Abstract
Human T-lymphotropic virus type 1 (HTLV-1) persists by driving clonal proliferation of infected T lymphocytes. A high proviral load predisposes to HTLV-1–associated diseases. Yet the reasons for the variation within and between persons in the abundance of HTLV-1–infected clones remain unknown. We devised a high-throughput protocol to map the genomic location and quantify the abundance of > 91 000 unique insertion sites of the provirus from 61 HTLV-1+ persons and > 2100 sites from in vitro infection. We show that a typical HTLV-1–infected host carries between 500 and 5000 unique insertion sites. We demonstrate that negative selection dominates during chronic infection, favoring establishment of proviruses integrated in transcriptionally silenced DNA: this selection is significantly stronger in asymptomatic carriers. We define a parameter, the oligoclonality index, to quantify clonality. The high proviral load characteristic of HTLV-1–associated inflammatory disease results from a larger number of unique insertion sites than in asymptomatic carriers and not, as previously thought, from a difference in clonality. The abundance of established HTLV-1 clones is determined by genomic features of the host DNA flanking the provirus. HTLV-1 clonal expansion in vivo is favored by orientation of the provirus in the same sense as the nearest host gene.
Introduction
Human T-lymphotropic virus type 1 (HTLV-1) causes adult T-cell leukemia-lymphoma (ATLL), HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP), uveitis, and infective dermatitis. It is estimated that 15 to 20 million persons live with HTLV-1 infection worldwide. A small proportion (up to 7%, depending on the area) of HTLV-1–infected persons develop disease, whereas the majority remain asymptomatic carriers (ACs). Infection occurs via breastfeeding, transfusion of infected cellular blood products, or sexual intercourse. Symptoms appear after a long period (years or decades) of clinical latency.1 The HTLV-1 proviral load (PVL) remains stable within each infected person and correlates with the outcome of infection. However, the PVL varies widely among infected people, even within a particular diagnostic group.2-4 The sequence of HTLV-1 is also stable within a person,5,6 indicating that the PVL is maintained in vivo mainly by mitosis of infected cells during the chronic phase of the infection. This interpretation is supported by the observation that individual clones of infected cells can persist in patients for several years.7-9 Thus, it has been hypothesized that infectious transmission of HTLV-1 is important early in infection across the virologic synapse,10 whereas mitotic replication is responsible for maintaining proviral load once a persistent infection has been established and reached an equilibrium with the immune response.11 In approximately 5% of infected people, persistent clonal proliferation culminates in malignant transformation in the disease ATLL.7,8 The leukemic clones carry generally one (complete or defective) provirus per cell.12-14
There has been a longstanding debate on the question of whether HTLV-1 is latent or persistently expressed in vivo. Persistent expression is strongly suggested by the extensive evidence that the strong, chronically activated cytotoxic T lymphocyte (CTL) response to HTLV-1 limits the proviral load and reduces the risk of HAM/TSP.11 Furthermore, there is both experimental evidence15 and theoretical justification16 for selective proliferation of HTLV-1–expressing T cells in vivo. However, with currently available methods, it is usually impossible to detect either HTLV-1 mRNA proteins or virions in fresh uncultured peripheral blood mononuclear cells (PBMCs).
Within a given HTLV-1–infected person, 2 features differentiate the clones of infected cells: (1) the antigenic specificity of the T-cell clones (ie, the T-cell receptor); and (2) the proviral insertion site in the host genome. Proviral integration in the genome is not random, and each retrovirus has distinct target site preferences.17,18 These different targeting preferences are determined by several factors, notably the properties of the viral integrase,19 cellular protein partners,20 and the chromatin structure at the point of integration.21,22 HTLV-1 integration was found to favor genes, transcriptional start sites, and CpG islands both in vitro and in vivo.18,23-26
We hypothesize that the genomic integration site of the provirus determines the level of proviral expression: in turn, the proviral gene products determine both the proliferation rate and the susceptibility to CTL-mediated lysis of that infected T-cell clone. That is, each HTLV-1–infected T-cell clone will have unique characteristics of the dynamics of proviral expression and immune destruction. Consistent with this hypothesis, we obtained evidence26 that integration of HTLV-1 in transcriptionally active regions of the genome is associated with spontaneous proviral expression in vitro and with the inflammatory disease HAM/TSP. However, because of the insensitivity and low throughput of conventional techniques, previous studies7,8,25,26 were limited to the analysis of the insertion sites of the most abundant clones: typically, fewer than 20 unique integration sites (UISs) of the provirus were identified in each sample. More recent studies on HIV22 or vector-based gene therapy27 used high-throughput sequencing, which allowed identification of a much larger number of UISs but lacked the ability to quantify accurately the abundance of the insertion sites because of preferential polymerase chain reaction (PCR) amplification of short DNA products. The use of restriction enzymes to cut the genomic DNA leads to the danger of preferential amplification and detection of UISs that lie close to a restriction site. These drawbacks preclude the use of these techniques to quantify accurately the abundance of HTLV-1–infected T-cell clones: because oligoclonal proliferation is a central feature of HTLV-1 persistence and pathogenesis, this remains a severe limitation. We have developed a new approach based on ligation-mediated PCR and massively parallel sequencing that allows simultaneously the mapping and the quantification of the abundance of each UIS. Here we report the analysis of more than 91 000 UISs from a total of 61 HTLV-1+ persons and more than 2100 UISs from in vitro infected T cells. By a quantitative analysis of integration site oligoclonality and bioinformatic analysis of host DNA flanking the integration sites, we infer selection forces that act in vivo to shape the distribution of the abundance of HTLV-1 clones and determine the outcome of the infection.
Methods
Blood samples
Sixty-one persons infected with HTLV-1 (14 ACs: mean age, 55 years; 1 patient with uveitis; 26 patients with HAM/TSP: mean age, 62 years; and 20 patients with ATLL: mean age, 53 years) attended the clinic at the National Center for Human Retrovirology (Imperial College Healthcare NHS Trust, St Mary's Hospital, London), and donated blood samples having given informed consent in accordance with the Declaration of Helsinki. All experimental protocols were approved by the United Kingdom Home Office. PBMCs were isolated using Histopaque-1077 (Sigma-Aldrich). Cells were washed and cryopreserved in fetal calf serum (Invitrogen) with 10% dimethyl sulfoxide (Sigma-Aldrich). DNA was extracted from PBMCs using DNeasy Blood and Tissue kit (QIAGEN). HTLV-1 proviral load was measured as previously described.26
Mapping of UISs and quantification of UIS abundance
A total of 10 μg of DNA was sheared by sonication with a Covaris S2 instrument. DNA ends were end-repaired using T4 DNA polymerase, DNA polymerase I Klenow fragment, and T4 polynucleotide kinase (New England Biolabs). Addition of an adenosine at the 3′ ends of the DNA was performed using Klenow fragment 3′ to 5′ exo- (New England Biolabs). A partially double-stranded DNA linker was ligated to the DNA ends using a Quick ligation kit (New England Biolabs). Twenty-six different linkers were constructed, each one with a specific 6-bp tag to allow multiplexing of DNA samples during the sequencing. The ligated product was amplified by a first PCR between the B3 and B4 primers using Phusion DNA polymerase (Finnzyme). To perform PCR2, 1/150th of the cleaned PCR1 product was amplified between the P5B5 and P7. DNA was finally cleaned using a Qiaquick PCR purification kit. A DNA “library” was constructed by pooling the different PCR2 products (each one possessing a specific tag). Quantification of the libraries was made by quantitative PCR using primers P5 and P7 and a LightCycler SYBR Green-1 kit (Roche Diagnostics). Standard curves were generated using a library quantified on a titration flow cell previously run on a Genome Analyzer II (Illumina). Stock libraries were diluted accordingly and clustered on the flow cell. Paired-end reads (read 1 and read 2, each 50 bp) plus a 6-bp tag read (read 3) were acquired on the GA II and the insertion sites, and shear sites were deduced. For each UIS, we counted the number of amplicons of different length (ie, different shear site). The absolute abundance of a given UIS (number of a particular insertion site per 10 000 PBMCs) was calculated from the number of amplicons of different length and the measurement of the proviral load. A complete description of the reaction conditions, primer and linker sequences, and the mapping and quantification procedures is given in supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
In vitro infection of T cells with HTLV-1
The HTLV-1–producing cell line MT-228 was labeled with anti-CD4 antibody-coupled microbeads (Miltenyi) and γ-irradiated (137Cs, 40 000 cGy). The cells were then cocultured with Jurkat cells for 3 hours at 37°C at a 1:1 ratio. The MT-2 cells were removed by magnetic depletion. After one week of culture, all remaining MT-2 cells had died. After 2 weeks of culture, DNA was extracted from the Jurkat cells and the HTLV-1 integration sites were amplified as described in “Mapping of UISs and quantification of UIS abundance.” To verify that the observed proviral integrations were novel and were not contaminating MT-2 sites, DNA was also extracted from MT-2 cells, and the resulting integrations were used to search the set of novel integrations for MT-2 sites. No contaminating MT-2 sites were found.
Oligoclonality index and estimation of the total number of UISs in the entire body of the host
The oligoclonality index is based on the Gini coefficient29 and is calculated as described in the supplemental data. Estimating the total number of UIS in the entire body of an infected patient is analogous to a classic problem in animal ecology known as the unseen species problem. The objective is to estimate the number of unique species in a large, complex population based on the number of unique species observed in a random, finite sample. We used the Chao1-bc estimator as an estimator of the lower bound of species richness,30 calculated as described in the supplemental data.
Statistics
Statistical tests were performed using GraphPad Prism 5 software and were 2-tailed when possible.
Results
Selective amplification and quantification of proviral insertion sites
The PCR strategy is outlined in Figure 1. DNA was extracted from uncultured PBMCs of HTLV-1+ persons and sheared by sonication. A linker containing a tag was ligated, and nested PCR was performed between the end of the HTLV-1 long terminal repeat and the linker. Nested PCR products were pooled to construct the library. A paired-end read (read 1 and read 2) plus a tag read were acquired on an Illumina Genome Analyzer II. Read 1 and read 2 were mapped against the human genome (build hg18) and the proviral insertion site and the shear site were deduced. For each UIS, we counted the number of amplicons of different length (ie, different shear sites). The absolute abundance of a given UIS (number of a particular insertion site per 10 000 PBMCs) was calculated from the number of amplicons of different length and the measurement of the proviral load. A complete description of the procedure is given in supplemental data.
UIS mapping and quantification of abundance. (A) Genomic DNA was extracted from PBMCs and sonicated. The end of the 3′-long terminal repeat and a fragment of genomic DNA were amplified by ligation-mediated PCR and the products sequenced on an Illumina Genome Analyser. (B) In this example, a genomic DNA sample contains 4 proviral copies from infected T-cell clone X and 1 copy from clone Y. Because the DNA shear site is random, the amplicon from each cell in clone X has a different shear site. The abundance of each UIS is quantified by counting the number of different shear sites for that UIS.
UIS mapping and quantification of abundance. (A) Genomic DNA was extracted from PBMCs and sonicated. The end of the 3′-long terminal repeat and a fragment of genomic DNA were amplified by ligation-mediated PCR and the products sequenced on an Illumina Genome Analyser. (B) In this example, a genomic DNA sample contains 4 proviral copies from infected T-cell clone X and 1 copy from clone Y. Because the DNA shear site is random, the amplicon from each cell in clone X has a different shear site. The abundance of each UIS is quantified by counting the number of different shear sites for that UIS.
HTLV-1 clonal structure in natural infection in vivo
Cellular clonality is usually described qualitatively as polyclonal, oligoclonal, or monoclonal. However, the terms “polyclonal” and “oligoclonal” are not rigorously defined or quantified; an objective parameter of clonality is required. To quantify clonality, we used the method of Gini (1914) to calculate an oligoclonality index (OCI: Supplemental data; supplemental Figure 2A). Our experimental protocol also allows the estimation of the total number of UISs present in a particular person, using the Chao1-bc estimator (supplemental data).
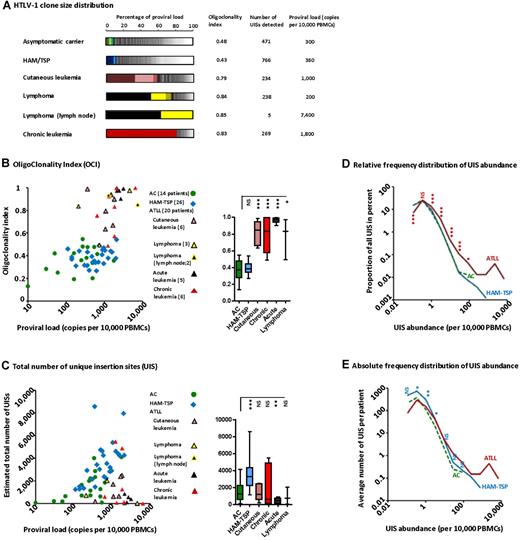
The oligoclonality of the UISs (the distribution of the abundance of each clone) in representative HTLV-1+ persons is depicted in Figure 2A. Each segment in the histogram represents a single UIS; the size of the segment is proportional to the relative abundance of that UIS. UIS oligoclonality in patients with nonmalignant infection (ie, ACs and patients with HAM/TSP) was relatively uniform (the segments in the histograms are of similar size) and so the oligoclonality index is low in these subjects. The OCI for the patients with ATLL was significantly greater, in some cases close to 1 (1.0 indicates perfect monoclonality). Relatively abundant UISs are illustrated by the big segments in the histograms. The 2 dominant UISs present in the blood of the patient with lymphoma were the same as the UISs present in the lymph node tumor (black and bright-yellow segments). The patient with chronic leukemia had a single large UIS (red segment) that constituted more than 80% of the total proviral load.
HTLV-1 clonal structure in naturally infected patients. (A) The clonal distribution in each genomic DNA sample is depicted by a histogram. Each segment represents one UIS; the width of the segment is proportional to the relative abundance of that UIS. The 3 most abundant UISs are colored. (B) The OCI did not correlate with PVL in ACs or in patients with HAM/TSP, but this correlation was significant in patients with ATLL (Spearman rank, P = .0065, R = 0.57). The mean coefficient of variation of OCI was 4.3% (N = 11 samples). The OCI was greater in patients with ATLL than in patients with nonmalignant HTLV-1 infection (box-plot insert, Mann-Whitney, P < .0001). (C) The total number of UISs calculated using the Chao1-bc estimator. The total number of UISs correlated positively with PVL, both in ACs (Spearman rank, P = .035, R = 0.50) and in patients with HAM/TSP (Spearman rank, P = .003, R = 0.57). The mean coefficient of variation of Chao1-bc estimator was 9.5% (N = 11 samples). The adjacent box-plot shows that the number of UISs was significantly greater in patients with HAM/TSP than in ACs (unpaired t test with Welch correction, P = .0002). (D) Low-abundance UISs made up the large majority of all UISs, regardless of disease status. Only ATLL patients had very large UISs (right-hand extremity of the curve). The relative frequency distribution of UIS abundance in ATLL patients showed a shift to the right: asterisks denote the significance of the difference in the proportion of UISs at a given abundance between ATLL patients and ACs (χ2 test). (E) ACs had fewer UISs in each abundance category compared with patients with HAM/TSP: asterisks denote the significance of the difference in the mean number of UISs of a given abundance between patients with HAM/TSP and ACs (Mann-Whitney). ***P < .001. **P < .01. *P < .05. NS indicates not significant (P > .05).
HTLV-1 clonal structure in naturally infected patients. (A) The clonal distribution in each genomic DNA sample is depicted by a histogram. Each segment represents one UIS; the width of the segment is proportional to the relative abundance of that UIS. The 3 most abundant UISs are colored. (B) The OCI did not correlate with PVL in ACs or in patients with HAM/TSP, but this correlation was significant in patients with ATLL (Spearman rank, P = .0065, R = 0.57). The mean coefficient of variation of OCI was 4.3% (N = 11 samples). The OCI was greater in patients with ATLL than in patients with nonmalignant HTLV-1 infection (box-plot insert, Mann-Whitney, P < .0001). (C) The total number of UISs calculated using the Chao1-bc estimator. The total number of UISs correlated positively with PVL, both in ACs (Spearman rank, P = .035, R = 0.50) and in patients with HAM/TSP (Spearman rank, P = .003, R = 0.57). The mean coefficient of variation of Chao1-bc estimator was 9.5% (N = 11 samples). The adjacent box-plot shows that the number of UISs was significantly greater in patients with HAM/TSP than in ACs (unpaired t test with Welch correction, P = .0002). (D) Low-abundance UISs made up the large majority of all UISs, regardless of disease status. Only ATLL patients had very large UISs (right-hand extremity of the curve). The relative frequency distribution of UIS abundance in ATLL patients showed a shift to the right: asterisks denote the significance of the difference in the proportion of UISs at a given abundance between ATLL patients and ACs (χ2 test). (E) ACs had fewer UISs in each abundance category compared with patients with HAM/TSP: asterisks denote the significance of the difference in the mean number of UISs of a given abundance between patients with HAM/TSP and ACs (Mann-Whitney). ***P < .001. **P < .01. *P < .05. NS indicates not significant (P > .05).
The OCI differed between patients with malignant and nonmalignant HTLV-1 infection (Figure 2B box plot, Mann-Whitney, P < .0001). However, the OCI did not distinguish between ACs and patients with HAM/TSP. In addition, there was no correlation between OCI and proviral load in either ACs or patients with HAM-TSP. The evenness of the UIS distribution can also be quantified by the Shannon evenness index. Similar to the OCI, the Shannon evenness index distinguished between patients with malignant and nonmalignant HTLV-1 infection (supplemental Figure 2C).
A higher proviral load in nonmalignant patients is attributable to a higher number of UISs
The total number of UISs differed among patients with nonmalignant infection (Figure 2C box plot, unpaired t test with Welch correction, P = .0002): the mean estimated total number of UISs was 1489 in ACs (mean PVL = 360 copies/10 000 PBMCs) compared with 3512 in patients with HAM-TSP (mean PVL =748 copies/10 000 PBMCs). There was a positive correlation between the estimated total number of UISs and proviral load in patients with nonmalignant infection (Figure 2C Spearman rank, P = .035 for ACs and P = .003 for patients with HAM-TSP). There was no correlation between the oligoclonality index and the estimated total number of UISs in patients with nonmalignant infection (supplemental Figure 2B). The relative frequency distribution of UIS abundance in ACs exactly overlay the distribution in patients with HAM-TSP (Figure 2D) in UISs of low to medium abundance; some HAM/TSP patients possessed additional large clones. However, ACs had a lower absolute number of UISs in each abundance category compared with patients with HAM-TSP (Figure 2E). These data show that the form of the UIS frequency distribution, as quantified by the OCI, did not differ between ACs and patients with HAM/TSP. Rather, the higher proviral load observed in patients with HAM/TSP was attributable to a larger number of UISs.
The majority of UISs were small, regardless of disease status (Figure 2D). However, there was a right-hand shift in the frequency distribution of UIS abundance in ATLL. This observation could be explained either by less efficient immune-mediated elimination of infected cells in ATLL patients or by superinfection of large infected T-cell clones.
Temporal evolution of HTLV-1 clonal structure in natural infection
We quantified the PVL and the OCI in PBMCs in each of 11 patients at 2 time points, separated by 5 to 9 years. The cohort included 1 AC, 1 patient with uveitis, and 9 patients with HAM/TSP. In agreement with previous observations, the PVL in subjects with nonmalignant infection remained relatively constant within each host, with minor fluctuations.2,4 In contrast, the PVL varied among different subjects by more than 100-fold (Figure 3A). There was an increase in the mean OCI (3 replicates of each sample) over time (Figure 3B, paired t test, P = .017). These observations show that the infection was not perfectly at equilibrium at the clonal level because the total lymphocyte count remained within normal limits (no difference between time points 1 and 2, paired t test, P = 1).
Temporal evolution of HTLV-1 clonal structure in natural infection. (A) Proviral load in PBMCs in 11 patients during follow-up for 5 to 9 years. (B) Clonality analysis was made in triplicate at time 1 (t1, A, □) and time 2 (t2, A, ○). Oligoclonality index increased with time in patients with nonmalignant infection (paired t test, P = .017). In March 2007, the oligoclonality index of patient TBK (black line) reached the range typical of ATLL (Figure 2A-B) and lymphoma-type ATLL was subsequently diagnosed in June 2009. (C) □ represents the percentage of the PVL at time 1 that was constituted by UISs, which were detected again at time 2; and ○, the percentage of PVL at time 2 constituted by UISs that had been detected at time 1. (D) The majority of large UISs (those that constituted the top quartile of the PVL) at time 2 were already large (top quartile of PVL) at time 1 (solid black bars). (E) Newly detected UISs at time 2 were mainly small UISs (black fraction of the bars) and on average made up less than 20% of the total PVL. (F) Temporal variation in UIS abundance. S1 represents the abundance of a given UIS at time 1; and S2, the abundance at time 2. Low-abundance UISs became less abundant, whereas high-abundance UISs grew. Asterisks denote the significance of difference of the observed ratio (S1/S2) from 1.0 (t test). Sample size: 0.1 and below, n = 3979; 0.1 to 1, n = 12 463; 1 to 5, n = 1016; 5 to 10, n = 52; and 10 and above, n = 21. ***P < .001. *P < .05. NS indicates not significant (P > .05).
Temporal evolution of HTLV-1 clonal structure in natural infection. (A) Proviral load in PBMCs in 11 patients during follow-up for 5 to 9 years. (B) Clonality analysis was made in triplicate at time 1 (t1, A, □) and time 2 (t2, A, ○). Oligoclonality index increased with time in patients with nonmalignant infection (paired t test, P = .017). In March 2007, the oligoclonality index of patient TBK (black line) reached the range typical of ATLL (Figure 2A-B) and lymphoma-type ATLL was subsequently diagnosed in June 2009. (C) □ represents the percentage of the PVL at time 1 that was constituted by UISs, which were detected again at time 2; and ○, the percentage of PVL at time 2 constituted by UISs that had been detected at time 1. (D) The majority of large UISs (those that constituted the top quartile of the PVL) at time 2 were already large (top quartile of PVL) at time 1 (solid black bars). (E) Newly detected UISs at time 2 were mainly small UISs (black fraction of the bars) and on average made up less than 20% of the total PVL. (F) Temporal variation in UIS abundance. S1 represents the abundance of a given UIS at time 1; and S2, the abundance at time 2. Low-abundance UISs became less abundant, whereas high-abundance UISs grew. Asterisks denote the significance of difference of the observed ratio (S1/S2) from 1.0 (t test). Sample size: 0.1 and below, n = 3979; 0.1 to 1, n = 12 463; 1 to 5, n = 1016; 5 to 10, n = 52; and 10 and above, n = 21. ***P < .001. *P < .05. NS indicates not significant (P > .05).
In one patient with HAM/TSP (coded TBK), the OCI increased strongly during the period of observation, reaching the range characteristic of ATLL at the second time point. The observed increase in the OCI was the result of the expansion of clones that were already abundant at time point 1 (supplemental Figure 3). Remarkably, this second time point preceded the diagnosis of ATLL by 26 months. Moreover, the patient developed lymphoma-type ATLL: that is, the increase in the OCI occurred in the absence of a significant change in PVL (Figure 3A; PVL rose from 1990 to 2080 copies/10 000 PBMCs), whereas the peripheral blood lymphocyte count remained within normal limits.
Regardless of abundance, the great majority of UISs were long-lived (Figure 3C). Abundant UISs were already abundant at the first time point, approximately 8 years earlier (Figure 3D), whereas newly observed UISs were mainly of low abundance (Figure 3E). These observations are consistent with the conclusion7,9 that the PVL is maintained chiefly by proliferation of infected T cells during the chronic phase of infection. Further detail on the rise in oligoclonality index over time is revealed in Figure 3F: small- and medium-abundance UISs shrank, whereas large UISs expanded. These data show that, within a relatively constant PVL, the clonal structure continuously evolved, becoming more oligoclonal over time.
Initial pattern of HTLV-1 integration
The dataset of UISs generated in vitro was analyzed for proximity to RefSeq genes, CpG islands, and various epigenetic marks observed in genome-wide studies of primary human CD4+ T cells31,32 (Supplemental data). To identify the genomic character of the insertion sites favored at the stage of HTLV-1 proviral integration, we compared the distribution of insertion sites resulting from short-term in vitro infection with random sites generated in silico.
In each parameter presented in Figure 4 (“In vitro” panels), the frequency of UISs integrated near the respective feature in vitro was significantly greater than the frequency observed in the randomly generated in silico sites. These observations demonstrate that initial HTLV-1 integration is not random but strongly favors proximity to genes, promoters (as identified by CpG islands), and epigenetic marks associated with the control of gene expression (both activation and repression). The findings confirm the results of previous studies18,26 and extend the number of in vitro HTLV-1 UISs analyzed to more than 2100.
Genetic and epigenetic environment around the proviral insertion site. UISs identified in vivo were organized according to the disease status of the subject (AC, HAM/TSP, and ATLL) and UIS abundance (number of UISs per 10 000 PBMCs). In the ATLL category, the last column named “Major UIS” referred to the most abundant UIS in each person (ie, the proviral insertion site present in the putative malignant clone). “In vitro” refers to insertion sites isolated after coculture of uninfected T cells with an HTLV-1–infected cell line. The y-axis represents the departure from the random distribution. The in vitro results were compared with sites that were randomly generated in silico (horizontal asterisks below “vs random”). The UISs of lowest and highest abundance in each disease status group were compared with the in vitro sites (vertical asterisks to the right of “vs in vitro”). The trends associated with the UIS abundance were also tested for significance (asterisks below the black arrows). (A) “Pr” is the proportion of insertion sites lying within 10 kb of a CpG island or a RefSeq gene. Enrichment toward a given mark is calculated as the log ratio of “Pr” over “Pr random” (proportion expected in case of perfect random integration). Insertion sites isolated in vitro were enriched in the vicinity of CpG islands and genes compared with random (χ2 test). Increasing UIS abundance was correlated with proximity to CpG islands and genes (χ2 test for trend). The UISs of lowest abundance in each disease group were significantly less frequently integrated near CpG islands and genes than were the in vitro sites (χ2 test). (B-D) “N” is the number of a given epigenetic mark in a 10-kb window (± 5 kb) around the insertion site. “N random” is the number of that mark in the case of perfectly randomly distributed insertion sites. Enrichment of a given epigenetic mark was calculated as log (N/N random). In vitro insertion sites were found to lie in an environment enriched for both active and repressive epigenetic marks compared with random (B-D; unpaired t test with Welch correction). UIS abundance was negatively correlated with the density of gene-silencing marks (B, Pearson correlation test). UIS abundance was positively correlated with the density of marks associated with active transcription start sites (TSS), promoters, and transcribed units (C-D, Pearson correlation test). The UISs of highest abundance in each disease group were less frequently associated with gene-silencing marks than were the UISs in vitro (panel B, unpaired t test with Welch correction). The UISs of lowest abundance were less frequently associated with activating marks than the in vitro sites (C-D, unpaired t test with Welch correction). Sample size: In vitro, n = 2135; AC < 0.1, n = 4544; AC 0.1 to 1, n = 8649; AC 1 to 10, n = 727; HAM-TSP < 0.1, n = 26 200; HAM-TSP 0.1 to 1, n = 36 377; HAM-TSP 1 to 10, n = 2931; HAM-TSP > 10, n = 39; ATLL 0.1 to 1, n = 9827; ATLL 1 to 10, n = 1659; ATLL > 10, n = 69; ATLL major UIS, n = 19. ***P < .001. **P < .01. *P < .05. NS indicates not significant (P > .05).
Genetic and epigenetic environment around the proviral insertion site. UISs identified in vivo were organized according to the disease status of the subject (AC, HAM/TSP, and ATLL) and UIS abundance (number of UISs per 10 000 PBMCs). In the ATLL category, the last column named “Major UIS” referred to the most abundant UIS in each person (ie, the proviral insertion site present in the putative malignant clone). “In vitro” refers to insertion sites isolated after coculture of uninfected T cells with an HTLV-1–infected cell line. The y-axis represents the departure from the random distribution. The in vitro results were compared with sites that were randomly generated in silico (horizontal asterisks below “vs random”). The UISs of lowest and highest abundance in each disease status group were compared with the in vitro sites (vertical asterisks to the right of “vs in vitro”). The trends associated with the UIS abundance were also tested for significance (asterisks below the black arrows). (A) “Pr” is the proportion of insertion sites lying within 10 kb of a CpG island or a RefSeq gene. Enrichment toward a given mark is calculated as the log ratio of “Pr” over “Pr random” (proportion expected in case of perfect random integration). Insertion sites isolated in vitro were enriched in the vicinity of CpG islands and genes compared with random (χ2 test). Increasing UIS abundance was correlated with proximity to CpG islands and genes (χ2 test for trend). The UISs of lowest abundance in each disease group were significantly less frequently integrated near CpG islands and genes than were the in vitro sites (χ2 test). (B-D) “N” is the number of a given epigenetic mark in a 10-kb window (± 5 kb) around the insertion site. “N random” is the number of that mark in the case of perfectly randomly distributed insertion sites. Enrichment of a given epigenetic mark was calculated as log (N/N random). In vitro insertion sites were found to lie in an environment enriched for both active and repressive epigenetic marks compared with random (B-D; unpaired t test with Welch correction). UIS abundance was negatively correlated with the density of gene-silencing marks (B, Pearson correlation test). UIS abundance was positively correlated with the density of marks associated with active transcription start sites (TSS), promoters, and transcribed units (C-D, Pearson correlation test). The UISs of highest abundance in each disease group were less frequently associated with gene-silencing marks than were the UISs in vitro (panel B, unpaired t test with Welch correction). The UISs of lowest abundance were less frequently associated with activating marks than the in vitro sites (C-D, unpaired t test with Welch correction). Sample size: In vitro, n = 2135; AC < 0.1, n = 4544; AC 0.1 to 1, n = 8649; AC 1 to 10, n = 727; HAM-TSP < 0.1, n = 26 200; HAM-TSP 0.1 to 1, n = 36 377; HAM-TSP 1 to 10, n = 2931; HAM-TSP > 10, n = 39; ATLL 0.1 to 1, n = 9827; ATLL 1 to 10, n = 1659; ATLL > 10, n = 69; ATLL major UIS, n = 19. ***P < .001. **P < .01. *P < .05. NS indicates not significant (P > .05).
We postulate that the same genomic features favored in short-term in vitro infection will be favored by initial integration of HTLV-1 in vivo. Comparison of integration sites in vitro with those observed in vivo, after years of persistent infection, will then reveal the results of subsequent selection in vivo. We wish to identify specific genomic features that favor the establishment of HTLV-1–infected T-cell clones (ie, UISs that survive immune surveillance and persist for years) and specific genomic features that favor the expansion of certain UISs (ie, established UISs whose abundance increases with time).
Genetic and epigenetic environment associated with clonal establishment
The bias toward insertion in transcribed regions that we observed in vitro contrasted strongly with the pattern observed in vivo in the UISs of lowest abundance. UISs of lowest abundance observed in vivo were significantly less strongly associated with proximity to CpG islands, proximity to genes, and activating epigenetic marks than the in vitro sites (Figure 4A,C-D). As shown before, the UISs of low abundance constituted the majority of UISs in each patient (Figure 2D). We conclude that infected cells carrying a provirus inserted near markers of transcriptional silencing probably establish but will not expand.
These observations suggest strong selection in vivo against proviral expression. The force that is most likely to exert this selection is the host immune response.11
Genetic and epigenetic environment associated with clonal expansion
Proximity to both CpG islands and genes was positively correlated with UIS abundance, regardless of clinical status (AC, HAM/TSP, and ATLL; Figure 4A). The observation that the mean frequency of proximity to CpG islands and genes in vitro was not significantly different from the frequency in the UISs of highest abundance in vivo, whereas the frequency in the UISs of lowest abundance in vivo was significantly less than the frequency in vitro implies that integration near a CpG island or host gene has a permissive effect on the successful proliferation of the HTLV-1–infected T cell. In other words, integration near a CpG island or gene confers a proliferative advantage on that clone, and UISs that lie distant from CpG island or genes may become established but do not expand.
Both the activating and repressive marks depicted in Figure 4 are associated with host transcription units. However, there was a negative correlation between UIS abundance and repressive epigenetic marks (Figure 4B), whereas there was a positive correlation between UIS abundance and markers of active transcription (Figure 4C-D). These observations imply that, in vivo, UISs present near gene-silencing epigenetic marks proliferate less well and that active transcription per se contributes to the expansion of the HTLV-1–carrying T cells. That is, it is not simply the open chromatin structure associated with transcription units, but rather active ongoing transcription that favors expansion. Supplemental Figure 4 illustrates the genomic environment of the major UIS in 3 different patients.
The frequency of epigenetic markers of active transcription near low abundance UISs was significantly lower in vivo than in vitro (Figure 4C-D). In contrast, the frequency of such markers near the UISs of highest abundance in vivo was not significantly different from that in vitro, except in asymptomatic carriers, in whom the frequency remained significantly lower than that observed in vitro. This result implies effective counter-selection of actively transcribed proviruses in ACs in vivo but less effective counter-selection in the patients with HAM/TSP or with ATLL. This selection is probably exerted by the host immune response, especially the class 1 major histocompatibility complex-restricted CTL response.11
Same-sense transcriptional orientation of the provirus favors HTLV-1 clonal expansion
The transcriptional activity of the host genome in the vicinity of an integrated provirus might influence the level of proviral transcription either indirectly, because the unfolded chromatin is accessible to transcription factor complexes, or directly, by specific interactions between host promoters or enhancers and the provirus. Such direct interactions might, moreover, depend on the relative transcriptional orientation of the provirus and nearby host promoters.33,34
The proportion of proviruses in vivo that were inserted inside a gene was higher in abundant UISs than in UISs of low abundance (Figure 5A). In addition, the percentage of proviruses integrated inside a gene was significantly smaller among low-abundance UISs than among the insertion sites identified in vitro. These observations support the hypothesis that proviral expression favors expansion.
Frequency of proviral insertion in genes and relative orientation of the provirus. (A) The proportion of proviruses inserted inside a RefSeq gene increased with UIS abundance (asterisks below the open black triangle; χ2 test for trend). In low-abundance UISs, the proportion of proviruses inserted inside a gene was smaller than in UISs identified in vitro (vertical asterisks to the right of “vs in vitro”; χ2 test). (B) When the provirus was inserted inside a RefSeq gene, it was integrated more frequently in the same orientation as the host gene in UISs identified in vivo; in contrast, the orientation of UISs identified in vitro was random (vertical asterisks to the right of “vs in vitro”; χ2 test). Increasing UIS abundance was associated with an increased percentage of proviruses oriented in the same transcriptional sense as the host gene (asterisks below the ▵; χ2 test for trend). Sample size: In vitro, n = 2135; AC < 0.1, n = 4544; AC 0.1 to 1, n = 8649; AC 1 to 10, n = 727; HAM-TSP < 0.1, n = 26 200; HAM-TSP 0.1 to 1, n = 36 377; HAM-TSP 1 to 10, n = 2931; HAM-TSP > 10, n = 39; ATLL 0.1 to 1, n = 9827; ATLL 1 to 10, n = 1659; ATLL > 10, n = 69; ATLL Major UIS, n = 19. ***P < .001. **P < .01. *P < .05. NS indicates not significant (P > .05).
Frequency of proviral insertion in genes and relative orientation of the provirus. (A) The proportion of proviruses inserted inside a RefSeq gene increased with UIS abundance (asterisks below the open black triangle; χ2 test for trend). In low-abundance UISs, the proportion of proviruses inserted inside a gene was smaller than in UISs identified in vitro (vertical asterisks to the right of “vs in vitro”; χ2 test). (B) When the provirus was inserted inside a RefSeq gene, it was integrated more frequently in the same orientation as the host gene in UISs identified in vivo; in contrast, the orientation of UISs identified in vitro was random (vertical asterisks to the right of “vs in vitro”; χ2 test). Increasing UIS abundance was associated with an increased percentage of proviruses oriented in the same transcriptional sense as the host gene (asterisks below the ▵; χ2 test for trend). Sample size: In vitro, n = 2135; AC < 0.1, n = 4544; AC 0.1 to 1, n = 8649; AC 1 to 10, n = 727; HAM-TSP < 0.1, n = 26 200; HAM-TSP 0.1 to 1, n = 36 377; HAM-TSP 1 to 10, n = 2931; HAM-TSP > 10, n = 39; ATLL 0.1 to 1, n = 9827; ATLL 1 to 10, n = 1659; ATLL > 10, n = 69; ATLL Major UIS, n = 19. ***P < .001. **P < .01. *P < .05. NS indicates not significant (P > .05).
When the provirus was inserted inside a gene, it was present (Figure 5B) in the same transcriptional orientation as the host gene (“same-sense” orientation) significantly more frequently in UISs identified in vivo than in UISs identified in vitro (in vitro, 50% of proviruses were in same-sense orientation, as expected by chance). Furthermore, the frequency of same-sense orientation was positively correlated with UIS abundance (Figure 5B). This effect was strongest (75% same-sense orientation) in the major UISs in patients with ATLL, in accordance with the observations of Doi et al.25 We hypothesize that transcription of the host gene favors infected T-cell proliferation by increasing expression of the provirus when integrated inside the gene and in the same transcriptional sense.
Discussion
The results of the present study suggest the following picture of HTLV-1 proviral integration and subsequent selection in vivo:
First phase: cell-to-cell dissemination
Soon after initial infection, before the host has mounted an effective immune response to HTLV-1, the virus disseminates by cell-to-cell transmission via the virologic synapse.10 A large number of distinct UISs are generated with a nonrandom integration of the provirus that strongly favors insertion next to genes, CpG islands, and epigenetic marks associated with the control of gene expression (Figure 4 in vitro panels). We postulate that this pattern of integration is the consequence either of greater accessibility of these genomic regions or of cooperation between cellular proteins and the pre-integration complex as shown for HIV.20,35
Second phase: chronic infection under immune surveillance
A person with nonmalignant HTLV-1 infection typically possesses a total of between approximately 500 and 5000 UISs in the chronic phase. Not only the large clones but also the majority of the UISs were maintained for years (Figure 3C-D). These observations do not support the use of antiretroviral drugs as a treatment against HTLV-1–associated diseases. Treatment with a combination of azidothymidine and interferon-α can decrease the proviral load in patients with ATLL, perhaps by inhibiting cellular proliferation or angiogenesis.36,37 The long-term stability of the UIS populations suggests that the majority of these clones were generated by viral dissemination during the early phase of infection. This conclusion is consistent with previous evidence from studies of adult seroconverters38 and animal models.39,40 By comparing the genomic features associated with UISs identified in vitro with those of the UISs isolated in vivo (and particularly those of low abundance that make up the majority of all UISs), regardless of disease status, we show that proviruses inserted in silenced DNA were more likely to establish long-lived infected T-cell clones (Figure 4). We hypothesize that lower proviral expression associated with transcriptional silencing of surrounding host DNA allows the infected cell to escape elimination by the immune response. We show that the difference in proviral load among patients with nonmalignant infection results mainly from a difference in the total number of UISs (ie, ACs have a lower proviral load because they have fewer infected clones; Figure 2C,E). This observation is consistent with the conclusion that ACs limit viral dissemination (and thus the establishment of T-cell clone populations) during primary infection more efficiently than persons who later develop HAM/TSP. Alternatively, progression from asymptomatic status to HAM/TSP may be accompanied by an increase in PVL and the total number of UISs. These results do not exclude the possibility that new UISs arise continuously at a low rate during persistent infection; further experimental and mathematical work is required to estimate the contribution of new UISs to the proviral load.
Among the established HTLV-1–infected cell populations, active transcription of the DNA flanking the provirus favors expansion of the infected lymphocytes (Figure 4A,C-D). Integration of the provirus in the same transcriptional orientation as the nearby host gene also drives selective expansion (Figure 5B). In ACs, the frequency of epigenetic marks associated with active transcription that was observed near integration sites was significantly lower in vivo than in vitro at all levels of UIS abundance (Figure 4C-D). In contrast, the frequency of such integration sites among the UISs of highest abundance in patients with HAM/TSP or ATLL was not significantly different from the frequency observed in vitro. That is, cells carrying proviruses integrated near activating epigenetic marks establish and expand more frequently in patients with HAM/TSP or ATLL than in ACs. These observations indicate that there was stronger selection against such integration sites in ACs than in patients with the HTLV-1–associated diseases. We postulate that the strong cell-mediated host immune response accounts for this observed negative selection of expressed proviruses. The model is depicted in supplemental Figure 5 by comparing the evolution of oligoclonality and the outcome of HTLV-1 infection between patients who differ in the efficiency of their anti-HTLV-1 immune response.
Even among the UISs of highest abundance, the frequency of proviruses integrated near genomic features associated with transcriptional activity (Figure 4A,C-D) never exceeded the frequency observed in proviral integration in vitro. We conclude that negative selection, probably because of the CTL response to HTLV-1, dominates during chronic infection and shapes the distribution of abundance of HTLV-1 clones in vivo. We also conclude that this negative selection is stronger in ACs than in patients with HAM/TSP or ATLL. This conclusion is consistent with evidence from studies of HTLV-1 genetics41 and host genetics42,43 in the chronic phase infection that ACs mount a more effective anti-HTLV-1 immune response than do patients with HAM/TSP.11 In contrast, ATLL patients are less able to restrict the abundance of the infected clones (Figure 2D), again perhaps because they mount a weaker anti-HTLV-1 immune response. The effectiveness of the CTL response to HTLV-1 is determined by the affinity with which host major histocompatibility complex class 1 alleles bind antigenic peptides derived from the viral antigen HBZ (HTLV-1 bZIP).44 In addition, the high frequency of FoxP3+ cells observed in HTLV-1 infection may limit both the effectiveness of the host immune response and the proliferation of HTLV-1–infected cells.45,46
The present results show that, despite a constant proviral load, the oligoclonality index increased significantly over time (Figure 3B), suggesting that there was competition among infected T-cell clones within each host. That is, expanding HTLV-1+ clones grow at the expense of minor clones, perhaps by competing for a finite resource (supplemental Figure 3). A combination of experimental and mathematical approaches will be needed to analyze the mechanism and the dynamics of this putative competition.
The proliferative advantage given to the infected cell by a provirus in same-sense orientation may result from increased transcription of the HTLV-1 tax gene: the Tax protein drives T-cell proliferation and can lead to cellular transformation.47 However, continued proviral expression may not be required in an established leukemic T-cell clone: proviral expression can be completely abrogated in leukemic clones by 5′-long terminal repeat methylation or by deletion or mutation of viral genes.12,48,49 The observation that in abundant UISs the HTLV-1 provirus was predominantly oriented in the same transcription sense as the host gene contrasts with the orientation of endogenous retroviruses, which are predominantly oriented in the opposite sense when inserted inside a gene.50 Further work will be necessary to identify the mechanisms by which host transcriptional activity influences proviral expression and to test the hypothesis that the ontology of the surrounding host genes also determines the extent of clonal expansion.
The technique described here could be adapted to analyze the genomic distribution of other proviruses or nonviral integrated elements and to monitor patients who undergo vector-based gene therapy because of its ability to quantify the UISs and to detect and quantify abnormally expanded clones.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the members of the Genomics Laboratory of the MRC Clinical Sciences Center, Hammersmith, London (Laurence Game, Adam Giess, Michael Jones, Nathalie Lambie, Frederique Maheo, and Elizabeth Webb) for their skill and commitment to this work and Masao Matsuoka (Institute for Virus Research, Kyoto University, Kyoto, Japan) for providing some of the ATLL samples.
This work was supported by the Wellcome Trust.
Wellcome Trust
Authorship
Contribution: N.A.G. and C.R.M.B. conceived and designed the experiments and wrote the paper; N.A.G. and N.M. analyzed the data; N.M., F.D.B., and C.B. provided the software tools to interrogate genomic databases; N.A.G., C.R.M.B., N.G., R.C., A.M., and D.B. developed the sequencing technique; and G.P.T. recruited the patients.
Conflict-of-interest disclosure: All authors at Illumina are employees of Illumina Inc, a public company that develops and markets systems for genetic analysis. The remaining authors declare no competing financial interests.
Correspondence: Charles R. M. Bangham, Department of Immunology, Wright-Fleming Institute, Imperial College London, London, W2 1PG, United Kingdom; e-mail: c.bangham@imperial.ac.uk; or Nicolas A. Gillet, Molecular and Cellular Epigenetics, Interdisciplinary Cluster for Applied Genoproteomics (GIGA) of University of Liège (ULg), avenue de l'Hôpital B34 Sart-Tilman, 4000 Liège, Belgium; e-mail: n.gillet@ulg.ac.be.
References
Author notes
N.M. and A.M. contributed equally to this study.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal