8α−) and pDCs (CD11c+B220+) were unaffected in miR155−/− mice (Table 1). The differentiation of BM-DCs and BM-pDCs was also unchanged in BM cultures from miR155−/− mice. The deficiency in miR155 does therefore not lead to a general defect in DC development.
DC numbers in miR155−/− mice
| . | WT C57BL/6 . | miR155−/− . | P . |
|---|---|---|---|
| Total splenocytes | 189.106 ± 57.106 | 128.106 ± 27.106 | .1752 |
| CD11c+CD8α− cDCs, % | 1.06 ± 0.13 | 0.86 ± 0.17 | .1730 |
| CD11c+CD8α+ cDCs, % | 0.37 ± 0.07 | 0.34 ± 0.06 | .5257 |
| CD11c+B220+ pDCs, % | 1.00 ± 0.13 | 1.33 ± 0.18 | .0610 |
| . | WT C57BL/6 . | miR155−/− . | P . |
|---|---|---|---|
| Total splenocytes | 189.106 ± 57.106 | 128.106 ± 27.106 | .1752 |
| CD11c+CD8α− cDCs, % | 1.06 ± 0.13 | 0.86 ± 0.17 | .1730 |
| CD11c+CD8α+ cDCs, % | 0.37 ± 0.07 | 0.34 ± 0.06 | .5257 |
| CD11c+B220+ pDCs, % | 1.00 ± 0.13 | 1.33 ± 0.18 | .0610 |
Means ± SDs were derived from 3 mice of each genotype.
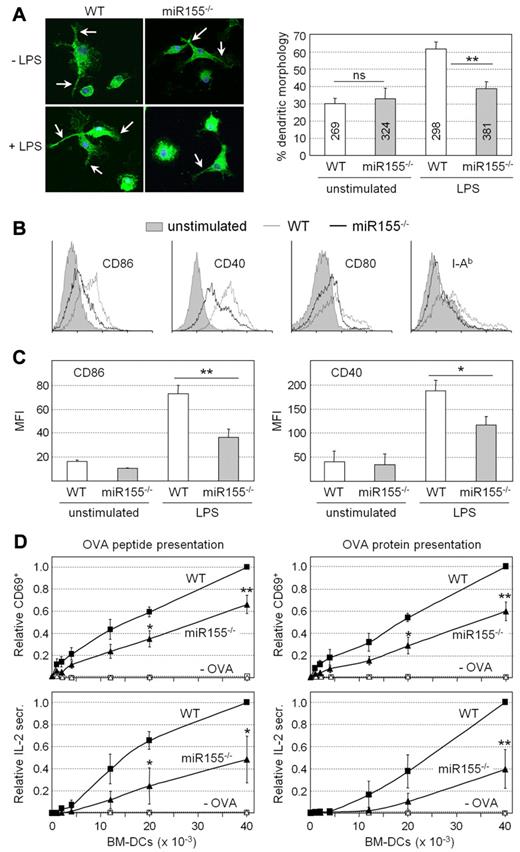
Unstimulated wild-type (WT) and miR155−/− BM-DCs exhibited similar frequencies of cells displaying a dendritic morphology characterized by long cellular protrusions (Figure 1A). However, the fraction of cells exhibiting this morphology after LPS treatment was significantly lower for miR155−/− BM-DCs (Figure 1A). LPS-induced increases in the cell-surface expression of MHCII (I-Ab) and costimulatory molecules (CD86, CD40, and CD80) were attenuated in BM-DCs from miR155−/− mice (Figure 1B). This was particularly evident for CD86 and CD40 (Figure 1C). BM-DCs from both WT and miR155-deficient mice exhibited strong induction of IL12-p40, IL12-p35, IL1β, IL6, and TNFα mRNA expression after exposure to LPS (supplemental Figure 6). However, the induction of IL12-p40, IL12-p35, and TNFα mRNAs tended to be slightly attenuated in miR155−/− BM-DCs. These findings indicate that miR155−/− BM-DCs exhibit selective defects in key processes associated with DC maturation.
Phenotypic and functional defects exhibited by miR155−/− DCs. (A) Unstimulated and LPS-treated BM-DCs prepared from WT and miR155−/− mice were stained with antibodies against CD11c, and the frequencies of cells exhibiting a characteristic dendritic morphology were determined. Representative images are shown at the left; dendritic protrusions are indicated with arrows. The bar graph represents the means and SDs derived from 3 independent BM-DC preparations; ns, not significant; **P < .01. The numbers of cells examined are indicated for each bar. (B) Cell-surface CD86, CD40, CD80, and I-Ab expression was analyzed by flow cytometry for unstimulated and LPS-treated BM-DCs from WT and miR155−/− mice. Histograms are representative of 3 experiments. (C) The mean fluorescence intensity (MFI) for cell-surface CD86 and CD40 expression was determined by flow cytometry for unstimulated and LPS-treated BM-DCs from WT and miR155−/− mice. The means and SDs derived from 3 independent experiments are shown; *P < .05; **P < .01. (D) LPS-treated BM-DCs from WT and miR155−/− mice were loaded with OVA peptide (left panels) or OVA protein (right panels) and cocultured with OVA-specific CD4+ T cells purified from TCR-transgenic OTII mice. BM-DCs that had not been loaded with antigen (−OVA) were used as negative controls. T-cell activation was determined by the analysis of cell-surface CD69 expression (top panels, relative frequencies of CD69+ cells) or secretion of IL2 into the supernatants (bottom panels, relative IL2 secretion). The means and SDs derived from 3 independent experiments are shown; *P < .05; **P < .01.
Phenotypic and functional defects exhibited by miR155−/− DCs. (A) Unstimulated and LPS-treated BM-DCs prepared from WT and miR155−/− mice were stained with antibodies against CD11c, and the frequencies of cells exhibiting a characteristic dendritic morphology were determined. Representative images are shown at the left; dendritic protrusions are indicated with arrows. The bar graph represents the means and SDs derived from 3 independent BM-DC preparations; ns, not significant; **P < .01. The numbers of cells examined are indicated for each bar. (B) Cell-surface CD86, CD40, CD80, and I-Ab expression was analyzed by flow cytometry for unstimulated and LPS-treated BM-DCs from WT and miR155−/− mice. Histograms are representative of 3 experiments. (C) The mean fluorescence intensity (MFI) for cell-surface CD86 and CD40 expression was determined by flow cytometry for unstimulated and LPS-treated BM-DCs from WT and miR155−/− mice. The means and SDs derived from 3 independent experiments are shown; *P < .05; **P < .01. (D) LPS-treated BM-DCs from WT and miR155−/− mice were loaded with OVA peptide (left panels) or OVA protein (right panels) and cocultured with OVA-specific CD4+ T cells purified from TCR-transgenic OTII mice. BM-DCs that had not been loaded with antigen (−OVA) were used as negative controls. T-cell activation was determined by the analysis of cell-surface CD69 expression (top panels, relative frequencies of CD69+ cells) or secretion of IL2 into the supernatants (bottom panels, relative IL2 secretion). The means and SDs derived from 3 independent experiments are shown; *P < .05; **P < .01.
Antigen-specific T cell–activation assays were performed to determine the functional consequences of the impaired maturation of miR155-deficient BM-DCs. OVA-specific CD4+ T cells from TCR-transgenic OTII mice were stimulated with LPS-treated WT or miR155−/− BM-DCs that had been loaded or not with OVA peptide or OVA protein. OTII T-cell activation was assessed by examining CD69 expression and IL2 secretion. OVA-specific OTII T-cell activation induced by miR155-deficient BM-DCs was significantly impaired (Figure 1D). Similar results were obtained by assessing T-cell activation and proliferation induced by the presentation of MOG35-55 to MOG-specific CD4+ T cells purified from TCR-transgenic 2D2 mice. MOG35-55-specfic 2D2 T-cell activation and proliferation induced by miR155-deficient BM-DCs were significantly reduced (supplemental Figure 7). BM-DCs from miR155−/− mice therefore exhibit marked defects in their ability to promote antigen-specific T-cell activation.
Identification of mRNAs regulated by miR155 in DCs
Microarray experiments were performed to document differences between the global gene-expression profiles of mature WT and miR155−/− BM-DCs. Direct targets of miR155 were expected to be enriched among mRNAs that are up-regulated in the miR155-deficient BM-DCs relative to WT DCs. mRNAs that were significantly increased in mature miR155−/− BM-DCs were therefore analyzed for the presence of potential miR155-binding sites. Six to 8 nucleotide sequences showing complementarity to the “seed” region situated at the 5′ end (positions 2-9) of miR155 were significantly enriched in the 3′UTRs of mRNAs that were up-regulated in mature miR155−/− BM-cDCs (Table 2). The enrichment of miR155-binding sites was also confirmed using 3 additional prediction models relying on favorable binding energy, sequence context, or evolutionary conservation (Table 2). As expected, no significant enrichment of miR155 targets was observed in mRNAs that were down-regulated in mature miR155−/− BM-DCs or in mRNAs that were increased or decreased in immature miR155−/− BM-DCs (Table 2). We next scanned the mRNAs that were up-regulated in miR155−/− BM-DCs for the presence of target sites for all mouse miRNAs included in the miRBase database (Version 14). miR155 was the only miRNA for which target site enrichment was observed using each of the 4 prediction models (Figure 2A). These results indicate that the altered mRNA expression profile of miR155−/− BM-DCs exhibits a clear miR155 signature.
Analysis of miR155 signature in miR155−/− BM-DCs
| . | Immature BM-DCs . | Mature BM-DCs* . | ||||||
|---|---|---|---|---|---|---|---|---|
| Targets in up-regulated mRNAs† . | Targets in down-regulated mRNAs† . | Targets in up-regulated mRNAs† . | Targets in down-regulated mRNAs† . | |||||
| % . | P . | % . | P . | % . | P‡ . | % . | P . | |
| 5% significance threshold for differential mRNA expression§ | ||||||||
| Seeds | 22 | .9 | 22 | .9 | 37 | 10−6 | 19 | .98 |
| Conserved seeds | 9.7 | .6 | 4.7 | 1.0 | 15 | .004 | 4 | 1.00 |
| Targetscan 4 | 4.5 | 1.0 | 6.5 | .8 | 12 | .003 | 4 | .99 |
| ΔG duplex | 1.2 | .7 | 1.0 | .7 | 2.4 | .082 | .4 | .97 |
| 1% significance threshold for differential mRNA expression¶ | ||||||||
| Seeds | 22 | .7 | 28 | .4 | 42 | .001 | 27 | .33 |
| Conserved seeds | 4.2 | .9 | 4.8 | .9 | 17 | .024 | 5.1 | .9 |
| Targetscan 4 | 5.3 | .7 | 7.1 | .6 | 13 | .08 | 10 | .24 |
| ΔG duplex | .0 | 1.0 | 1.6 | .7 | 4.3 | .14 | 0.0 | 1.0 |
| . | Immature BM-DCs . | Mature BM-DCs* . | ||||||
|---|---|---|---|---|---|---|---|---|
| Targets in up-regulated mRNAs† . | Targets in down-regulated mRNAs† . | Targets in up-regulated mRNAs† . | Targets in down-regulated mRNAs† . | |||||
| % . | P . | % . | P . | % . | P‡ . | % . | P . | |
| 5% significance threshold for differential mRNA expression§ | ||||||||
| Seeds | 22 | .9 | 22 | .9 | 37 | 10−6 | 19 | .98 |
| Conserved seeds | 9.7 | .6 | 4.7 | 1.0 | 15 | .004 | 4 | 1.00 |
| Targetscan 4 | 4.5 | 1.0 | 6.5 | .8 | 12 | .003 | 4 | .99 |
| ΔG duplex | 1.2 | .7 | 1.0 | .7 | 2.4 | .082 | .4 | .97 |
| 1% significance threshold for differential mRNA expression¶ | ||||||||
| Seeds | 22 | .7 | 28 | .4 | 42 | .001 | 27 | .33 |
| Conserved seeds | 4.2 | .9 | 4.8 | .9 | 17 | .024 | 5.1 | .9 |
| Targetscan 4 | 5.3 | .7 | 7.1 | .6 | 13 | .08 | 10 | .24 |
| ΔG duplex | .0 | 1.0 | 1.6 | .7 | 4.3 | .14 | 0.0 | 1.0 |
Treated with LPS for 24 hours.
Expression in miR155−/− BM-DCs relative to WT BM-DCs.
One-sided Fisher test (bold indicates P < .05).
541 up-regulated mRNAs in miR155−/− DCs (t test).
139 up-regulated mRNAs in miR155−/− DCs (t test).
Identification of mRNAs that are regulated by miR155 in DCs. (A) Microarray experiments were performed to compare the gene-expression profiles of mature LPS-treated BM-DCs from WT and miR155−/− mice. The 3′UTRs of mRNAs that were significantly up-regulated in miR155−/− DCs were analyzed for the presence of potential target sites of all mouse miRNAs using 4 prediction models (all seeds, conserved seeds, Targetscan 4, and ΔG duplex). The graphs represent the fold enrichment of target sites for each miRNA. The position of miR155 and its ranking with respect to target-site enrichment are indicated for each graph. (B) Microarray data for mature LPS-treated BM-DCs from WT and miR155−/− mice are represented as a scatter plot showing average normalized signal intensities derived from 3 independent experiments. Each dot represents a probe set corresponding to one mRNA. Only dots corresponding to mRNAs exhibiting greater than a 1.5-fold difference in expression between the 2 genotypes are shown. Dots corresponding to Picalm, c-Fos, Pu.1, Ship, and Smad5 mRNAs are indicated. (C) Microarray experiments were performed to compare the gene-expression profiles of control and BIC-transduced DC2114 cells. Results are represented as a scatter plot showing average normalized signal intensities derived from 3 independent experiments. Each dot represents a probe set corresponding to one mRNA. Only dots corresponding to mRNAs exhibiting greater than a 1.5-fold difference in expression between control and BIC-transduced cells are indicated. The dot corresponding to c-Fos mRNA is indicated.
Identification of mRNAs that are regulated by miR155 in DCs. (A) Microarray experiments were performed to compare the gene-expression profiles of mature LPS-treated BM-DCs from WT and miR155−/− mice. The 3′UTRs of mRNAs that were significantly up-regulated in miR155−/− DCs were analyzed for the presence of potential target sites of all mouse miRNAs using 4 prediction models (all seeds, conserved seeds, Targetscan 4, and ΔG duplex). The graphs represent the fold enrichment of target sites for each miRNA. The position of miR155 and its ranking with respect to target-site enrichment are indicated for each graph. (B) Microarray data for mature LPS-treated BM-DCs from WT and miR155−/− mice are represented as a scatter plot showing average normalized signal intensities derived from 3 independent experiments. Each dot represents a probe set corresponding to one mRNA. Only dots corresponding to mRNAs exhibiting greater than a 1.5-fold difference in expression between the 2 genotypes are shown. Dots corresponding to Picalm, c-Fos, Pu.1, Ship, and Smad5 mRNAs are indicated. (C) Microarray experiments were performed to compare the gene-expression profiles of control and BIC-transduced DC2114 cells. Results are represented as a scatter plot showing average normalized signal intensities derived from 3 independent experiments. Each dot represents a probe set corresponding to one mRNA. Only dots corresponding to mRNAs exhibiting greater than a 1.5-fold difference in expression between control and BIC-transduced cells are indicated. The dot corresponding to c-Fos mRNA is indicated.
The expression levels of several mRNAs that were known or suspected to be regulated by miR155 or a viral ortholog (miR-K12-1) encoded by Kaposi-sarcoma–associated herpes virus14,18,26,27 were increased in mature miR155−/− BM-DCs (Figure 2B). Increased expression was statistically significant for some of these mRNAs (Picalm, Pu.1, and Smad5) but not for others (c-Fos and Ship), suggesting that down-regulation of target mRNAs by miR155 is variable in efficiency. This is consistent with the fact that repression by miRNAs can occur mainly at the translational level, with little or no reduction in mRNA abundance.
As a complementary approach to identifying targets of miR155, we studied the impact of overexpressing miR155 in DCs. DC2114 cells were transduced with a lentiviral BIC expression vector that drives miR155 expression to a level comparable to that observed in DC2114 cells stimulated with CpG + αCD40 (supplemental Figure 8A). Examinations of cell-surface maturation markers revealed that enforced miR155 expression in DC2114 cells did not trigger spontaneous maturation or hinder maturation induced by CpG + αCD40 (supplemental Figure 8B). Microarray experiments were performed to document differences between the global gene-expression patterns exhibited by DC2114 cells transduced with the BIC expression vector and a control vector. Only minor changes in gene expression were induced by miR155 overexpression (Figure 2C). Among the target mRNAs that were up-regulated in miR155−/− BM-DCs (Figure 2B), only c-Fos mRNA was reduced in DC2114 cells overexpressing miR155 (Figure 2C).
Repression of c-Fos expression by miR155
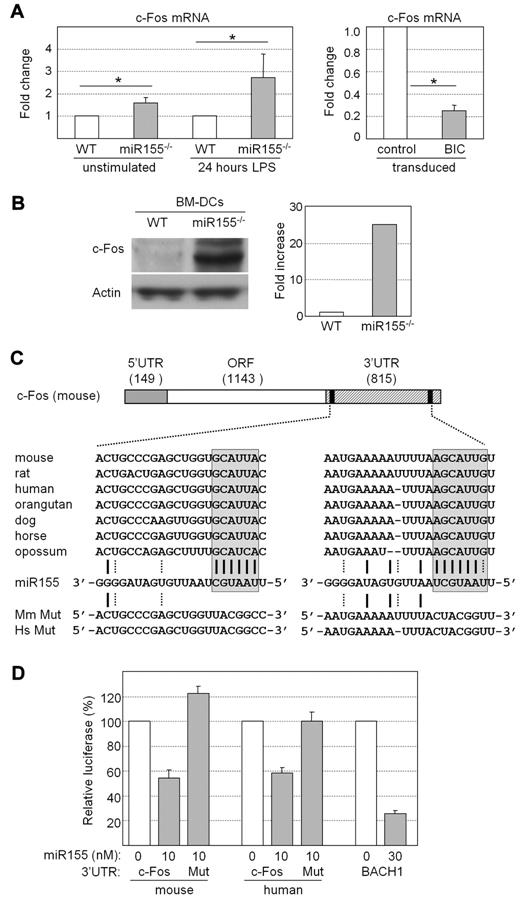
The microarray data suggested that c-Fos mRNA could be a critical target of miR155 in DCs. Real-time RT-PCR experiments confirmed that c-Fos mRNA abundance was indeed significantly increased in miR155−/− BM-DCs, whereas it was markedly decreased in DC2114 cells transduced with the BIC expression vector (Figure 3A). Furthermore, transduction with the BIC expression vector also induced a decrease in c-Fos mRNA levels in WT and miR155−/− BM-DCs, as well as in a control c-Fos–expressing mouse epithelial cell line (MLE12) (supplemental Figure 9A-C).
c-Fos expression is regulated by miR155. (A) Expression of c-Fos mRNA was analyzed by real-time RT-PCR in unstimulated and LPS-stimulated BM-DCs from WT or miR155−/− mice (left) and in DC2114 cells transduced with empty vector or the BIC expression vector (right). Results are represented as relative c-Fos mRNA expression. The means and SDs derived from 3 independent experiments are shown; *P < .05. (B) Expression of c-Fos protein was analyzed by Western blotting in BM-DCs prepared from WT and miR155−/− mice. Actin was used as internal control. A gel representative of 3 independent experiments is shown (left). c-Fos signals were quantified and normalized relative to actin (right). The results represent the mean fold increase derived from 2 independent experiments. (C) Schematic representation of mouse c-Fos mRNA. The sizes in nucleotides of the 5′UTR, open reading frame (ORF), and 3′UTR are indicated. The 3′UTR contains 2 predicted binding sites (black boxes) for miR155. The sequence of mouse miR155 is shown aligned with its predicted target sites in the 3′UTR of c-Fos mRNAs from the indicated species. A-U and G-C base pairs are represented by solid lines; G-U base pairs are represented by dotted lines. The miR155 seed region and its complementary sequences in c-Fos mRNAs are enclosed by boxes. Sequences of the mutated (Mut) 3′UTRs of human (Hs) and mouse (Mm) c-Fos mRNA are indicated. (D) Luciferase reporter constructs containing the WT or mutated 3′UTRs of human or mouse c-Fos mRNA were transfected into 293T cells. A reporter construct containing the 3′UTR of BACH1 mRNA, a known target of miR155, was used as positive control. The constructs were transfected together with the indicated amounts of human or mouse miR155. Luciferase activity was measured 24 hours after transfection, normalized with respect to the activity obtained with a control reporter vector, and is expressed as relative luciferase activity. The means and SDs derived from 3 independent experiments are shown.
c-Fos expression is regulated by miR155. (A) Expression of c-Fos mRNA was analyzed by real-time RT-PCR in unstimulated and LPS-stimulated BM-DCs from WT or miR155−/− mice (left) and in DC2114 cells transduced with empty vector or the BIC expression vector (right). Results are represented as relative c-Fos mRNA expression. The means and SDs derived from 3 independent experiments are shown; *P < .05. (B) Expression of c-Fos protein was analyzed by Western blotting in BM-DCs prepared from WT and miR155−/− mice. Actin was used as internal control. A gel representative of 3 independent experiments is shown (left). c-Fos signals were quantified and normalized relative to actin (right). The results represent the mean fold increase derived from 2 independent experiments. (C) Schematic representation of mouse c-Fos mRNA. The sizes in nucleotides of the 5′UTR, open reading frame (ORF), and 3′UTR are indicated. The 3′UTR contains 2 predicted binding sites (black boxes) for miR155. The sequence of mouse miR155 is shown aligned with its predicted target sites in the 3′UTR of c-Fos mRNAs from the indicated species. A-U and G-C base pairs are represented by solid lines; G-U base pairs are represented by dotted lines. The miR155 seed region and its complementary sequences in c-Fos mRNAs are enclosed by boxes. Sequences of the mutated (Mut) 3′UTRs of human (Hs) and mouse (Mm) c-Fos mRNA are indicated. (D) Luciferase reporter constructs containing the WT or mutated 3′UTRs of human or mouse c-Fos mRNA were transfected into 293T cells. A reporter construct containing the 3′UTR of BACH1 mRNA, a known target of miR155, was used as positive control. The constructs were transfected together with the indicated amounts of human or mouse miR155. Luciferase activity was measured 24 hours after transfection, normalized with respect to the activity obtained with a control reporter vector, and is expressed as relative luciferase activity. The means and SDs derived from 3 independent experiments are shown.
Western blot experiments demonstrated that the low levels of c-Fos detected in WT BM-DCs was strongly increased in miR155−/− BM-DCs (Figure 3B). The fact that c-Fos protein was increased to a greater extent than c-Fos mRNA (25-fold versus 2- to 3-fold) suggested that miR155 represses c-Fos expression mainly at the level of translation.
Computational approaches for identifying miRNA target sequences based on well-established criteria, including complementarity to the miRNA seed region, favorable sequence context, stability of the miRNA-mRNA duplex, and conservation across multiple species,28-31 predicted 2 miR155-binding sites in the 3′UTR of c-Fos mRNA in all species examined (Figure 3C). Conservation was strongest in the segments showing complementary to the seed region of miR155 (Figure 3C).
The complete 3′UTRs of mouse and human c-Fos mRNA were inserted into reporter vectors downstream of the Renilla luciferase gene. As controls we used vectors in which the seed regions of the 2 predicted miR155-binding sites within the c-Fos 3′UTRs were mutated. These constructs were transfected into 293T cells with or without the corresponding human or mouse miR155 precursors. Cotransfection of the nonmutated construct with the miR155 precursors resulted in a significant reduction in luciferase activity (Figure 3D). In contrast, no reduction was observed when the mutated constructs were cotransfected with the miR155 precursors. These results confirmed that the predicted miR155-binding sites in the 3′UTR of c-Fos mRNA were indeed targeted directly by miR155.
c-Fos expression is silenced during DC maturation
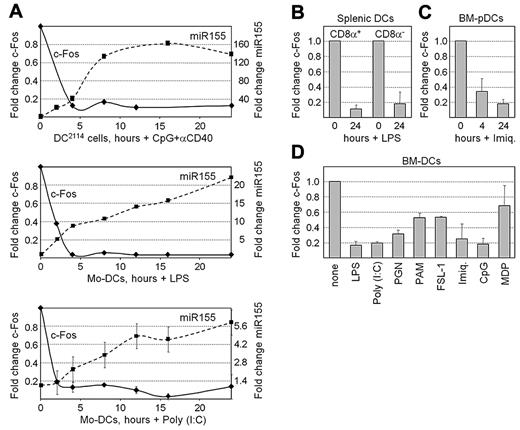
Time-course RT-PCR experiments were performed to quantify the changes in c-Fos mRNA expression that occur in mouse DC2114 cells stimulated with CpG + αCD40 and in human Mo-DCs treated with LPS or poly (I:C). In each system, maturation was accompanied by a rapid decrease in c-Fos mRNA abundance that was correlated with the concomitant increase in miR155 expression (Figure 4A). c-Fos expression was also decreased in CD8α+ and CD8α− splenic DCs after exposure to LPS (Figure 4B). Analysis of microarray expression-profiling experiments indicated that c-Fos expression is also silenced in BM-pDCs stimulated with imiquimod (Figure 4C). Finally, a reduction in c-Fos mRNA expression was in BM-DCs by all stimuli that activated miR155 expression, including LPS, poly (I:C), PGN, PAM3CSK4, FSL-1, imiquimod, CpG, and MDP (Figure 4D). Silencing of c-Fos expression is thus a conserved and general feature of DC maturation.
c-Fos expression is silenced during DC maturation. (A) Time-course RT-PCR experiments were performed to quantify the expression of c-Fos mRNA and miR155 in mouse DC2114 cells stimulated with CpG + αCD40 and human Mo-DCs stimulated with LPS or poly (I:C). Results are represented as the fold change in the expression of c-Fos mRNA (left axes) and miR155 (right axes). Representative experiments are shown for the top 2 panels. The means and SDs derived from 2 experiments are shown for the bottom panel. (B) c-Fos mRNA was quantified by real-time RT-PCR in unstimulated and LPS-treated CD8α+ and CD8α− splenic DCs. Results are represented as the fold change in c-Fos mRNA expression. The means and SDs derived from 2 independent experiments are shown. (C) c-Fos expression was analyzed in microarray data derived from BM-pDCs stimulated for 0, 4, and 24 hours with imiquimod. Results are represented as the fold change in signal intensities for c-Fos mRNA. The means and SDs derived from 3 experiments are shown. (D) c-Fos mRNA was quantified by real-time RT-PCR in unstimulated mouse BM-DCs and BM-DCs stimulated with LPS, poly (I:C), PGN, PAM3CSK4, FSL-1, imiquimod, CpG, or MDP. Results are represented as the fold change in c-Fos mRNA expression. The means and SDs derived from 2 independent experiments are shown.
c-Fos expression is silenced during DC maturation. (A) Time-course RT-PCR experiments were performed to quantify the expression of c-Fos mRNA and miR155 in mouse DC2114 cells stimulated with CpG + αCD40 and human Mo-DCs stimulated with LPS or poly (I:C). Results are represented as the fold change in the expression of c-Fos mRNA (left axes) and miR155 (right axes). Representative experiments are shown for the top 2 panels. The means and SDs derived from 2 experiments are shown for the bottom panel. (B) c-Fos mRNA was quantified by real-time RT-PCR in unstimulated and LPS-treated CD8α+ and CD8α− splenic DCs. Results are represented as the fold change in c-Fos mRNA expression. The means and SDs derived from 2 independent experiments are shown. (C) c-Fos expression was analyzed in microarray data derived from BM-pDCs stimulated for 0, 4, and 24 hours with imiquimod. Results are represented as the fold change in signal intensities for c-Fos mRNA. The means and SDs derived from 3 experiments are shown. (D) c-Fos mRNA was quantified by real-time RT-PCR in unstimulated mouse BM-DCs and BM-DCs stimulated with LPS, poly (I:C), PGN, PAM3CSK4, FSL-1, imiquimod, CpG, or MDP. Results are represented as the fold change in c-Fos mRNA expression. The means and SDs derived from 2 independent experiments are shown.
Deregulated c-Fos expression recapitulates miR155 deficiency in DCs
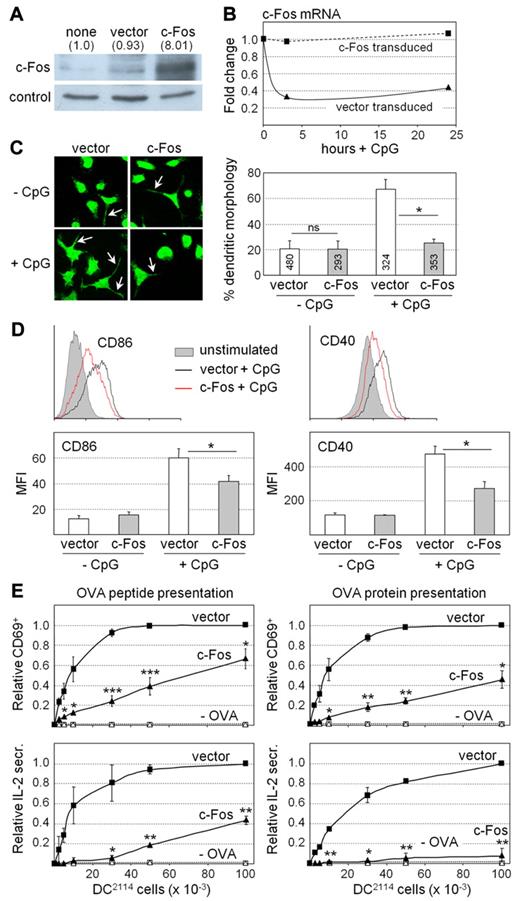
To determine whether impaired silencing of c-Fos expression might be responsible for the defects exhibited by miR155−/− DCs (Figure 1), we analyzed the consequences of deregulated c-Fos expression in DCs. DC2114 cells were transduced with a lentiviral vector expressing c-Fos under the control of heterologous promoter and 3′UTR regions. High transduction frequencies (supplemental Figure 10A) and efficient c-Fos expression (Figure 5A) were obtained. c-Fos overexpression in the transduced cells (Figure 5A) attained a level similar to that observed in miR155−/− BM-DCs (Figure 3B). In contrast to the endogenous c-Fos mRNA in control vector–transduced DC2114 cells, c-Fos mRNA levels were not decreased during maturation in c-Fos–transduced cells (Figure 5B). Unstimulated DC2114 cells transduced with c-Fos and control vectors exhibited similar frequencies of cells displaying a dendritic morphology (Figure 5C). However, the increase in the fraction of cells carrying dendritic protrusions after stimulation with CpG was significantly lower for c-Fos–transduced cells (Figure 5C). Enhanced CD86 and CD40 expression after activation with CpG was also significantly reduced in c-Fos–transduced DC2114 cells (Figure 5D). Furthermore, c-Fos–transduced DC2114 cells exhibited a strongly reduced ability to induce antigen-specific T-cell activation. CD69 expression and IL2 secretion by OTII cells was strongly reduced when c-Fos–transduced DC2114 cells were used as stimulators (Figure 5E). This was true irrespective of whether the DC2114 cells were loaded with OVA peptide or OVA protein (Figure 5E). Similarly, CD69 expression and IL2 secretion by 2D2 cells was strongly reduced when they were cocultured with c-Fos–transduced DC2114 cells loaded with MOG35-55 peptide (supplemental Figure 10B). Finally, although c-Fos–transduced DC2114 cells retained the ability to up-regulate the expression of proinflammatory cytokine mRNAs during maturation, this induction tended to be reduced 2- to 3-fold relative to untransduced DC2114 cells (supplemental Figure 11). These results indicate that deregulated c-Fos expression in DC2114 cells leads to phenotypic and functional defects that are strikingly similar to those observed in miR155−/− BM-DCs.
Deregulation of c-Fos expression induces phenotypic and functional defects in DCs. (A) Expression of c-Fos protein was analyzed by Western blotting in untransduced DC2114 cells and DC2114 cells transduced with an empty expression vector or a c-Fos expression vector. Actin was used as internal control. A representative gel is shown. c-Fos signals were quantified and normalized relative to actin. Changes (-fold) are indicated above the gel. (B) c-Fos mRNA was quantified by real-time RT-PCR in CpG-treated DC2114 cells transduced with an empty expression vector or a c-Fos expression vector. A representative graph of 2 independent experiments is shown. (C) Unstimulated and CpG-treated DC2114 cells transduced with an empty expression vector or a c-Fos expression vector were examined by immunofluoresence microscopy, and frequencies of cells exhibiting a characteristic dendritic morphology were determined. Representative images are shown at the left; dendritic protrusions are indicated with arrows. The bar graph represents the means and SDs derived from 3 independent transductions; ns, not significant; *P < .05. (D) Cell-surface CD86 and CD40 expression was analyzed by flow cytometry for unstimulated and CpG-stimulated DC2114 cells. The histograms (top) are representative of at least 3 experiments. The graphs (bottom) represent the mean fluorescence intensities (MFI) for cell-surface CD86 and CD40 expression, and show the means and SDs derived from at least 3 independent experiments; *P < .05. (E) CpG-treated DC2114 cells transduced with an empty vector or a c-Fos expression vector were loaded with OVA peptide (left panels) or OVA protein (right panels) and cocultured with OVA-specific CD4+ T cells purified from TCR-transgenic OTII mice. DC2114 cells that had not been loaded with antigen (−OVA) were used as negative controls. T-cell activation was determined by the analysis of cell-surface CD69 expression (top panels, relative frequencies of CD69+ cells) or secretion of IL2 into the supernatants (bottom panels, relative IL2 secretion). The means and SDs derived from 3 independent experiments are shown; *P < .05; **P < .01; ***P < .001.
Deregulation of c-Fos expression induces phenotypic and functional defects in DCs. (A) Expression of c-Fos protein was analyzed by Western blotting in untransduced DC2114 cells and DC2114 cells transduced with an empty expression vector or a c-Fos expression vector. Actin was used as internal control. A representative gel is shown. c-Fos signals were quantified and normalized relative to actin. Changes (-fold) are indicated above the gel. (B) c-Fos mRNA was quantified by real-time RT-PCR in CpG-treated DC2114 cells transduced with an empty expression vector or a c-Fos expression vector. A representative graph of 2 independent experiments is shown. (C) Unstimulated and CpG-treated DC2114 cells transduced with an empty expression vector or a c-Fos expression vector were examined by immunofluoresence microscopy, and frequencies of cells exhibiting a characteristic dendritic morphology were determined. Representative images are shown at the left; dendritic protrusions are indicated with arrows. The bar graph represents the means and SDs derived from 3 independent transductions; ns, not significant; *P < .05. (D) Cell-surface CD86 and CD40 expression was analyzed by flow cytometry for unstimulated and CpG-stimulated DC2114 cells. The histograms (top) are representative of at least 3 experiments. The graphs (bottom) represent the mean fluorescence intensities (MFI) for cell-surface CD86 and CD40 expression, and show the means and SDs derived from at least 3 independent experiments; *P < .05. (E) CpG-treated DC2114 cells transduced with an empty vector or a c-Fos expression vector were loaded with OVA peptide (left panels) or OVA protein (right panels) and cocultured with OVA-specific CD4+ T cells purified from TCR-transgenic OTII mice. DC2114 cells that had not been loaded with antigen (−OVA) were used as negative controls. T-cell activation was determined by the analysis of cell-surface CD69 expression (top panels, relative frequencies of CD69+ cells) or secretion of IL2 into the supernatants (bottom panels, relative IL2 secretion). The means and SDs derived from 3 independent experiments are shown; *P < .05; **P < .01; ***P < .001.
Discussion
We demonstrate here that miR155 expression is induced by diverse maturation stimuli in human Mo-DCs and in all mouse DC subsets examined. Up-regulated miR155 expression thus appears to be a general feature of DC activation. miR155 was also found to be required for DC maturation. This function was emphasized by the finding that miR155−/− DCs exhibited marked defects in their acquisition of phenotypic and functional properties of mature DCs. These defects included a block in the appearance of a typical dendritic morphology, a reduction in the up-regulation of costimulatory molecules, particularly CD40 and CD86, and a strongly impaired ability to promote antigen-specific CD4+ T-cell activation and proliferation. Not all DC functions were perturbed, because the production of the proinflammatory cytokines IL1β, TNFα, IL12, and IL6 was affected only modestly. These findings extend the previous observation that miR155−/− BM-DCs are less efficient at inducing CD4+ T-cell activation.12 They are also consistent with the finding that conditional deletion of the gene coding for Dicer in Langerhans cells leads to impaired maturation of this DC subset. As observed for miR155−/− DCs, Dicer-deficient Langerhans cells exhibited reduced up-regulation of CD40 and CD86 expression and an impaired ability to induce antigen-specific CD4+ T-cell activation.32
c-Fos mRNA levels were found to decrease in a manner that was closely correlated with increased miR155 expression during the activation of human Mo-DCs and various mouse DC subtypes by diverse stimuli. Analysis of data derived from published microarray experiments33-35 confirmed that c-Fos mRNA levels are down-regulated during DC maturation. Furthermore, c-Fos expression was de-repressed in miR155−/− DCs, whereas it was reduced in DCs overexpressing miR155. Finally, the 3′UTR of c-Fos mRNA contains 2 miR155-binding sites and is a direct target of miR155. These results indicate that silencing of c-Fos expression by miR155 is a general mechanism associated with DC maturation.
The importance of miR155-mediated silencing of c-Fos expression in DCs was underscored by the striking similarity between the phenotypic and functional defects displayed by miR155−/− DCs and DCs in which c-Fos expression was removed from its endogenous regulatory controls. Constitutive c-Fos expression in DCs was sufficient to reproduce all of the defects documented for miR155−/− DCs. These findings imply that the abrogation of c-Fos repression by miR155 accounts for many of the functional defects exhibited by miR155−/− DCs.
Earlier studies suggested that c-Fos could inhibit proinflammatory cytokine production by DCs. siRNA-mediated c-Fos knock-down experiments and analyses performed with c-Fos−/− DCs indicated that c-Fos dampens IL12-p70 secretion by human Mo-DCs and mouse splenic DCs stimulated with the TLR2 ligand PAM3CSK4.36-38 TNFα, IL12-p70, and IL6 secretion by mature BM-DCs was also attenuated in a c-Fos–dependent manner by stimuli that raise the intracellular concentration of cAMP.39 Finally, overexpression of c-Fos in BM-DCs dampened IFNβ, IL12-p40, and IL12-p70 production in response to stimulation with CpG.40 The inhibition of proinflammatory cytokine production by c-Fos was of variable magnitude in different systems. It was not evident in mouse splenic DCs activated with zymosan and only weak in splenic and BM-DCs treated with LPS.37-39 Our results are consistent with these findings, because proinflammatory cytokine mRNA induction was attenuated only slightly in miR155−/− DCs activated with LPS, but was reduced more markedly in CpG-treated DC2114 cells overexpressing c-Fos. The strength of the repressive effect of c-Fos on proinflammatory cytokine production could be influenced by various parameters, including the type of DC, the nature and potency of the maturation signal, and the level of c-Fos expression. We have observed that there is a correlation between the level of c-Fos overexpression in DC2114 cells and the extent of inhibition of proinflammatory cytokine mRNA induction (data not shown).
Our results indicate that miR155 regulates both the stability and translation of c-Fos mRNA. Repression at the level of translation appears to be the dominant mechanism, because the increase in c-Fos protein in miR155−/− DCs was considerably greater than the increase in c-Fos mRNA abundance. We have also found that transcription of the c-Fos gene is silenced in Mo-DCs treated with LPS (unpublished data). c-Fos expression in DCs is thus regulated at the levels of transcription, mRNA stability, and translation, suggesting that tightly controlled silencing of c-Fos expression is critical for DC maturation and function.
It remains to be determined whether the repression of c-Fos by miR155 is a mechanism that is specific to DCs. miR155 expression is also induced during the activation of other cell types, including B cells and macrophages. We have observed that c-Fos mRNA levels are strongly decreased in B cells activated with CpG (unpublished data). Down-regulation of c-Fos mRNA expression has also been documented in macrophages stimulated with LPS.41 Repression of c-Fos expression by miR155 may thus be of more general importance for the activation of various cell types.
Two additional miRNAs have been implicated in the regulation of c-Fos expression. miR101 promotes apoptosis by inhibiting c-Fos expression in human hepatocellular carcinoma cells.42 c-Fos translation was also reported to be repressed by miR7b in the hypothalamus after chronic hyperosmolar stimulation.43 Our miRNA expression–profiling experiments indicated that miR101 and miR7b are not expressed at significant levels in immature human or mouse DCs and are not induced upon maturation. It is therefore unlikely that these 2 miRNAs collaborate with miR155 in regulating c-Fos expression in DCs. However, the identification of 3 miRNAs capable of targeting c-Fos mRNA in different systems suggests that the regulation of c-Fos expression by cell-type–restricted miRNAs is a widespread mechanism.
The 3′UTR of c-Fos mRNA contains adenylate- and uridylate-rich elements (ARE) known to promote mRNA degradation.44 A link was recently established between ARE-mediated mRNA decay and regulation by miRNAs.45 This raises the attractive possibility that c-Fos expression could be controlled in a cell-type–specific manner by collaboration between specific miRNAs and the ARE-mediated mRNA decay machinery.
c-Fos functions as one subunit of a group of dimeric transcription factors collectively referred to as activating protein 1 (AP-1). AP-1 proteins constitute a population of homo- and heterodimeric complexes containing subunits belonging to the Fos (c-Fos, FosB, Fra-1, and Fra-2), Jun (c-Jun, JunB, and JunD), and activating transcription factor families. AP-1 factors have been implicated in a wide range of processes, including proliferation, differentiation, apoptosis, responses to stress and environmental cues, oncogenic transformation, and metastasis.46,47 The composition of the population of AP-1 dimers is one of the critical parameters determining AP-1 function in different cell types. Our finding that continued c-Fos expression is detrimental for DC maturation suggests that AP-1 complexes containing c-Fos might repress genes that are implicated in DC maturation. This interpretation is consistent with the recent finding that c-Fos can inhibit TNFα expression by binding to the p65 subunit of NF-kB and inhibiting its recruitment to the Tnf promoter.39 However, a change in the expression of a specific AP-1 subunit could also affect the overall balance between the relative abundance of different AP-1 complexes. An alternative possibility could therefore be that silencing of c-Fos expression induces a shift in the equilibrium between different AP-1 dimers such that the formation of specific AP-1 complexes required for maturation is favored. Distinguishing between these and other possibilities will require a detailed characterization of the composition of the AP-1 population present in immature DCs and the changes that occur in this population when maturation is induced.
The BIC gene was first identified as a frequent integration site of avian leucosis virus in chicken B-cell lymphomas.8,9 It was subsequently found that miR155 expression is frequently up-regulated in B-cell lymphomas and in a wide range of other cancers.10,11 Furthermore, mice carrying a transgene that enforces miR155 expression in B cells are characterized by a preleukemic pre–B-cell proliferation that eventually develops into B-cell malignancy.48 The mechanisms underlying the oncogenic role of miR155 remain obscure, but our results raise the intriguing possibility that these mechanisms could involve deregulated c-Fos expression. Indeed, although oncogenic transformation and tumor progression were initially believed to be associated with increased c-Fos expression, there is growing evidence that c-Fos can also exert tumor-suppressive functions.49 It is therefore tempting to speculate that abrogation of the tumor suppressor influence of c-Fos could contribute to the oncogenic role of miR155 in the development of tumors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Emmanuèle Barras, Antoine Geinoz, Grégory Schneiter, Mahdia Benkhoucha, Patrice Lalive, David Suter, Bertrand Huard, and Mylène Docquier for technical help and to all members of the laboratory for valuable discussions. 2D2 mice were provided by B. Becher (Zurich, Switzerland) with the permission of V. Kuchroo (Boston, MA). OTII mice were provided by S. Amigorena (Paris, France).
Work in the laboratory of W.R. was supported by the Swiss National Science Foundation, the Geneva Cancer League, The Ernst and Lucy Schmidheiny Foundation, the Swiss Multiple Sclerosis Society, the National Center of Competence in Research on Neural Plasticity and Repair (NCCR-NEURO), and the European Union FP6 consortium Dendritic Cells for Novel Immunotherapies (DC-THERA). Work in the laboratory of H.A.-O. and E.M.Z. was supported by the Swiss National Science Foundation. M.-L.S.-R. was supported by the Alliance for Lupus Research.
Authorship
Contribution: I.D.-S. conceived, designed, and performed the experiments and wrote the paper; W.R. conceived and designed the experiments and wrote the paper; L.C. performed experiments; M.I. and Q.S.-E. contributed to the experimental design; H.A.-O. shared reagents and materials; C.E.V., O.S., P.D., and E.M.Z. performed bioinformatical analysis; and M.-L.S.-R. helped with the design of experiments and analysis of data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Walter Reith, Department of Pathology and Immunology, Faculty of Medicine, University of Geneva, 1 rue Michel-Servet, CH-1211 Geneva, Switzerland; e-mail: walter.reith@unige.ch.