Abstract
IL-15 has growth-promoting effects on select lymphoid subsets, including natural killer (NK) cells, NK T cells, intraepithelial lymphocytes (IELs), CD8 T cells, and γδ-T cells. Constitutive expression of murine IL-15 in IL-15–transgenic mice was reported to cause T-NK leukemia. We investigated whether IL-15 expression is sufficient for leukemic transformation using a human IL-15–transgenic (IL-15Tg) mouse model. We noted that 100% of the mice observed over a 2-year period (n > 150) developed fatal expansions of CD8 T cells with NK markers, and determined that these cells expressed IL-15 receptor alpha (IL-15Rα). The expression of IL-15Rα on CD8 T cells appears to be required for uncontrolled aggressive lymphoproliferation, because none of the IL-15Rα−/−–IL-15Tg mice that we followed for more than 2 years developed the fatal disease despite controlled expansion of CD8 T cells. In addition, in contrast to IL-15Tg mice, in which leukemia-like CD8 T cells expressed IL-15Rα persistently, acutely activated normal CD8 T cells only transiently expressed IL-15Rα. Inhibition of DNA methylation enabled sustained IL-15Rα expression induced by activation. We present a scenario for IL-15Tg mice in which CD8 T cells that acquire constitutive persistent IL-15Rα expression are at a selective advantage and become founder cells, outgrow other lymphocytes, and lead to the establishment of a leukemia-like condition.
Introduction
IL-15 belongs to a family of cell growth–promoting cytokines (IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21) that share the common γ subunit (γc, CD132).1-4 Despite this sharing of receptor components, each cytokine also possesses unique, nonredundant functions, as demonstrated by the study of gene-manipulated mice lacking the individual cytokine or its receptor components. Mice lacking IL-15 have a severely reduced number of natural killer (NK) cells, CD1d-restricted NK-T cells, γδ-T cells, intraepithelial lymphocytes (IELs), and select subsets of CD8 T cells.5 In particular, NK and CD8 T cells seem to be the primary targets of IL-15 action in vivo, because administration of recombinant IL-15 to mice led to a rapid expansion of NK and CD44hiCD8 T cells.6 Moreover, IL-15 has been suggested to augment the cytotoxic function of NK and CD8 T cells.7,8 These observations and studies of IL-15 in murine tumor models led us to initiate a clinical strategy that involved administration of recombinant IL-15 to humans to facilitate immunologic eradication of metastatic melanoma and metastatic renal cell cancer. In addition, IL-15 is known to enhance the survival of memory CD8 T cells,9,10 an action that could be exploited to enhance the efficacy of engineered vaccines that incorporate IL-15. Despite these promising prospects for clinical use of IL-15, one previous study raised concerns related to the safety of IL-15 use in humans, because constitutive expression of murine IL-15 in human IL-15–transgenic (IL-15Tg) mice led to the rapid development of fatal CD8/T-NK leukemia.11 Recently, the investigators involved in this study further characterized the model and demonstrated that 19 of 39 leukemic mice studied had NK leukemia and 20 had T-cell large granular lymphocyte leukemia.12 We generated a transgenic mouse that constitutively expresses human IL-15 (hIL-15) systemically.13 We report that a similar fatal leukemia–like condition of CD8 T cells with NK markers occurred at later time points in the lives of our IL-15Tg mice. We also report that the development of such a lymphoid proliferative disorder requires an additional event that is independent of constitutive IL-15 production, suggesting that 2 independent mechanisms are involved in the progress of uncontrolled and aggressive lymphoid expansion. In other words, the leukemia-like condition did not develop solely by autocrine production of IL-15. Furthermore, we observed that IL-15 receptor α (IL-15Rα) expression was absent in nonactivated CD8 T cells of young mice, presumably because of an epigenetic mechanism, and that only CD8 T cells that had experienced a breach in this barrier and acquired persistent expression of IL-15Rα became founder cells for the fatal T-cell-leukemia–like disease. Because this event seems to require a long time to develop, our work provides evidence that simple, short-term administration of recombinant IL-15 to humans would not present a meaningful risk for leukemia development.
Methods
Mice
Human IL-15Tg mice were generated previously by our group.13 We have established 2 strains (B1 and K2, expressing 100-150 pg/mL and 600-800 pg/mL of serum hIL-15, respectively), both of which succumb to fatal leukemia. Human IL-15RαTg mice were also generated by our group.14 IL-15Rα−/− mice (The Jackson Laboratory) were backcrossed to the C57/B6 strain for more than 15 additional generations. All animal experiments were performed according to an animal experiment protocol that was approved by the National Cancer Institute animal care and use committee.
Antibodies and reagents
Most antibodies (Abs) for flow cytometry experiments and the TMβ1 anti–mouse IL-2R/IL-15Rβ monoclonal Ab were purchased from eBioscience. 5-Azacytidine (5-Aza; Calbiochem) was dissolved at 10mM in 100% methanol. The hIL-15 ELISA kit, neutralizing anti–hIL-15 Ab (MAB247), and biotinylated anti-mouse IL-15Rα Ab (BAF551) for flow cytometry were from R&D Systems. Anti-STAT5a/b Ab (C-18; Santa Cruz Biotechnology) was used for Western blotting and supershift assays. Anti-phosphoSTAT5a Ab was from Cell Signaling Technology. Recombinant hIL-15 and mouse GM-CSF were purchased from Peprotech. IFN-γ was from R&D Systems and poly(I:C) and lipopolysaccharide (LPS) were from Sigma-Aldrich. OVA peptide (257-264; SIINFEKL) was purchased from AnaSpec and OVA tetramer from Beckman-Coulter.
Stimulation of cells
CD8 T cells that were purified from C57/B6 wild-type (WT) mice (MACS; Miltenyi Biotec) were activated by plate-coated anti-CD3 (2C11, 10 μg/mL) and anti-CD28 (5 μg/mL) Abs with or without hIL-15 (5nM) for 48 hours. BM-derived dendritic cells grown in GM-CSF (10 ng/mL) were stimulated with IFN-γ (20 ng/mL) and LPS (60 ng/mL) for 24 hours or loaded with OVA peptide (1 μg/mL) in the presence of LPS (5 ng/mL) overnight.
Cellular proliferation assay
Fifty thousand cells were incubated with the appropriate reagents (ie, cytokines or antibodies) in a flat-bottom, 96-well plate for 44 hours, followed by 4 hours of incubation with 1 μCi 3H-thymidine (GE Healthcare). The incorporated β-emissions were measured using a β-counter (PerkinElmer).
Immunoblotting
Fifteen micrograms of cellular lysate in radioimmunoprecipitation assay buffer was resolved using an 8% SDS gel (Invitrogen), transferred to polyvinylidene fluoride membranes, and then probed with an anti–phospho-STAT5a Ab (0.2 μg/mL).
Electrophoresis mobility shift assay (EMSA)
Complementary single-stranded oligonucleotides were commercially synthesized (Invitrogen) with a 10 extra base pairs flanking the STAT5 target sequence from the il2 gene.15 The primers used were 5′-agttattagaaatTTCAAGGAAgtgacaacagag-3′ (STAT5 WT), 5′agttattagaaatTTCAACCTTgtgacaacaga-3′ (STAT5 mutant, mutation underlined). Capital letters represent the STAT5 motif. Two micrograms of the complementary-strand oligonucleotides were annealed and 32P-labeled. A 20-μL mixture containing 10 μg of nuclear proteins (see “DNase I hypersensitivity assay”), 1 μg of poly(dI-dC) (Sigma), and a 32P-probe (30 000 cpm) were prepared in 15mM HEPES, 1.5mM MgCl2, 60mM KCl, 0.1mM EDTA, 0.5mM DTT, and 10% glycerol. For the supershift assay, 1 μL of Ab was added and incubated on ice for 15 minutes before addition of the probe.
RNA extraction and Northern blot analysis
Total cellular RNA was extracted using the Gentra RNA extraction kit (QIAGEN). Twenty micrograms of RNA was resolved in 1.2% formaldehyde agarose gel and transferred to a Hybond HL membrane (GE Healthcare). A 200-bp mouse IL-15Rα probe was amplified by PCR (SN: 5′-gcaggagctacagggacaggc-3′, AN: 5′-ccaggccaggaaagccatcac-3′). Because human and mouse IL-15Rα display the lowest homology at this region, the probe would not detect human IL-15Rα mRNA expressed from the human IL-15Rα transgene. The probe was labeled using 32P-dCTP (GE Healthcare), random 9-mer oligonucleotides, and the Klenow enzyme (Roche Applied Science).
Southern blotting analysis
Genomic DNA was prepared from cells using a Gentra DNA-extraction kit (QIAGEN). Ten micrograms of DNA was digested with HindIII or AleI restriction enzymes for 12 hours at 37°C, with HindIII addressing constant region 1 variations and AleI both constant region 1 and 2 variations. The digested samples were resolved in a 0.65% Tris-acetate EDTA agarose gel and transferred to a Hybond HL membrane using 10× saline-sodium citrate. The TCRβ rearrangement pattern was determined using a 32P-labeled probe that had been PCR-amplified from the C57/B6 mouse tcrb locus (primers, SN: 5′-gagaaatgtgactccacccaaggt-3′, AS: 5′-caggcctctgcactgatgttctgt-3′) by hybridization.16
DNase I hypersensitivity assay
Cytoplasmic compartments were stripped from cells by incubation in a cold lysis buffer (50mM KCl, 25mM HEPES, pH 7.8, 0.5% NP40, 1mM PMSF, 10 μg/mL of leupeptin, 20 μg/mL of aprotinin, and 0.1mM DTT) for 5 minutes on ice, centrifuged at 2000g, and washed twice in a wash buffer (lysis buffer without NP40). Isolated nuclei were digested with 1 U/mL of DNase I (Roche) at 37°C for 5 minutes before DNA extraction using the standard phenol-chloroform method.
ChIP
Genomic DNA prepared from cells was sonicated on ice using 15 pulses of 20 seconds each with a 20-second interval between each pulse. PCR primers were designed to flank the first CpG island (SN: 5′-ttgaaatctgaaggtcagcagcca-3′, AS: 5′- tgctttagcccccggattgcagag-3′). Chromatin immunoprecipitation (ChIP) was performed using the MethylCollector kit (Active Motif) according to manufacturer's instructions. Fifty nanograms of sonicated DNA was incubated with the His–anti-MBP2 Ab, and MBP2-bound fractions were recovered by elution. Two-hundredths (0.25 ng of DNA equivalent) of the total amount of DNA before and after Ab incubation was used for semiquantitative PCR (28 cycles of amplification).
Densitometry analysis
ImageJ 1.44m software (National Institutes of Health) was used for densitometry analysis.
Statistical analysis
P values were calculated using StatView 5.0.1 software (Adept Scientific). A standard Student t test was applied to the proliferation and weight measurement assays and a log-rank test to the survival plots.
Results
Development of a fatal CD8 T-cell leukemia in IL-15Tg mice
Fehniger et al first reported that constitutive expression of murine IL-15 in mice led to the development of a CD8/T-NK type leukemia by 5 months of age.11 Conversely, the IL-15Tg mice expressing hIL-15 (under the human EF1α promoter)13 that we generated only manifested a benign expansion of lymphoid subsets at 5 months. These expanded cells included CD44hiCD8, NK, and novel, non-IEL CD8NK+ T cells.17 However, the 2 IL-15Tg mouse strains that we established displayed considerably shorter life spans than their control littermates (Figure 1A). This was true even though the strains13 differed in serum hIL-15 concentrations (600-800 pg/mL in the K2 strain vs 100-150 pg/mL in the B1 strain), integration number, and positions of the hIL-15 genes (data not shown). In 100% of IL-15Tg mice examined (n > 150 for both strains), the benign expansions of CD8 T cells were followed by an acute explosive expansion of CD8 T cells, which we called the terminal stage. At the terminal stage, CD44hiCD8 T cells reached ∼ 100% of lymphoid compartments and completely displaced other lymphocytes, exhibiting a level of > 1.2 × 105 cells/mm3 in the circulation (Figure 1B). We subjected these expanded cells to a standard in vitro culture without any supplements and established 2 CD8 T-cell clones: B1-001, referred to hereafter as B1, and K2-001, referred to hereafter as K2.
The human IL-15Tg mice developed a fatal lymphoproliferative disorder. (A) Survival plots of IL-15Tg mice on different backgrounds. IL-15Tg (B1) and IL-15Rα−/−IL-15Tg mice were compared in terms of their life span with IL-15Tg mice showing a shortened survival (n = 12, P < .001). IL-15Rα–IL-15 double-Tg and IL-15Tg mice were compared as well (n = 15, P < .001). IL-15Rα−/−-IL-15Tg mice lived as long as their littermates, whereas IL-15Rα–IL-15 double-Tg mice died at a very young age because of uncontrolled expansion of CD8 T cells. (B) Expansion of CD44hiCD8 T cells in the IL-15Tg mice at the benign and terminal stages of leukemia. Splenocytes from a WT mouse (i), a young IL-15Tg (3-month-old) mouse (ii), a 13-month-old IL-15Tg mouse (iii), a 13-month-old IL-15Rα−/−IL-15Tg mouse (iv), and a 3-month-old IL-15Rα–IL-15 double-Tg mouse (v) were stained with anti-CD3, anti-CD44, and anti-CD8 Abs and analyzed by flow cytometry. The majority (> 95%) of the splenocytes expressed CD3 and CD8 in the terminal-stage leukemic IL-15Tg mice (iii), whereas in the benign-phase IL-15Tg mice (ii) and IL-15Rα−/−IL-15Tg mice (iv), CD8 T cells maintained an altered but stable homeostasis (with 80% and 44% of CD8+ cells in the CD3-expressing population, respectively). In the terminal-phase leukemic IL-15Tg mice (iii), T cells almost completely displaced other types of cells from the spleen. In the IL-15Rα–IL-15 double-Tg mice (v), even young mice (< 3 months old) displayed displacement of peripheral lymphocytes with CD8 T cells (> 92% of the T-cell compartment). Data represent results from 4-6 mice per group (see supplemental Figure 2 for a summary of all data). (C) Development of lymphoma/leukemia in WT mice transplanted with B1 (B1-001) IL-15Tg cells. Four weeks after the IV injection of 1 × 106 B1 leukemic cell line cells, the WT recipient mice developed tumor masses in their spleens (left, indicated with an arrow). As a surrogate marker to monitor leukemic cell expansion, the concentrations of hIL-15 in the sera of transplanted mice were determined by a specific IL-15 ELISA (right, n = 8). (D) Constitutive phosphorylation and DNA binding of STAT5 in leukemic cells. In the left panel, constitutive tyrosine-phosphorylation of the STAT5a molecules in the ex vivo B1 leukemic cells (left lane) was demonstrated using an anti-pSTAT5a Ab by immunoblotting. An anti-STAT5a/b Ab was used to quantitate the amount of the STAT protein in each lane. In the right panel, STAT5 shows constitutive DNA binding in the ex vivo B1 leukemic cells as demonstrated using an EMSA assay. The target oligonucleotides were 5′-agttattagaaatTTCAAGGAAgtgacaacagag-3′ (STAT5 WT), 5′agttattagaaatTTCAACCTTgtgacaacaga-3′ (STAT5 mutant, mutation underlined), which were annealed, 32P-labeled, and used as the probe. An anti-STAT5a/b Ab was used to supershift the DNA-protein complexes. The lanes indicate without (i) and with (ii) anti-STAT5a/b Ab. (E) Confirmation of the IL-15 dependency of the leukemic cells. Inclusion of the anti–hIL-15 Ab (MAB247; R&D Systems) abrogated the constitutive proliferation of IL-15Tg leukemic cells as assessed by [3H]-thymidine incorporation, suggesting that these cells depend on an autocrine supply of IL-15 for their growth and survival. The addition of anti-mouse IL-2/15Rβ Ab (TMβ1) led to a partial inhibition with B1/K2 clones, whereas the inhibition by TMβ1 was complete, with normal CD8 T cells stimulated with hIL-15. The data represent 3 independent experiments (n = 3 each).
The human IL-15Tg mice developed a fatal lymphoproliferative disorder. (A) Survival plots of IL-15Tg mice on different backgrounds. IL-15Tg (B1) and IL-15Rα−/−IL-15Tg mice were compared in terms of their life span with IL-15Tg mice showing a shortened survival (n = 12, P < .001). IL-15Rα–IL-15 double-Tg and IL-15Tg mice were compared as well (n = 15, P < .001). IL-15Rα−/−-IL-15Tg mice lived as long as their littermates, whereas IL-15Rα–IL-15 double-Tg mice died at a very young age because of uncontrolled expansion of CD8 T cells. (B) Expansion of CD44hiCD8 T cells in the IL-15Tg mice at the benign and terminal stages of leukemia. Splenocytes from a WT mouse (i), a young IL-15Tg (3-month-old) mouse (ii), a 13-month-old IL-15Tg mouse (iii), a 13-month-old IL-15Rα−/−IL-15Tg mouse (iv), and a 3-month-old IL-15Rα–IL-15 double-Tg mouse (v) were stained with anti-CD3, anti-CD44, and anti-CD8 Abs and analyzed by flow cytometry. The majority (> 95%) of the splenocytes expressed CD3 and CD8 in the terminal-stage leukemic IL-15Tg mice (iii), whereas in the benign-phase IL-15Tg mice (ii) and IL-15Rα−/−IL-15Tg mice (iv), CD8 T cells maintained an altered but stable homeostasis (with 80% and 44% of CD8+ cells in the CD3-expressing population, respectively). In the terminal-phase leukemic IL-15Tg mice (iii), T cells almost completely displaced other types of cells from the spleen. In the IL-15Rα–IL-15 double-Tg mice (v), even young mice (< 3 months old) displayed displacement of peripheral lymphocytes with CD8 T cells (> 92% of the T-cell compartment). Data represent results from 4-6 mice per group (see supplemental Figure 2 for a summary of all data). (C) Development of lymphoma/leukemia in WT mice transplanted with B1 (B1-001) IL-15Tg cells. Four weeks after the IV injection of 1 × 106 B1 leukemic cell line cells, the WT recipient mice developed tumor masses in their spleens (left, indicated with an arrow). As a surrogate marker to monitor leukemic cell expansion, the concentrations of hIL-15 in the sera of transplanted mice were determined by a specific IL-15 ELISA (right, n = 8). (D) Constitutive phosphorylation and DNA binding of STAT5 in leukemic cells. In the left panel, constitutive tyrosine-phosphorylation of the STAT5a molecules in the ex vivo B1 leukemic cells (left lane) was demonstrated using an anti-pSTAT5a Ab by immunoblotting. An anti-STAT5a/b Ab was used to quantitate the amount of the STAT protein in each lane. In the right panel, STAT5 shows constitutive DNA binding in the ex vivo B1 leukemic cells as demonstrated using an EMSA assay. The target oligonucleotides were 5′-agttattagaaatTTCAAGGAAgtgacaacagag-3′ (STAT5 WT), 5′agttattagaaatTTCAACCTTgtgacaacaga-3′ (STAT5 mutant, mutation underlined), which were annealed, 32P-labeled, and used as the probe. An anti-STAT5a/b Ab was used to supershift the DNA-protein complexes. The lanes indicate without (i) and with (ii) anti-STAT5a/b Ab. (E) Confirmation of the IL-15 dependency of the leukemic cells. Inclusion of the anti–hIL-15 Ab (MAB247; R&D Systems) abrogated the constitutive proliferation of IL-15Tg leukemic cells as assessed by [3H]-thymidine incorporation, suggesting that these cells depend on an autocrine supply of IL-15 for their growth and survival. The addition of anti-mouse IL-2/15Rβ Ab (TMβ1) led to a partial inhibition with B1/K2 clones, whereas the inhibition by TMβ1 was complete, with normal CD8 T cells stimulated with hIL-15. The data represent 3 independent experiments (n = 3 each).
CD8 T-leukemic cells are transferrable into WT mice
We demonstrated that these leukemic cells and cell lines (B1 and K2) could be transplanted into WT syngeneic mice and cause a secondary leukemia. Because the IL-15Tg mice were generated on the Ly5.2+ C57/B6 background, congenic hosts (Ly5.1+ C57/B6) were used in transfer experiments. We injected 1 × 106 cells of cultured leukemic clones (B1 and K2) or CD8 T cells recovered from IL-15Tg mice in the terminal phase. These recipient mice died within 3-5 weeks of injection (B1 cells, n = 8; Figure 1C) or by 24 weeks (K2 cells). Flow cytometric results of the Ly5.2-gated cells (ie, representing leukemic cells; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) indicated that these cells retained the initial surface phenotypes in recipients (CD2−, CD69hi, NK1.1+ Ly49ADG+, NKG2D+, KLRG1+). These cells demonstrated constitutive tyrosine phosphorylation of Jak3 kinase (data not shown) and the STAT5a molecule, and STAT5 showed constitutive binding to DNA in B1 leukemic cells in an EMSA assay (Figure 1D). The inclusion of anti–IL-15 Ab (MAB247) to ex vivo cultures abrogated proliferative responses (Figure 1E), confirming that these cells were still dependent on IL-15 for their survival and growth. Interestingly, the addition of anti–murine IL-2/IL-15Rβ Ab (TMβ1) had only marginal effects on the proliferation of leukemic cells, but effectively blocked the proliferation of normal activated CD8 T cells in response to IL-15. This observation was intriguing, because in cell lines that express IL-15Rα as the heterotrimeric IL-15R (α/β/γc) in cis, IL-15–induced proliferations were not effectively inhibited by the addition of TMβ1 Ab (Y.T., unpublished observation), which led us to speculate that the leukemic cells might express IL-15Rα on their surfaces. Thus, we have created a true cytokine-dependent and transferrable leukemic system that is a versatile resource for the mechanistic study of lymphoid malignant transformation.
Two phases of the CD8-cell expansion in IL-15Tg mice: a benign homeostatic phase and a terminal malignant phase
Mice of both IL-15Tg strains (B1 and K2) inherently manifest an expansion of NK cells and a preferential expansion of CD44hiCD8 T-cell subsets in their peripheral lymphoid organs.13 The thymic CD4/8 lineage decision was not affected.13 In the periphery, the CD4/CD8 ratio was altered to 1:1 in the B1 strain and 1:5 in the K2 strain. This altered ratio appeared stable for several months (Figure 1B and supplemental Figure 2), suggesting that excess cytokine only created an altered equilibrium between different lymphoid subsets (ie, CD4 vs CD8); we called this the benign altered homeostatic phase. When we compared the surface phenotypes of expanded CD8 T cells between the benign and the terminal phases of leukemia-like disorders, we identified one peculiar difference: only the CD8 T cells from the terminal stage expressed IL-15Rα (Figure 2A), a result consistent with the efficient inhibition by the anti–IL-15 Ab, but inefficient inhibition of their autocrine proliferation by the TMβ1 Ab.
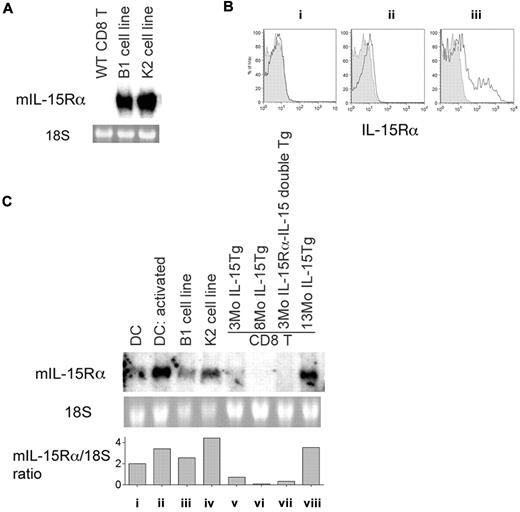
IL-15Rα is expressed on leukemic CD8 T cells from IL-15Tg mice. (A) Expression of mouse IL-15Rα mRNA in leukemic cell lines but not in normal CD8 T cells. Twenty micrograms of total RNA was resolved in a formaldehyde agarose gel and transferred to a nylon membrane, which was probed with a 32P-labeled mouse IL-15Rα fragment. As a loading control, the 18S ribosomal RNA was visualized with ethidium bromide staining. Origins of RNA were CD8 T cells from normal WT mice, from the B1 leukemia cell line, and from the K2 leukemia cell line. (B) Detection of IL-15Rα expression by an anti-mouse IL-15Rα antibody in CD8 T cells from IL-15Tg mice. IL-15Rα expression on CD8 T cells from IL-15Tg mice of different ages (solid lines) with staining of CD8 T cells from an IL-15Rα−/− mouse shown as a control (shaded area). Results represent 3-month-old (i), 9-month-old (ii), and leukemic 19-month-old (iii) IL-15Tg mice, with 3-6 mice in each age group. The averages of IL-15Rα mean fluorescent intensity in IL-15Rα−/− mice and IL-15Tg mice in groups i, ii, and iii were 4.2 ± 0.9, 6.8 ± 0.8, 7.6 ± 1.4, and 33.4 ± 10.2, respectively. (C) Acquisition of IL-15Rα expression in the leukemic phase, but not in the benign phase, by CD8 T cells in IL-15Tg mice. RNA was extracted from CD8 T cells purified from spleens, and 10 μg total RNA was loaded onto each lane and probed with a 32P-mouse IL-15Rα fragment. Origins of RNA were resting dendritic cells (DCs) (i), activated (poly I:C) DCs (ii), cells from the B1 cell line (iii), cells from the K2 cell line (iv), CD8 T cells from a young (3-month-old) IL-15Tg mouse (v), CD8 T cells from an 8-month-old IL-15Tg mouse (vi), IL-15Rα–IL-15 double-Tg cells from a 3-month-old mouse (vii), and CD8 T cells from a leukemic 13-month-old IL-15Tg mouse (viii). Densitometry analysis was performed, and the levels of IL-15Rα RNA expression normalized to that of 18S ribosomal RNA are indicated.
IL-15Rα is expressed on leukemic CD8 T cells from IL-15Tg mice. (A) Expression of mouse IL-15Rα mRNA in leukemic cell lines but not in normal CD8 T cells. Twenty micrograms of total RNA was resolved in a formaldehyde agarose gel and transferred to a nylon membrane, which was probed with a 32P-labeled mouse IL-15Rα fragment. As a loading control, the 18S ribosomal RNA was visualized with ethidium bromide staining. Origins of RNA were CD8 T cells from normal WT mice, from the B1 leukemia cell line, and from the K2 leukemia cell line. (B) Detection of IL-15Rα expression by an anti-mouse IL-15Rα antibody in CD8 T cells from IL-15Tg mice. IL-15Rα expression on CD8 T cells from IL-15Tg mice of different ages (solid lines) with staining of CD8 T cells from an IL-15Rα−/− mouse shown as a control (shaded area). Results represent 3-month-old (i), 9-month-old (ii), and leukemic 19-month-old (iii) IL-15Tg mice, with 3-6 mice in each age group. The averages of IL-15Rα mean fluorescent intensity in IL-15Rα−/− mice and IL-15Tg mice in groups i, ii, and iii were 4.2 ± 0.9, 6.8 ± 0.8, 7.6 ± 1.4, and 33.4 ± 10.2, respectively. (C) Acquisition of IL-15Rα expression in the leukemic phase, but not in the benign phase, by CD8 T cells in IL-15Tg mice. RNA was extracted from CD8 T cells purified from spleens, and 10 μg total RNA was loaded onto each lane and probed with a 32P-mouse IL-15Rα fragment. Origins of RNA were resting dendritic cells (DCs) (i), activated (poly I:C) DCs (ii), cells from the B1 cell line (iii), cells from the K2 cell line (iv), CD8 T cells from a young (3-month-old) IL-15Tg mouse (v), CD8 T cells from an 8-month-old IL-15Tg mouse (vi), IL-15Rα–IL-15 double-Tg cells from a 3-month-old mouse (vii), and CD8 T cells from a leukemic 13-month-old IL-15Tg mouse (viii). Densitometry analysis was performed, and the levels of IL-15Rα RNA expression normalized to that of 18S ribosomal RNA are indicated.
Progressive and aggressive lymphoproliferative disorder in IL-15Tg mice was associated with IL-15Rα expression on CD8 T cells
The relatively long duration of the benign expansion phase in IL-15Tg mice prompted us to postulate that a critical secondary event might accompany the constitutive presence of IL-15 for aggressive, uncontrolled lymphoproliferation to occur. We also postulated that such an event might have some connection with the IL-15R system. Because established leukemic cells expressed the IL-15Rα molecule (Figure 2A), and because the growth of leukemia-like CD8 T cells from IL-15Tg mice was only partially inhibited by the IL-2R/IL-15Rβ antibody, we investigated whether IL-15Rα was expressed by CD8 T cells from various stages of IL-15Tg mice. Flow cytometry (Figure 2B; n = 3-6) and conventional Northern blot analyses were used to analyze IL-15Rα expression levels. Figure 2B-C shows selective expression of murine IL-15Rα molecules and transcripts, respectively, in CD8 T cells purified from the lymphoid organs of IL-15Tg mice of different ages. Only cells from terminal-stage mice expressed high levels of IL-15Rα transcripts and IL-15Rα protein.
Requirement of IL-15Rα expression for IL-15–mediated, uncontrolled lymphoid expansion
As shown above, a close connection between IL-15Rα expression and the leukemic nature of CD8 T cells was observed. To further address this issue, we used a strategy to force either overexpression or null expression of IL-15α in mice on the IL-15Tg background and examined the effects of such manipulations on leukemogenesis. We crossed the K2 strain of IL-15Tg mice with IL-15Rα−/− mice. Our first concern was that the abrogation of IL-15Rα could lead to nonexpression of IL-15 in mice based on reported observations that these 2 molecules need to be coexpressed for stable expression of IL-15.18-20 However, we detected ∼ 350 pg/mL of IL-15 in their sera, presumably because the engineered IL-15 was human in origin and that hIL-15 production/secretion is less dependent on the coexpression of IL-15Rα (see “Discussion”). We were also concerned that lymphocytes would not be capable of responding to circulating IL-15 in the absence of IL-15Rα. This possibility was excluded because IL-15Rα−/−IL-15Tg mice maintained biased but controlled CD8 expansion (Figure 1B). All of the generated IL-15Rα−/−IL-15Tg mice (n = 31) survived for more than 15 months (Figure 1A) without manifesting aggressive signs of uncontrolled lymphoproliferation, suggesting that IL-15Rα expression by CD8 T cells was critically required for pathologic transformation in mice.
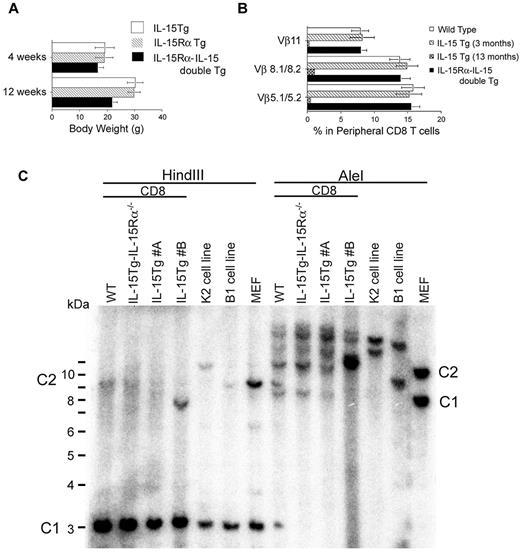
We then examined the consequence of aberrant expression of IL-15Rα on CD8 T cells in IL-15Tg mice by crossing IL-15Tg mice with human IL-15RαTg mice.14 We could not obtain viable litters when we crossed the high IL-15–producing strain K2 with IL-15RαTg mice. However, the lower IL-15–expressing strain, B1, when crossed with IL-15RαTg mice, gave birth to litters and the frequency of double-Tg mice followed a Mendelian distribution. The double-Tg litters were smaller in size than their single-Tg littermates (Figure 3A), and exhibited gross abnormalities such as rough coats, runted postures, and death at an early age (average life span ∼ 2 months, n = 15; Figure 1A). Peripheral lymphoid tissues of such mice displayed massive lymphocytosis, with CD44hiCD8 cells occupying > 95% of the peripheral lymphoid compartment (Figure 1B). Curiously, even at the terminal stages, the CD8 T cells from these IL-15Rα–IL-15 double-Tg mice remained polyclonal (Figure 3B).
Monoclonality of certain IL-15Tg leukemic cells and polyclonal CD8 T-cell expansion was demonstrated in IL-15Rα–IL-15 double-Tg mice. (A) Smaller body weights of IL-15Rα–IL-15 double-Tg mice compared with IL-15Tg and IL-15RαTg mice. The body weights of IL-15Tg, IL-15RαTg, and IL-15Rα–IL-15 double-Tg litters (n = 5 for each group) were compared. At 12 weeks and later time points (not shown), there was a significant difference between the double- and single-Tg groups (P < .05). (B) Flow cytometric assessment of the clonality of CD8 T cells in IL-15Rα–IL-15 double-Tg mice. To assess whether the CD8 T cells in IL-15Rα–IL-15 double-Tg mice maintained polyclonality based on their usage of the TCR repertoire, cells were analyzed by flow cytometry using anti–Vβ5.1/5.2, anti–Vβ8.1/8.2, and anti–Vβ11 Abs. If polyclonal, the percentages of staining for the Vβ should be 8%-15%, whereas the disappearance of this subset indicates the loss of polyclonality in the 13-month-old IL-15Tg mouse. (C) Genomic Southern blot analysis to assess the clonality of the CD8 T cells from IL-15Tg mice. DNA was extracted from CD8 T cells purified from the spleen and lymph nodes of WT mice, IL-15Rα−/−IL-15Tg mice, a terminal-stage IL-15Tg mouse #A, a terminal-stage IL-15Tg mouse #B, or cells from the K2 leukemia cell line or the B1 leukemic cell line and a germline control (mouse embryonic fibroblast). Ten micrograms of each DNA was digested with HindIII or AleI enzymes and loaded to each lane. A 32P-labeled TCRβ constant-region fragment was used as a probe. The positions of constant region 1 and 2 in the germline control are indicated as C1 and C2, respectively.
Monoclonality of certain IL-15Tg leukemic cells and polyclonal CD8 T-cell expansion was demonstrated in IL-15Rα–IL-15 double-Tg mice. (A) Smaller body weights of IL-15Rα–IL-15 double-Tg mice compared with IL-15Tg and IL-15RαTg mice. The body weights of IL-15Tg, IL-15RαTg, and IL-15Rα–IL-15 double-Tg litters (n = 5 for each group) were compared. At 12 weeks and later time points (not shown), there was a significant difference between the double- and single-Tg groups (P < .05). (B) Flow cytometric assessment of the clonality of CD8 T cells in IL-15Rα–IL-15 double-Tg mice. To assess whether the CD8 T cells in IL-15Rα–IL-15 double-Tg mice maintained polyclonality based on their usage of the TCR repertoire, cells were analyzed by flow cytometry using anti–Vβ5.1/5.2, anti–Vβ8.1/8.2, and anti–Vβ11 Abs. If polyclonal, the percentages of staining for the Vβ should be 8%-15%, whereas the disappearance of this subset indicates the loss of polyclonality in the 13-month-old IL-15Tg mouse. (C) Genomic Southern blot analysis to assess the clonality of the CD8 T cells from IL-15Tg mice. DNA was extracted from CD8 T cells purified from the spleen and lymph nodes of WT mice, IL-15Rα−/−IL-15Tg mice, a terminal-stage IL-15Tg mouse #A, a terminal-stage IL-15Tg mouse #B, or cells from the K2 leukemia cell line or the B1 leukemic cell line and a germline control (mouse embryonic fibroblast). Ten micrograms of each DNA was digested with HindIII or AleI enzymes and loaded to each lane. A 32P-labeled TCRβ constant-region fragment was used as a probe. The positions of constant region 1 and 2 in the germline control are indicated as C1 and C2, respectively.
Furthermore, in IL-15 single-Tg mice, we observed by Southern blotting both monoclonal leukemia and polyclonal lymphoid expansion, which were undistinguishable in disease course (Figure 3C). These findings suggest that monoclonality of the CD8 T cells was not a prerequisite for fatal expansion of CD8 T cells. However, the acquisition of IL-15Rα expression by the leukemic founder CD8 T cells does seem to be critical for this uncontrolled fatal expansion in the high IL-15 environment. In fact, we observed accelerated CD8 T-cell expansion in IL-15Tg mice, leading to their quick death, after the emergence CD8 T cells with further up-regulated IL-15Rα expression. Although the coexpression of IL-15 and IL-15Rα appears to be necessary for leukemic transformation, it does not appear to be sufficient for the transition to monoclonality.
Potential epigenetic silencing of IL-15Rα gene expression in normal resting CD8 T cells
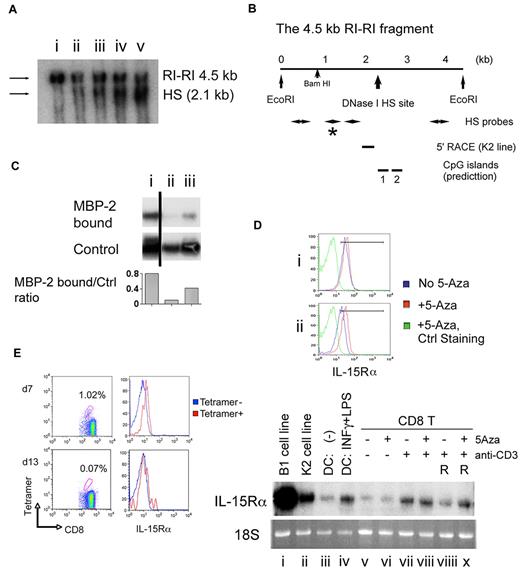
As a first step to delineating the mechanisms that distinguish normal and leukemic CD8 T cells in terms of their IL-15Rα expression, we performed a DNase I hypersensitivity assay.21 A single hypersensitivity site (HS) was identified in the immediate upstream region of the proposed transcription initiation site for the il15rα gene (supplemental Figure 3) in CD8 T cells from leukemic IL-15Tg mice (Figure 4A lanes ii-iii) and the leukemic cell lines (K2 and B1 clones; Figure 4A lanes iv-v). This HS was not identified in normal, resting CD8 T cells (Figure 4 A lane i). Interestingly, when we subjected this region to an algorithmic analysis (Methprimer22 ) that detects the presence of potential CpG islands, we found an overlap between the DNase I HS and one of the predicted CpG islands (Figure 4B). We then hypothesized that shedding methyl-binding proteins from this region might underlie the persistently relaxed chromatin conformation. To test this hypothesis, we performed a ChiP assay and observed that this region of chromatin from resting normal CD8 T cells was bound by the MBP-2 protein, whereas the region was unbound by MBP-2 protein in CD8 T cells that were freshly purified from leukemic IL-15Tg mice or K2 leukemic clone cells. These observations suggest the possible explanation that demethylation of the CpG island (Figure 4C) renders chromatin into a relaxed formation in leukemic CD8 T cells, which may lead to the recruitment of various enhancers and promoters of the transcription machinery to the il15rα gene.
An epigenetic mechanism controls the transcription of the il15rα gene in normal and leukemic CD8 T cells into negative and positive directions. (A) Presence of a DNase I HS site in IL-15Tg leukemic cells and its position in the transcription control site of the il15rα gene. Intact nuclei were isolated from purified CD8 T cells from terminally leukemic IL-15Tg mice (n = 2) and from K2 and B1 leukemic cells, and subjected to 1 U/mL of DNase I for 15 minutes at 37°C. The genomic DNA was extracted subsequently and digested by the EcoRI enzyme. The digested DNA was subjected to 0.7% Tris-acetate EDTA gel electrophoresis, blotted, and hybridized with a 32P-labeled fragment (as shown by the asterisk in panel B) from the il15rα transcription initiation site. As a control, CD8 T cells purified from WT mice were activated by plate-coated anti-CD3/anti-CD28 Abs and hIL-15 (5nM; PeproTech) for 48 hours before DNase I treatment. The sources of genomic DNAs were: normal CD8 T cells (i), CD8 T cells from leukemic IL-15Tg mouse #1 (ii), CD8 T cells from leukemic IL-15Tg mouse #2 (iii), cells from the K2 leukemic cell line (iv), and cells from the B1 leukemic cell line (v). The size of the upper band (EcoRI-EcoRI fragment, intact) was 4.5 kb, and the lower (DNase I cleaved) band migrated at 2.1 kb. (B) Determination of the positions of the HS site and CpG islands that were predicted by a computer algorithm. In the 4.5-kb EcoRI-EcoRI fragment, as shown in supplemental Figure 2, the HS (2.1 kb downstream of the 5′ EcoRI site) site was localized at the 3′ end of exon 1 (∼ 350 bp). The MethPrimer algorithm predicted the presence of 2 CpG islands: CpG1 was localized 2.1-2.25 kb downstream of the 5′ EcoRI site, and the CpG2 2.8-2.9 kb downstream of the 5′ EcoRI site. (C) ChIP revealed a potential hypermethylation of this gene region in resting normal CD8 T cells. A small aliquot of the sonicated genomic DNA (0.25 ng) from before and after the enrichment of the MBP2-bound DNA fractions was used. PCR was performed using a primer set that flanks the first CpG island. The reaction was terminated after 28 cycles of amplification in the semilog amplification phase for a quantitative comparison. Sources of DNA were: resting CD8 T cells (i), CD8 T cells from a leukemic IL-15Tg mouse (ii), and cells from the K2 leukemic cell line (iii). The black bar indicates that distant lanes from the same experiment were combined. Densitometry analysis was performed and the amount of MBP-2 bound DNA normalized to that of control DNA is indicated. (D) In vitro activation of normal CD8 T cells temporally induced IL-15Rα, and treatment with 5-Aza enabled sustained IL-15Rα expression. CD8 T cells from WT mice were incubated with plate-coated anti-CD3/anti-CD28 Abs in the presence or absence of 5-Aza (6μM) for 48 hours, and then stained with anti–IL-15Rα Ab (BAF551; R&D Systems) and FITC-streptavidin secondary Ab (eBioscience). Fractions of the cells were transferred to fresh medium with IL-2 (5nM) and cultured for an additional 4 days. At the same time, total RNA was extracted from the cells and subjected to Northern blot analysis using a 32P-labeled anti-mouse IL-15Rα fragment as the probe and to quantitative RT-PCR (supplemental Figure 4) to examine the expression of mouse IL-15Rα transcripts. Top panel shows IL-15Rα staining of activated (i) and rested (ii) CD8 T cells with and without 5-Aza. The data represent 4 independent experiments (see supplemental Table 1 for a summary of all data). Bottom panel shows Northern blotting. As a loading control, the 18S ribosomal RNA was visualized with ethidium bromide staining. Origins of RNA were the B1 (i) and K2 (ii) leukemia cell lines, ex vivo cultured DCs without (iii) and with (iv) activation by IFN-γ and LPS, CD8 T cells from WT mice cultured in the medium for 6 hours (v) or treated with 5-Aza for 6 hours (vi), TCR activated without (vii) and with (viii) 5-Aza for 48 hours, followed by IL-2 culture for 4 days without (viiii) and with (x) 5-Aza. (E) Antigenic challenge of WT mice only transiently induced IL-15Rα on specific CD8 T cells. WT mice were challenged once with OVA peptide by injecting peptide-loaded DCs subcutaneously to both inguinal regions (1.7 × 105 cells each) and IP (6.7 × 105 cells). Seven days later, IL-15Rα expression levels on the antigen-specific (tetramer-positive) activated CD8 T cells and nonactivated (tetramer-negative) CD8 T cells were analyzed by flow cytometry. The IL-15Rα expression levels were assessed on days 7 and 13. The data represent 2 independent experiments (n = 3 each; see supplemental Table 2 for a summary of all data).
An epigenetic mechanism controls the transcription of the il15rα gene in normal and leukemic CD8 T cells into negative and positive directions. (A) Presence of a DNase I HS site in IL-15Tg leukemic cells and its position in the transcription control site of the il15rα gene. Intact nuclei were isolated from purified CD8 T cells from terminally leukemic IL-15Tg mice (n = 2) and from K2 and B1 leukemic cells, and subjected to 1 U/mL of DNase I for 15 minutes at 37°C. The genomic DNA was extracted subsequently and digested by the EcoRI enzyme. The digested DNA was subjected to 0.7% Tris-acetate EDTA gel electrophoresis, blotted, and hybridized with a 32P-labeled fragment (as shown by the asterisk in panel B) from the il15rα transcription initiation site. As a control, CD8 T cells purified from WT mice were activated by plate-coated anti-CD3/anti-CD28 Abs and hIL-15 (5nM; PeproTech) for 48 hours before DNase I treatment. The sources of genomic DNAs were: normal CD8 T cells (i), CD8 T cells from leukemic IL-15Tg mouse #1 (ii), CD8 T cells from leukemic IL-15Tg mouse #2 (iii), cells from the K2 leukemic cell line (iv), and cells from the B1 leukemic cell line (v). The size of the upper band (EcoRI-EcoRI fragment, intact) was 4.5 kb, and the lower (DNase I cleaved) band migrated at 2.1 kb. (B) Determination of the positions of the HS site and CpG islands that were predicted by a computer algorithm. In the 4.5-kb EcoRI-EcoRI fragment, as shown in supplemental Figure 2, the HS (2.1 kb downstream of the 5′ EcoRI site) site was localized at the 3′ end of exon 1 (∼ 350 bp). The MethPrimer algorithm predicted the presence of 2 CpG islands: CpG1 was localized 2.1-2.25 kb downstream of the 5′ EcoRI site, and the CpG2 2.8-2.9 kb downstream of the 5′ EcoRI site. (C) ChIP revealed a potential hypermethylation of this gene region in resting normal CD8 T cells. A small aliquot of the sonicated genomic DNA (0.25 ng) from before and after the enrichment of the MBP2-bound DNA fractions was used. PCR was performed using a primer set that flanks the first CpG island. The reaction was terminated after 28 cycles of amplification in the semilog amplification phase for a quantitative comparison. Sources of DNA were: resting CD8 T cells (i), CD8 T cells from a leukemic IL-15Tg mouse (ii), and cells from the K2 leukemic cell line (iii). The black bar indicates that distant lanes from the same experiment were combined. Densitometry analysis was performed and the amount of MBP-2 bound DNA normalized to that of control DNA is indicated. (D) In vitro activation of normal CD8 T cells temporally induced IL-15Rα, and treatment with 5-Aza enabled sustained IL-15Rα expression. CD8 T cells from WT mice were incubated with plate-coated anti-CD3/anti-CD28 Abs in the presence or absence of 5-Aza (6μM) for 48 hours, and then stained with anti–IL-15Rα Ab (BAF551; R&D Systems) and FITC-streptavidin secondary Ab (eBioscience). Fractions of the cells were transferred to fresh medium with IL-2 (5nM) and cultured for an additional 4 days. At the same time, total RNA was extracted from the cells and subjected to Northern blot analysis using a 32P-labeled anti-mouse IL-15Rα fragment as the probe and to quantitative RT-PCR (supplemental Figure 4) to examine the expression of mouse IL-15Rα transcripts. Top panel shows IL-15Rα staining of activated (i) and rested (ii) CD8 T cells with and without 5-Aza. The data represent 4 independent experiments (see supplemental Table 1 for a summary of all data). Bottom panel shows Northern blotting. As a loading control, the 18S ribosomal RNA was visualized with ethidium bromide staining. Origins of RNA were the B1 (i) and K2 (ii) leukemia cell lines, ex vivo cultured DCs without (iii) and with (iv) activation by IFN-γ and LPS, CD8 T cells from WT mice cultured in the medium for 6 hours (v) or treated with 5-Aza for 6 hours (vi), TCR activated without (vii) and with (viii) 5-Aza for 48 hours, followed by IL-2 culture for 4 days without (viiii) and with (x) 5-Aza. (E) Antigenic challenge of WT mice only transiently induced IL-15Rα on specific CD8 T cells. WT mice were challenged once with OVA peptide by injecting peptide-loaded DCs subcutaneously to both inguinal regions (1.7 × 105 cells each) and IP (6.7 × 105 cells). Seven days later, IL-15Rα expression levels on the antigen-specific (tetramer-positive) activated CD8 T cells and nonactivated (tetramer-negative) CD8 T cells were analyzed by flow cytometry. The IL-15Rα expression levels were assessed on days 7 and 13. The data represent 2 independent experiments (n = 3 each; see supplemental Table 2 for a summary of all data).
Treatment with 5-Aza induced IL-15Rα expression in anti–CD3/CD28-stimulated normal CD8 T cells
Finally, we tested whether DNA hypermethylation repressed IL-15Rα expression by normal CD8 T cells upon anti-CD3/CD28 stimulation. We treated normal purified CD8 T cells with 5-Aza upon stimulation with plate-coated anti-CD3 and anti-CD28 Abs for 2 days and after resting for an additional 4 days. As shown in Figure 4D, supplemental Figure 4, and supplemental Table 1, activated CD8 T cells induced IL-15Rα; however, without 5-Aza treatment, IL-15Rα expression was transient and underwent down-regulation when stimulation ceased. Interestingly, 5-Aza–treated CD8 T cells sustained their expression of IL-15Rα after activation. We also confirmed the transience of IL-15Rα induction on activation in vivo by challenging mice with a peptide antigen loaded on dendritic cells (Figure 4E and supplemental Table 2). These findings do not allow us to conclude that the DNA methylation status of the HS in the il15rα gene unilaterally controlled the on and off states of IL-15Rα transcription; however, it seems reasonable to hypothesize that an epigenetic mechanism involving DNA methylation plays an important role in regulating the homeostatic proliferation of CD8 T cells and activation-induced clonal expansion.
Discussion
In this study, we investigated conditions under which a continuous action of IL-15 in vivo could lead to the development of a fatal lymphoid expansion. We suggest that an independent second event, namely acquisition of expression of IL-15Rα by CD8 T cells, must occur for the leukemia-like condition to become established. The lymphoproliferative disorder in our mice took 12-15 months to develop into a fatal, uncontrolled stage. This observation is consistent with our hypothesis that acquisition of IL-15Rα expression by CD8 T cells is under tight regulation that may involve an epigenetic mechanism. Once generated, such persistently IL-15Rα–positive IL-15–producing CD8 T cells could be at a selective advantage and might quickly expand and overwhelm other lymphoid cells, leading to a leukemic or leukemia-like condition. When the expression of IL-15Rα was also forced onto all CD8 T cells by transgenesis (the IL-15Rα–IL-15 double-Tg mice), peripheral CD8 T cells became activated and replicated uncontrollably in vivo, resulting in an aggressive lymphoproliferative disorder and rapid death. Although this is a highly experimental condition unlike human leukemia, it suggests that the coexpression of IL-15 and IL-15Rα by the same CD8 T cells is sufficient to cause an uncontrolled expansion of CD8 T cells in vivo.
Fehniger et al first reported leukemia in IL-15Tg mice overexpressing murine IL-15,11 and their recent extensive phenotype analyses concluded that such mice developed leukemia of either the NK or the T (T-NK)–cell type.12 Our leukemic cells and clones strongly expressed NK antigens, including NK1.1, NKG2D, and select Ly49 isoforms. Nevertheless, our cells had undergone rearrangements of the TCRβ locus. CD8 T cells with leukemic potential in our mice were phenotypically different from the CD1d-restricted NK-T cells (CD4+Vα14+) that were originally described by Bendelac et al.23 We recently analyzed distinct NK1.1+CD8+ cells in the benign phase of our IL-15Tg mice.17 These cells represent a new subset of CD8 T cells, because they express CD8αα homodimers.24 However, the leukemia-like CD8 T cells we observed in our Tg mice expressed CD8αβ heterodimers and are clearly different from the CD8αα NK+ T cells. This observation suggests that not all expanded cells in the presence of IL-15 eventually develop uncontrolled growth and leukemia.
Whereas the potential role of IL-15Rα in CD8 leukemogenesis is intriguing, the expansion of benign CD8 T cells did not require IL-15Rα expression in cis or in trans, because we observed a benign expansion of CD8 T cells in IL-15Rα−/−IL-15Tg mice (Figure 1B lane iv). T-cell leukemia never developed in these mice, suggesting that a simple IL-15–mediated autocrine growth does not lead to malignant transformation. This observation prompted us to postulate that the transduced signal may be qualitatively different in the absence or presence of IL-15Rα. In fact, we observed that the presence of IL-15Rα on signaling T cells enables prolonged signaling.14,25 In light of the recent conceptual circuitry simulation of cellular signaling mechanism using systems biology, it is intriguing to postulate the presence of a persistent, coherent feed-forward loop26 that filters out transient activation signals and only allows persistent signals to reach the nucleus for subsequent cellular growth and mitosis. In normal CD8 T cells without persistent IL-15Rα expression, it is possible that even constitutively produced IL-15 would not reach the threshold sufficient to cause meaningfully persistent signals to cells, leading only to intermittent signaling that would cause modest propagation by inhibiting cell death by neglect. However, such a transient signal might be filtered out by the cohesive, feed-forward loop device and might fail to cause malignant transformation. These new insights create new questions as to why continued growth-factor action only sometimes leads to continuous and uncontrolled cell growth that leads to cancer, circumventing control by negative regulators.
Lack of development of a fatal lymphoproliferative disorder in IL-15Rα−/−IL-15Tg mice clearly suggests that the coexpression of IL-15 and IL-15Rα appear to be necessary for leukemic transformation. However, it does not appear to be sufficient for inducing the transition to monoclonality, in that aggressive CD8 T cells in our terminal-stage IL-15Tg mice expressing both IL-15 and IL-15Rα exhibited both monoclonal and polyclonal expansion (Southern blot, Figure 3C). It remains possible that an additional event(s) might be required for this terminal event. In their murine IL-15Tg leukemia model, Yokoyama et al12 observed chromosomal abnormalities, namely trisomy of chromosome 15 and/or 17.
The coexpression of IL-15 and IL-15Rα in leukemic cells presents another interesting issue. Three studies18-20 have presented an interesting hypothesis that stable production and secretion of IL-15 requires coexpression of IL-15Rα. This proposal that IL-15Rα expression is required for stable IL-15 excretion is in perfect accord with our previous proposition that IL-15 and IL-15Rα exist in vivo as a heterodimeric cytokine, either as a soluble dimer6 or as cell-bound dimer molecules,25 and that we should consider the combination of IL-15 (p14) and IL-15Rα (p35) to be similar to another cytokine, IL-12. These proposals reaffirm another proposal of ours and others that IL-15 in vivo is most frequently presented in trans,14,25,27 as opposed to binding to preexisting α/β/γc cis receptor complexes. According to the new proposal, we should expect an almost complete loss of hIL-15 production in IL-15Rα−/−IL-15Tg mice (ie, in the complete absence of IL-15Rα). However, we saw only some reduction of IL-15 levels in the sera (from ∼ 500-850 pg/mL to 350 pg/mL), accompanied by a moderate expansion of CD8 T cells (Figure 2C lane iv). This observation may seem to contradict recently published results, but we observed that hIL-15, as opposed to murine IL-15, is relatively easily expressed and secreted by human HEK293T cells upon cDNA transfection, and that coexpression of human IL-15Rα is almost dispensable for successful secretion of hIL-15. On the other hand, in accordance with Mortier et al,19 mouse IL-15 was extremely poorly produced from HEK293T cells without cotransfection of mouse IL-15Rα. Therefore, the use of hIL-15 in our case would provide a reasonable explanation for the successful hIL-15 expression in mice. In the study by Fehniger et al,11 in which murine IL-15 was expressed in a leukemia model, it is possible that their leukemic cells, like ours, expressed IL-15Rα.
By blocking chromatin DNA methylation, we observed sustained IL-15Rα expression on acutely activated normal CD8 T cells, whereas without the blockade, IL-15Rα was down-regulated soon after the activation subsided (Figure 4D-E). This observation suggests a silencing mechanism that limits the period of IL-15Rα expression in normal, acutely activated CD8 T cells.
An interesting debate concerns whether long-lived memory CD8 T lymphocytes express IL-15Rα. Our leukemic CD8 T cells, which resemble the CD8 memory T cells by virtue of surface-marker expression (Figure 1B and supplemental Figure 1), do express IL-15Rα (as shown by flow cytometry and Northern analysis). A specific mechanism that suppresses persistent silencing of epigenetic control might be required to enable the constitutive expression of IL-15Rα in leukemic cells. It would be reasonable to assume that the development of memory CD8 T cells from their precursors involves a similar epigenetic change in which a set of genes, including the il15rα gene, are turned on or off in a coordinated manner. Accordingly, disruption of normal changes in the chromatin methylation status could have a profound effect on the generation of memory CD8 T cells in normal immune responses.
The clinical use of recombinant hIL-15 for human diseases has just been given approval by the US Food and Drug Administration, and presently proposed therapeutic applications include the treatment of cancer.28 The original report by Fehniger et al11 cast some shadow on the safety of such clinical IL-15 administration. Our report largely mitigates such concerns, because a brief treatment period involving IL-15 administration should not cause leukemia or lymphoproliferative disorders as long as IL-15 is the only reagent administered. However, we wish to propose some caution regarding recent proposals involving the induced stable coexpression of IL-15 and IL-15Rα in humans for more efficient action of the cytokine in therapeutic interventions18,29 because if a CD8 T cell acquires the ability to persistently coexpress IL-15 and IL-15Rα, it may become a leukemia founder cell. The extremely rapid and aggressive development of fatal expansion of CD8 T cells in IL-15Rα–IL-15 double-Tg mice should prompt a caution against this strategy. Therefore, our current opinion is that the simple administration of recombinant IL-15 is the safest method of augmenting IL-15 function in humans.
In summary, we suggest a critical role for IL-15Rα expression by CD8 T cells that is required for these cells to develop into an uncontrolled, lymphoproliferative disorder such as fatal leukemia in IL-15Tg mice. In light of the IL-15 transpresentation paradigm that we proposed years ago,25 this is an unusual situation in which the conventional, 3-chain IL-15R expressed in cis would be crucial for pathologic leukemia development. We also propose a potential epigenetic mechanism that silences IL-15Rα transcription in naive CD8 T cells. However, persistent unlocking of this mechanism may constitute an important process upon transition of naive CD8 T cells to memory CD8 T cells. For future clinical applications involving IL-15 administration or transduction of IL-15 cDNA, we propose that it would be prudent to administer only recombinant hIL-15 protein to humans, as opposed to more radical gene therapies, particularly those that combine the expression of IL-15 and IL-15Rα, to enable more efficient IL-15 action in vivo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We express our sincere thanks to Drs Jacqueline M. Sasaki and Joanna Marks-Konczalik, former laboratory members, who made founding and seminal contributions to the current work. We also appreciate the conceptual contribution by Dr Sigrid Dubois, Metabolism Branch, National Cancer Institute, National Institutes of Health, Bethesda, MD.
This work was supported by intramural research funds of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD.
National Institutes of Health
Authorship
Contribution: N.S., S.F., W.J., M.N.P., and Y.T. performed experiments; N.S., H.S., R.N.B., T.A.W., and Y.T. designed the project; and N.S., R.N.B., T.A.W., and Y.T. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas A. Waldmann, Metabolism Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bldg 10, Rm 4N115, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: tawald@mail.nih.gov or Yutaka Tagaya, 725 W Lombard St, S-620, Baltimore, MD 21201; e-mail: ytagaya@ihv.umaryland.edu.

![Figure 1. The human IL-15Tg mice developed a fatal lymphoproliferative disorder. (A) Survival plots of IL-15Tg mice on different backgrounds. IL-15Tg (B1) and IL-15Rα−/−IL-15Tg mice were compared in terms of their life span with IL-15Tg mice showing a shortened survival (n = 12, P < .001). IL-15Rα–IL-15 double-Tg and IL-15Tg mice were compared as well (n = 15, P < .001). IL-15Rα−/−-IL-15Tg mice lived as long as their littermates, whereas IL-15Rα–IL-15 double-Tg mice died at a very young age because of uncontrolled expansion of CD8 T cells. (B) Expansion of CD44hiCD8 T cells in the IL-15Tg mice at the benign and terminal stages of leukemia. Splenocytes from a WT mouse (i), a young IL-15Tg (3-month-old) mouse (ii), a 13-month-old IL-15Tg mouse (iii), a 13-month-old IL-15Rα−/−IL-15Tg mouse (iv), and a 3-month-old IL-15Rα–IL-15 double-Tg mouse (v) were stained with anti-CD3, anti-CD44, and anti-CD8 Abs and analyzed by flow cytometry. The majority (> 95%) of the splenocytes expressed CD3 and CD8 in the terminal-stage leukemic IL-15Tg mice (iii), whereas in the benign-phase IL-15Tg mice (ii) and IL-15Rα−/−IL-15Tg mice (iv), CD8 T cells maintained an altered but stable homeostasis (with 80% and 44% of CD8+ cells in the CD3-expressing population, respectively). In the terminal-phase leukemic IL-15Tg mice (iii), T cells almost completely displaced other types of cells from the spleen. In the IL-15Rα–IL-15 double-Tg mice (v), even young mice (< 3 months old) displayed displacement of peripheral lymphocytes with CD8 T cells (> 92% of the T-cell compartment). Data represent results from 4-6 mice per group (see supplemental Figure 2 for a summary of all data). (C) Development of lymphoma/leukemia in WT mice transplanted with B1 (B1-001) IL-15Tg cells. Four weeks after the IV injection of 1 × 106 B1 leukemic cell line cells, the WT recipient mice developed tumor masses in their spleens (left, indicated with an arrow). As a surrogate marker to monitor leukemic cell expansion, the concentrations of hIL-15 in the sera of transplanted mice were determined by a specific IL-15 ELISA (right, n = 8). (D) Constitutive phosphorylation and DNA binding of STAT5 in leukemic cells. In the left panel, constitutive tyrosine-phosphorylation of the STAT5a molecules in the ex vivo B1 leukemic cells (left lane) was demonstrated using an anti-pSTAT5a Ab by immunoblotting. An anti-STAT5a/b Ab was used to quantitate the amount of the STAT protein in each lane. In the right panel, STAT5 shows constitutive DNA binding in the ex vivo B1 leukemic cells as demonstrated using an EMSA assay. The target oligonucleotides were 5′-agttattagaaatTTCAAGGAAgtgacaacagag-3′ (STAT5 WT), 5′agttattagaaatTTCAACCTTgtgacaacaga-3′ (STAT5 mutant, mutation underlined), which were annealed, 32P-labeled, and used as the probe. An anti-STAT5a/b Ab was used to supershift the DNA-protein complexes. The lanes indicate without (i) and with (ii) anti-STAT5a/b Ab. (E) Confirmation of the IL-15 dependency of the leukemic cells. Inclusion of the anti–hIL-15 Ab (MAB247; R&D Systems) abrogated the constitutive proliferation of IL-15Tg leukemic cells as assessed by [3H]-thymidine incorporation, suggesting that these cells depend on an autocrine supply of IL-15 for their growth and survival. The addition of anti-mouse IL-2/15Rβ Ab (TMβ1) led to a partial inhibition with B1/K2 clones, whereas the inhibition by TMβ1 was complete, with normal CD8 T cells stimulated with hIL-15. The data represent 3 independent experiments (n = 3 each).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/15/10.1182_blood-2010-09-307504/4/m_zh89991168980001.jpeg?Expires=1770054775&Signature=YZd1IDX9DvxdFloj45w3RYM8ZCuxlaIiYCD9LfD84abwnxVbOkiZrn8DUwLr4079PwYXz12WE7q8Kz0Bjbjzoc~XVO7~M1fuqPvDsri66V8pQq16JPXMX4cOeK2zLkfu5TsR3RRyvhWavUXCgBhrQ~GX3uNg5AlquGe8stmBw-SCsBJO7IgpdDQHdln2x3nGLyCnXqstCB6Wkd3r7Wj0OwhpVy3xguitN5nTn6YKlKQqp6vSNJSymGuwkBWJCRCklezc9FsjxtVEjjE6Tgg0lv0FPDGXN9Z8FQPRjkP~a69d8mUhhqGCs4LIagpQ5zwdbukcAkZWAKIhck3YUbaNgA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal