Abstract
Robust and rapid induction of interferon-β (IFN-β) in monocytes after pathogenic stimulation is a hallmark of innate immune responses. Here, we reveal the molecular mechanism underlying this key property that is exclusive to human blood monocytes. We found that IFN-β was produced rapidly in primary human monocytes as a result of cooperation between the myeloid-specific transcription factor IRF8 and the ubiquitous transcription factor IRF3. Knockdown of IRF8 in monocytes abrogated IFN-β transcription, whereas reintroduction of IRF8 into the IRF8−/− 32Dcl3 murine myeloid cell line reinstated IFN-β transcription. Moreover, we provide evidence that IRF8 constitutively binds to the ETS/IRF composite element of the IFN-β promoter region together with PU.1 in vivo. Furthermore we uncovered a requirement for IRF3, a master regulator of IFN-β production, as a previously un-indentified interaction partner of IRF8. We mapped the protein-protein interacting regions of IRF3 and IRF8, and found that their interaction was independent of the DNA-binding domain and the IRF association domain of IRF8 and IRF3, respectively. Therefore, we propose a model for the rapid induction of IFN-β in monocytes, whereby IRF8 and PU.1 form a scaffold complex on the IFN-β promoter to facilitate the recruitment of IRF3, thus enabling rapid IFN-β transcription.
Introduction
Production of interferon-β (IFN-β) by the host is an essential first line of defense against both viral and nonviral pathogens.1-3 As well as its involvement in innate immunity, IFN-β is an important initiator and modulator of the adaptive immune response.4-6 For example, IFN-β influences the differentiation, maturation, and migration of dendritic cells.7 In addition, it regulates antibody production and antigen-mediated apoptosis of B cells,8 alongside promoting the generation of CD4+ Th1 responses,9 and the expansion of antigen-specific CD8+ T cells.10 Interestingly, Stockinger et al have recently demonstrated that IFN-β production contributes to Listeria monocytogenes-induced lethality in mice.11 As such, IFN-β seems able to induce immunopathology as well as influence the resolution of infections, thus making it important to understand the mechanisms linking pathogen stimulation of immune cells with the production of IFN-β.
IFN-β transcription is initiated after cellular recognition of pathogen-associated molecular patterns by pattern recognition receptors.12 Many cell types express IFN-β after microbial exposure,13-15 although the underlying mechanisms vary. In murine embryonic fibroblasts, IFN-β induction is mediated by the activation of both IRF3 and IRF7.16,17 In contrast, induction of IFN-β in murine plasmacytoid dendritic cells occurs after CpG recognition by Toll-like receptor 9 and is entirely dependent on IRF7,18 whereas in murine macrophages transcription of IFN-β is dependent on IRF3 alone.19 IRF3 is a ubiquitous cytosolic transcription factor that translocates to the nucleus on activation,20 where it is able to induce IFN-β expression. However, this process takes more than 6 hours to produce detectable levels of IFN-β mRNA in HeLa cells, with levels peaking even later at 9 to 19 hours.21
Monocytes are myeloid cells of the immune system that play a key role in the rapid innate response to pathogens, including the production of IFN-β.11,22 However, the molecular mechanisms underlying the rapid induction of IFN-β in monocytes have not been elucidated, and the actions of ubiquitous IRF3 alone cannot account for the rapidity of IFN-β production seen after microbial stimulation. We asked whether the myeloid-specific transcription factor IRF823 was involved in rapid IFN-β production in monocytes. IRF8 coordinates cell growth and differentiation in lymphoid and myeloid cells, including monocytes.24 Previous studies demonstrated that IRF8 has a role in the feedback phase of IFN-β induction in murine dendritic cells,25 but it is unknown whether IRF8 is also involved in the rapid induction of IFN-β in human monocytes.
In this study, we demonstrated that, after pathogenic stimulation, primary human blood monocytes express IFN-β much more rapidly than nonmyeloid cell types. We related this finding to our data showing that IRF8 forms a complex with another myeloid transcription factor, PU.1,26 and together constitutively bind the IFN-β promoter region in vivo. Knockdown of either IRF8 or PU.1 profoundly reduced IFN-β transcription. Murine 32Dcl3 cells deficient in IRF8 regained their ability to rapidly transcribe IFN-β after reintroduction of IRF8. Finally, we revealed that IRF3, the master regulator of IFN-β transcription, is a novel and important interaction partner of IRF8.
Methods
Human blood monocyte isolation
Within 4 hours of donation at the Singapore Health Science Authority, peripheral blood mononuclear cells were isolated from buffy coat by centrifugation through Ficoll-paque (GE Healthcare). Monocytes were purified using MACS CD14 Microbeads (Miltenyi Biotec) according to the manufacturer's protocol. Monocyte purity was assessed by flow cytometry and was more than 95% for all experiments. For microbial ligands or virus stimulation, cells were exposed to 100 ng/mL lipopolysaccharide (LPS; Escherichia coli 05:88; Sigma-Aldrich), 1 μg/mL Pam3CSK4 (InvivoGen), or Sendai virus (Charles River Laboratories) at a multiplicity of infection of 2.5. All protocols were approved by the Institutional Review Board of the Health Science Authority of Singapore.
Luciferase reporter assays
32Dcl3 cells were transfected with Nucleofector (Lonza Switzerland) following the manufacturer's instructions, and luciferase activity was detected using a luciferase assay kit (Promega). Cells (1 × 106) were transfected with the reporter (2 μg pGL3-IFN-β promoter-luc), effectors (2 μg IRF8 plasmids or 2 μg IRF3 plasmids), and the internal control reporter (30 ng pGL2 SV40 promoter/enhancer driving luciferase).
Electrophoretic mobility shift assay
Nuclear proteins were extracted from THP-1 and HEK293 cells using Nuclear Extraction-Protein Extraction Reagent Kit nuclear and cytoplasmic extraction reagents (Pierce Chemical) with a protease inhibitor cocktail (Sigma-Aldrich). An oligonucleotide probe containing the IFN-β promoter region was prepared and end labeled with biotin by Sigma-Aldrich. Detection of transcription factor-oligonucleotide complexes was performed using a LightShift chemiluminescent EMSA kit (Pierce Chemical), according to the method previously described.27 For supershift assays, anti-IRF8, anti-PU.1, or anti-IRF3 antibodies were added to the binding-reaction mixture. The IRF8/PU.1 binding probe from IgλL oligonucleotides sequence was as follows: 5′-CTAGCGAGAAATAAAAGGAAGTGAAACCAAGA-3′.28 The in vitro translated IRF8 and PU.1 proteins were synthesized using the TNT T7 transcription/translation system (Promega).
ChIP
Human monocytes were cross-linked with 1% formaldehyde at room temperature and then quenched using 125mM glycine. The crosslinked chromatin and associated protein was isolated and sonicated to produce chromatin fragments with an average size of 500 bp. Chromatin fragments were immunoprecipitated using anti-GST, anti-PU.1, or anti-IRF8 antibodies bound to beads. For the chromatin immunoprecipitation (ChIP) experiments, the method has been described.29 Real-time polymerase chain reaction (PCR) analyses were performed using the ABI Prism 7900 Sequence Detection System. Relative occupancy values were calculated by determining the apparent immunoprecipitation efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample) and normalized to the level observed at a control region, which was defined as 1.0. All primers used gave a single product of the expected size, as confirmed by agarose gel electrophoresis. These primers also gave no DNA product in the no-template control. The primers used for real-time PCR to quantitate the ChIP-enriched DNA are as indicated in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Coimmunoprecipitation
For the immunoprecipitation of expressed exogenous proteins, transfected HEK293 cells were lysed using 1% Triton X-100 in phosphate-buffered saline (pH 7.4). Cell lysate was precleared overnight at 4°C using 30 μL mouse IgG-Agarose (Sigma-Aldrich) and then incubated with 30 μL FLAG antibody-Sepharose (Sigma-Aldrich) for 3 hours at 4°C. Nonspecifically bound proteins were washed off using 0.1% Triton X-100/phosphate-buffered saline (pH 7.4). Immunoprecipitated proteins were then eluted with 2 times sodium dodecyl sulfate loading buffer before Western blotting.
For the immunoprecipitation of endogenous proteins, the experiment was performed according to a previous published protocol30 with some modification. After washing with 1 times Tris-buffered saline (pH 7.4), THP-1 cells (1 × 107per reaction) were lysed using 1% digitonin in 1 times Tris-buffered saline (pH 7.4) with mini-ethylenediaminetetraacetic acid-free protease inhibitor cocktail (Roche Diagnostics). Cell lysates were precleared overnight at 4°C using 30 μL mouse IgG-Agarose (Sigma-Aldrich) and then incubated with 2 μg of antibodies overnight at 4°C. Afterward, 30 μL protein-G Sepharose (GE Healthcare) was added and incubated for 1 hour at 4°C. The nonspecifically bound proteins were removed by 3 washes with 0.1% digitonin/1 times Tris-buffered saline (pH 7.4) with ethylenediaminetetraacetic acid-free protease inhibitor cocktail. Immunoprecipitated proteins were eluted using 2 times sodium dodecyl sulfate loading buffer.
Other methods
Other methods, including cell culture, plasmids, RNA isolation, enzyme-linked immunosorbent assay, protein extraction and Western blotting, and RNAi transfection of human monocytes, are in the Supplemental data.
Results
Human blood monocytes rapidly transcribe IFN-β after microbial stimulation
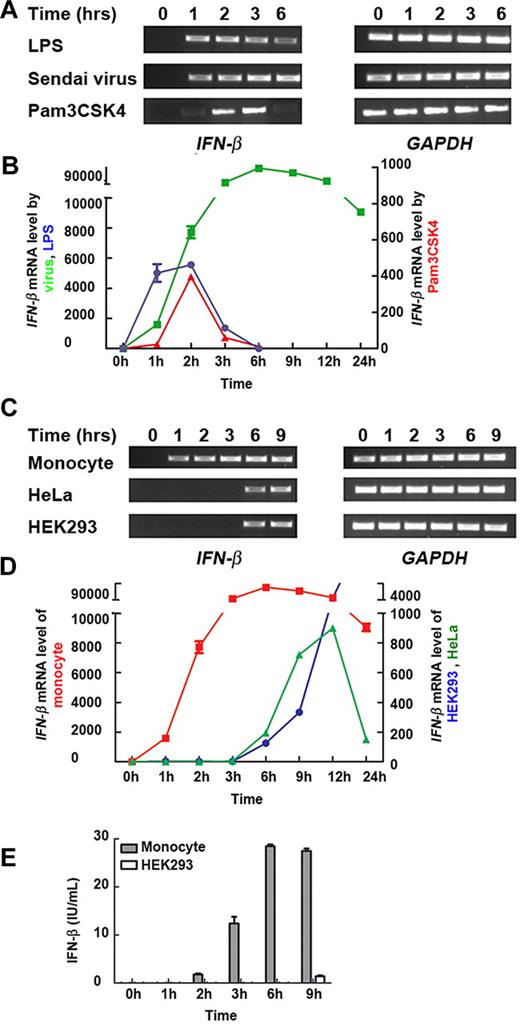
IFN-β is an essential component of the defense mechanism used by innate immune cells. We stimulated primary human blood monocytes with the Toll-like receptor ligands LPS, Pam3CSK4, or Sendai virus and measured the amount of IFN-β mRNA at various time points. IFN-β was induced as early as 1 hour after stimulation and rapidly reached maximal levels (Figure 1A-B), reflecting the remarkably quick monocyte response to a range of pathogenic stimuli.
Human monocytes transcribe IFN-β more rapidly than nonmyeloid cell types. (A) Primary monocytes were exposed to LPS, Pam3CSK4, or Sendai virus for 0, 1, 2, 3, and 6 hours. The level of IFN-β mRNA was measured by semiquantitative reverse transcription PCR. (B) The kinetics of IFN-β mRNA expression in pathogen-stimulated primary monocytes. Monocytes were exposed to LPS (blue), Sendai virus (green), or Pam3CSK4 (red). The level of IFN-β mRNA was then measured by real-time PCR. (C) Primary monocytes, and nonmyeloid HeLa and HEK293 cells were exposed to Sendai virus for 0, 1, 2, 3, 6, and 9 hours. The level of IFN-β mRNA was measured by semiquantitative reverse transcription PCR. (D) The kinetics of IFN-β mRNA expression in Sendai virus-exposed primary monocytes (red), HeLa cells (green), and HEK293 cells (blue). The level of IFN-β mRNA was then measured by real-time PCR. (E) Primary monocytes and HEK293 cells were stimulated by Sendai virus. The supernatants were tested by enzyme-linked immunosorbent assay for IFN-β production. (A-E) Similar results were obtained in 3 independent experiments with monocytes from 3 different donors, one of which is shown.
Human monocytes transcribe IFN-β more rapidly than nonmyeloid cell types. (A) Primary monocytes were exposed to LPS, Pam3CSK4, or Sendai virus for 0, 1, 2, 3, and 6 hours. The level of IFN-β mRNA was measured by semiquantitative reverse transcription PCR. (B) The kinetics of IFN-β mRNA expression in pathogen-stimulated primary monocytes. Monocytes were exposed to LPS (blue), Sendai virus (green), or Pam3CSK4 (red). The level of IFN-β mRNA was then measured by real-time PCR. (C) Primary monocytes, and nonmyeloid HeLa and HEK293 cells were exposed to Sendai virus for 0, 1, 2, 3, 6, and 9 hours. The level of IFN-β mRNA was measured by semiquantitative reverse transcription PCR. (D) The kinetics of IFN-β mRNA expression in Sendai virus-exposed primary monocytes (red), HeLa cells (green), and HEK293 cells (blue). The level of IFN-β mRNA was then measured by real-time PCR. (E) Primary monocytes and HEK293 cells were stimulated by Sendai virus. The supernatants were tested by enzyme-linked immunosorbent assay for IFN-β production. (A-E) Similar results were obtained in 3 independent experiments with monocytes from 3 different donors, one of which is shown.
To understand whether this rapid IFN-β response was a specific property of monocytes, we compared the kinetics of IFN-β mRNA production with that in HeLa and HEK293 cells in response to Sendai virus exposure. IFN-β transcription was detected in all cell types after incubation with Sendai virus. In line with published literature,21,31 the nonmyeloid HeLa and HEK293 cells expressed IFN-β mRNA from approximately 6 hours after stimulation, with a further increase at 9 hours (Figure 1C-D).
To support this, the kinetics of IFN-β protein production from Sendai virus-stimulated HEK293 cells and monocytes were measured by enzyme-linked immunosorbent assay (Figure 1E). IFN-β release from monocytes was much faster than HEK293 cells. After 2-hour incubation with virus, IFN-β was already detectable in the supernatants of monocytes, whereas 9 hours of incubation was required for the detectable IFN-β expression from HEK293 cells. This led us to ask how IFN-β induction in monocytes was regulated differently to permit such a unique response to the same pathogenic stimulus.
IRF8 is an essential transcriptional regulator of IFN-β expression in monocytes
We hypothesized that IRF8, a transcription factor found only in lymphoid and myeloid cells such as macrophages,24,32 might be involved. Unstimulated human monocytes expressed IRF8 at both the mRNA and protein level, and this level was increased after LPS stimulation (supplemental Figure 1A-B). We then asked whether IRF8 was essential for IFN-β expression in monocytes. Endogenous IRF8 mRNA (supplemental Figure 1C) and protein expression (Figure 2A bottom) was eliminated in cells using 2 siRNAs. In IRF8 knockdown monocytes stimulated with LPS, transcription of IFN-β was negligible compared with control cells (Figure 2A top); therefore, IRF8 was necessary for IFN-β transcription in monocytes.
IRF8 positively regulates IFN-β transcription in monocytes. (A) Primary monocytes were transfected with IRF8 siRNA or control siRNA. Western blot (bottom) shows the IRF8 protein level in total cell lysates. IFN-β transcription in monocytes stimulated with LPS for 2 hours, determined by real-time PCR (top). (B) The mapping of IRF8-binding sites across IFN-β genomic DNA using 5 different pairs of primers (bottom). Primary monocytes were stimulated with LPS for 1 hour before ChIP analysis. ChIP was performed using either anti-IRF8 or control anti-GST antibody. (A-B) Similar results were obtained in 4 independent experiments with monocytes from 4 different donors, one of which is shown. (C) The sequences of IFN-β WT(−99 to −71) probe containing the EIRE motif and WT(−83 to −55) probe containing the EICE motif are shown. (D) EMSA was performed with IFN-β WT(−99 to −71) probe (lanes 1-4) or WT(−83 to −55) probe (lanes 5-8). Lanes 1 and 5 indicate free probe; lanes 2 and 6, with nuclear extract from LPS stimulated THP-1 cells after 2 hours; lanes 3 and 7, addition of 20-fold molar excess of unlabeled WT(−99 to −71) competitor probe; and lanes 4 and 8, addition of 20-fold molar excess of unlabeled WT(−83 to −55) competitor probe. (E) EMSA was performed with IFN-β WT(−83 to −55) probe. Lane 1 indicates free probe; lanes 2 to 4, nuclear extract from LPS stimulated THP-1 cells after 2 hours; lane 3, control anti-GST antibody; and lane 4, anti-IRF8 antibody. (D-E) Similar results were obtained in 4 independent experiments, one of which is shown.
IRF8 positively regulates IFN-β transcription in monocytes. (A) Primary monocytes were transfected with IRF8 siRNA or control siRNA. Western blot (bottom) shows the IRF8 protein level in total cell lysates. IFN-β transcription in monocytes stimulated with LPS for 2 hours, determined by real-time PCR (top). (B) The mapping of IRF8-binding sites across IFN-β genomic DNA using 5 different pairs of primers (bottom). Primary monocytes were stimulated with LPS for 1 hour before ChIP analysis. ChIP was performed using either anti-IRF8 or control anti-GST antibody. (A-B) Similar results were obtained in 4 independent experiments with monocytes from 4 different donors, one of which is shown. (C) The sequences of IFN-β WT(−99 to −71) probe containing the EIRE motif and WT(−83 to −55) probe containing the EICE motif are shown. (D) EMSA was performed with IFN-β WT(−99 to −71) probe (lanes 1-4) or WT(−83 to −55) probe (lanes 5-8). Lanes 1 and 5 indicate free probe; lanes 2 and 6, with nuclear extract from LPS stimulated THP-1 cells after 2 hours; lanes 3 and 7, addition of 20-fold molar excess of unlabeled WT(−99 to −71) competitor probe; and lanes 4 and 8, addition of 20-fold molar excess of unlabeled WT(−83 to −55) competitor probe. (E) EMSA was performed with IFN-β WT(−83 to −55) probe. Lane 1 indicates free probe; lanes 2 to 4, nuclear extract from LPS stimulated THP-1 cells after 2 hours; lane 3, control anti-GST antibody; and lane 4, anti-IRF8 antibody. (D-E) Similar results were obtained in 4 independent experiments, one of which is shown.
IRF8-mediated transcriptional regulation depends on its binding to specific conserved DNA sequences in the promoter region of the gene. Given our observations of rapid IRF8-dependent transcription of IFN-β in monocytes, next we studied IRF8 binding to the IFN-β gene to reveal whether its effect was direct. ChIP assays were first performed in LPS-stimulated human monocytes29 using an antibody against IRF8 and a control antibody against GST. These assays revealed that IRF8 bound significantly more to the IFN-β locus compared with control DNA (ZNF524; supplemental Figure 2A). One possible molecular mechanism of promoting the rapid induction of genes is priming by constitutively bound transcription factors. We wondered whether this might be the case with IRF8 in monocytes. Using ChIP, we found that bound IRF8 could be detected before activation (supplemental Figure 2B), suggesting that IRF8 might prime the IFN-β gene for rapid induction. We also observed increased levels of IRF8 binding to the IFN-β locus in LPS-stimulated monocytes after 1 hour (supplemental Figure 2B).
To understand the molecular basis of IRF8-mediated IFN-β regulation, we wanted to further characterize the interaction between IRF8 and the IFN-β locus. To define the binding site of IRF8, we examined the occupancy of different genomic regions within the IFN-β gene (Figure 2B). PCR primers were designed to correspond to sequences within the promoter region, 5′ upstream region, and the internal sequence of the gene.33,34 IRF8's binding affinity for each region of the IFN-β gene was then assessed by anti-IRF8 ChIP. IRF8 exhibited maximal binding at the promoter region of the IFN-β gene (primer III, Figure 2B), relative to the 5′ upstream and internal regions (Figure 2B). The sequences of the putative IRF8-binding sites within the promoter region of human and mouse IFN-β are shown in supplemental Figure 2C. They contain 2 conserved elements (5′-GAAA-3′), both of which are known IRF8-binding sites.24 One is the ETS/IRF response element (EIRE, -GGAANNNNGAAA-, −83 to −80 bp), and the other is the ETS/IRF composite element (EICE, -GAAANNNGGAA-, −70 to −67 bp) (supplemental Figure 2C).
To identify the precise binding site of IRF8 within the IFN-β promoter region, we designed a set of DNA probes for use in electrophoretic mobility shift assays (EMSAs). Probes derived from the IFN-β promoter region included those with the aforementioned EIRE and EICE motifs (Figure 2C). In Figure 2D, the lanes contained probe only and accordingly showed only a single band of free DNA (Figure 2D lanes 1 and 5). DNA-protein complexes were observed in the nuclear extracts incubated with the IFN-β wild-type (WT)(−83 to −55) probe (Figure 2D lane 6), which contained the EICE motif. Conversely, no DNA-protein complexes were observed with the WT(−99 to −71) probe containing the EIRE motif (Figure 2D lane 2). As an additional control, unlabeled competitor oligonucleotides were added to the binding reactions in 20-fold excess of probe. The DNA-protein complex was competed out by a WT(−83 to −55) competitor (Figure 2D lane 8), but not a WT(−99 to −71) competitor (Figure 2D lane 7). Thus, nuclear protein binds to WT(−83 to −55) DNA rather than WT(−99 to −71) DNA. To confirm that the DNA-protein complex contained IRF8, a second EMSA was carried out with anti-IRF8 antibody. The probe alone formed no complex (Figure 2E lane 1), whereas a DNA-protein complex was observed with nuclear extract and the WT(−83 to −55) probe (Figure 2E lane 2). This DNA-protein complex band was supershifted when anti-IRF8 antibody was added (Figure 2E lane 4), but not in the presence of a control antibody (Figure 2E lane 3). This indicates that the -GAAA- motif (−70 to −67 bp) in IFN-β WT(−83 to −55) DNA represents a specific IRF8-binding site within the promoter region of the IFN-β gene.
IRF8 restores IFN-β transcription in 32Dcl3 (IRF8−/−) cells
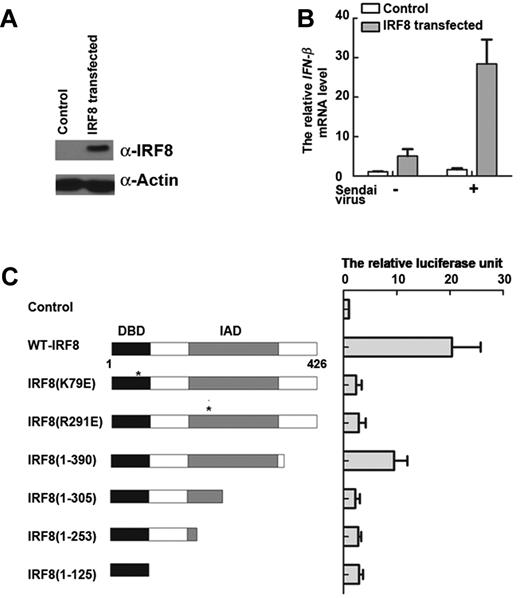
Our results so far have demonstrated that the transcription factor IRF8 is essential for IFN-β transcription in monocytes. To further substantiate this observation, we reintroduced IRF8 into cells lacking this particular transcription factor. 32Dcl3 is a murine myeloid progenitor cell line that naturally does not express IRF835 (supplemental Figure 3A). Cells were transfected with a plasmid encoding the IRF8 gene, and subsequent IRF8 protein expression was confirmed by Western blot (Figure 3A). Sendai virus was added to control or IRF8-reconstituted cells to provide a stimulus for IFN-β transcription. After 2 hours, IFN-β transcripts were detected by real-time PCR. Nontransfected 32Dcl3 cells failed to produce detectable levels of IFN-β, whereas IFN-β transcription was restored in the IRF8-transfected cells (Figure 3B). These findings further validate and extend our previous observation that IRF8 is a key determinant of IFN-β transcription in monocytic cells of both human and murine origin.
IRF8 contributes to the rapid expression of IFN-β. (A) 32Dcl3 cells were transfected with IRF8 plasmid. IRF8 protein levels in total cell lysates by Western blot. Similar results were obtained in 3 independent experiments, one of which is shown. (B) Reintroduction of IRF8 into 32Dcl3 (IRF8−/−) cells. IFN-β transcription in cells stimulated with Sendai virus for 2 hours, as determined by real-time PCR. (C) WT and mutant IRF8 activate expression of a luciferase reporter construct driven by the IFN-β promoter region in 32Dcl3 cells induced by Sendai virus after 2 hours. A schematic representation of IRF8 mutant constructs is shown on the left. The DBD and IAD are highlighted. Luciferase activity of the respective constructs is shown in the graph on the right. (B-C) Data are mean ± SD from 3 independent experiments.
IRF8 contributes to the rapid expression of IFN-β. (A) 32Dcl3 cells were transfected with IRF8 plasmid. IRF8 protein levels in total cell lysates by Western blot. Similar results were obtained in 3 independent experiments, one of which is shown. (B) Reintroduction of IRF8 into 32Dcl3 (IRF8−/−) cells. IFN-β transcription in cells stimulated with Sendai virus for 2 hours, as determined by real-time PCR. (C) WT and mutant IRF8 activate expression of a luciferase reporter construct driven by the IFN-β promoter region in 32Dcl3 cells induced by Sendai virus after 2 hours. A schematic representation of IRF8 mutant constructs is shown on the left. The DBD and IAD are highlighted. Luciferase activity of the respective constructs is shown in the graph on the right. (B-C) Data are mean ± SD from 3 independent experiments.
We made further use of the reconstituted 32Dcl3 model to understand the molecular mechanism of IRF8-mediated IFN-β transcription. IRF8 has a DNA-binding domain (DBD) in the N-terminus and an IRF association domain (IAD) in the C-terminus.20 We generated a range of IRF8 constructs mutated in each of these regions, transfected them into 32Dcl3 cells lacking IRF8, and carried out an IFN-β promoter luciferase assay (Figure 3C). WT-IRF8 induced a 20-fold increase in luciferase activity compared with that in nontransfected cells (Figure 3C). In contrast, there were only basal levels of activity for most of the truncated forms of IRF8, including mutants of IRF8(K79E), IRF8(R291E), IRF8(1-305), IRF8(1-253), and IRF8(1-125). Surprisingly, IRF8(1-390) exhibited a 10-fold increase in luciferase activity compared with basal levels, which might be explained by the fact that IRF8(1-390) has an intact DBD and IAD, lacking only 36 residues at the C-terminus (Figure 3C). Therefore, we showed that reintroduction of IRF8 into 32Dcl3 cells restored the ability to efficiently transcribe IFN-β in response to viral infection. Moreover, complete domains in IRF8 are important for IFN-β transcription; mutation in either the DBD or IAD profoundly affects the ability of IRF8 to initiate IFN-β transcription. These findings are consistent with other groups' studies showing that binding of IRF8 to the IL-12B gene depends not only on an intact DBD, but also the interaction with another partner via the IAD.36
IRF8 binds to the IFN-β promoter constitutively with PU.1 in monocytes
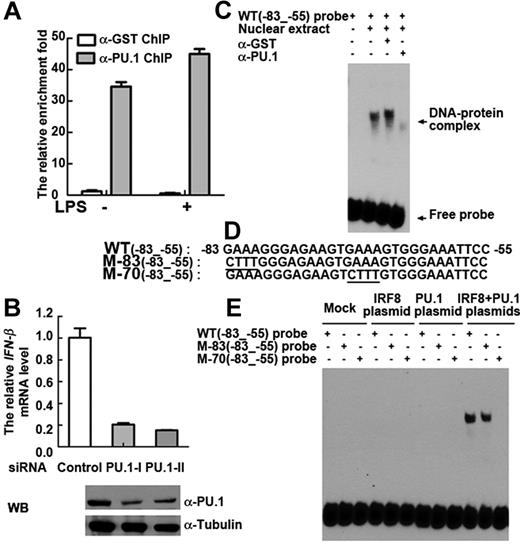
IRF8 occupies EICE (-GAAANNNGGAA-) by interacting with PU.1.37 The EMSA results in Figure 2D and E suggest that IRF8 bound to -GAAA- in the EICE motif of the IFN-β promoter; therefore, we hypothesized that PU.1 might be involved in mediating IRF8 binding and IFN-β transcription in monocytes. To ascertain whether PU.1 bound to the IFN-β promoter, we carried out ChIP assays and observed that, like IRF8, PU.1 constitutively binds to IFN-β genomic DNA (Figure 4A). PU.1 binding also increased after LPS stimulation in monocytes (Figure 4A). To assess the functional role of PU.1 in IFN-β induction, we used siRNAs to knock down PU.1 mRNA (supplemental Figure 3B) and protein expression (Figure 4B bottom). In PU.1 knockdown monocytes, mRNA transcripts of IFN-β were profoundly reduced (Figure 4B top). A further reduction in IFN-β transcription was seen when we concurrently knocked down both PU.1 and IRF8 in monocytes (supplemental Figure 3C-D). These findings suggest that the constitutive binding of PU.1 to the IFN-β promoter also contributes to LPS-induced rapid IFN-β gene transcription.
Occupancy of the IFN-β promoter by IRF8 is stabilized by PU.1. (A) ChIP analysis (with PU.1 antibody or control GST antibody) was performed in unstimulated and LPS-stimulated primary monocytes. ZNF524 primer was used as control. (B) Primary monocytes were transfected with PU.1 siRNA or control siRNA. Western blot (bottom) shows the PU.1 protein level in total cell lysates. Top: IFN-β gene expression in cells that were stimulated with LPS for 2 hours. (A-B) Similar results were obtained in 5 independent experiments with monocytes from 5 different donors, one of which is shown. (C) EMSA was performed with IFN-β WT(−83 to −55) probe. Lane 1 indicates free probe; lanes 2 to 4, nuclear extract from LPS stimulated THP-1 cells after 2 hours; lane 3, control GST antibody; and lane 4, PU.1 antibody. (D) The sequences of IFN-β wild-type WT(−83 to −55) probe and mutant M-83(−83 to −55) and mutant M-70(−83 to −55) probes are shown. (E) EMSA was performed with nuclear extracts from HEK293 cells transfected with mock control (lanes 1-3), IRF8 plasmid (lanes 4-6), PU.1 plasmid (lanes 7-9), or both IRF8 and PU.1 plasmids (lanes 10-12). WT(−83 to −55) probe and M-83(−83 to −55) and M-70(−83 to −55) probes were used. (C,E) Similar results were obtained in 3 independent experiments, one of which is shown.
Occupancy of the IFN-β promoter by IRF8 is stabilized by PU.1. (A) ChIP analysis (with PU.1 antibody or control GST antibody) was performed in unstimulated and LPS-stimulated primary monocytes. ZNF524 primer was used as control. (B) Primary monocytes were transfected with PU.1 siRNA or control siRNA. Western blot (bottom) shows the PU.1 protein level in total cell lysates. Top: IFN-β gene expression in cells that were stimulated with LPS for 2 hours. (A-B) Similar results were obtained in 5 independent experiments with monocytes from 5 different donors, one of which is shown. (C) EMSA was performed with IFN-β WT(−83 to −55) probe. Lane 1 indicates free probe; lanes 2 to 4, nuclear extract from LPS stimulated THP-1 cells after 2 hours; lane 3, control GST antibody; and lane 4, PU.1 antibody. (D) The sequences of IFN-β wild-type WT(−83 to −55) probe and mutant M-83(−83 to −55) and mutant M-70(−83 to −55) probes are shown. (E) EMSA was performed with nuclear extracts from HEK293 cells transfected with mock control (lanes 1-3), IRF8 plasmid (lanes 4-6), PU.1 plasmid (lanes 7-9), or both IRF8 and PU.1 plasmids (lanes 10-12). WT(−83 to −55) probe and M-83(−83 to −55) and M-70(−83 to −55) probes were used. (C,E) Similar results were obtained in 3 independent experiments, one of which is shown.
To further substantiate the functional relationship between IRF8 and PU.1 in IFN-β induction, we overexpressed both proteins in HEK293 cells, which are of epithelial origin and therefore naturally lack both IRF8 and PU.1.28 Overexpression of either IRF8 or PU.1 did not dramatically increase IFN-β transcription. However, overexpressing both proteins simultaneously induced a significant increase in IFN-β transcription (supplemental Figure 3E). To validate this observation of a cooperative synergism between IRF8 and PU.1, we carried out EMSA. Figure 4C shows EMSA results obtained using an anti-PU.1 antibody to visualize the interaction of PU.1 with the IFN-β promoter region. Although a high molecular weight band corresponding to a DNA-protein complex could be seen (Figure 4C, lane 2), the band intensity was decreased in the presence of the anti-PU.1 antibody (Figure 4C, lane 4), compared with control antibody (anti-GST). It is possible that the DNA-protein complex containing PU.1 was largely eliminated by the antibody to PU.1. Regardless of this technical point, the clear presence of a high molecular weight band demonstrates that PU.1 is an additional DNA binding factor at the IFN-β promoter.
To demonstrate the interdependence of IRF8 and PU.1 for DNA binding, WT and mutant probes from −83 to −55 bp of the IFN-β promoter were used (Figure 4D). An EMSA was performed with nuclear extracts derived from the IRF8- and PU.1-transfected HEK293 cells (Figure 4E). This showed that formation of the DNA-protein complex could be completely abolished by mutation of the sequence between the -GAAA- at −70 to −67 bp. In contrast, mutation of the -GAAA- sequence between −83 to −80 did not affect complex formation, demonstrating the specificity of the IRF8/PU.1 complex for the sequence between −70 to −67 bp. Interestingly, the nuclear extract from PU.1 plasmid transfected-HEK293 cells did not bind to the IFN-β probe (Figure 4E), which is in contrast to the reported finding for PU.1 binding to the IgGλL probe (supplemental Figure 4A).28
To confirm the nature of the proteins bound to the IFN-β promoter, in vitro translated proteins were used to repeat the results obtained using nuclear extracts. Consistent with the experiments using nuclear extracts, in vitro expressed IRF8 protein or PU.1 protein alone did not bind to the IFN-β probe, whereas coincubation of IRF8 and PU.1 proteins led to the formation of DNA-protein complexes (supplemental Figure 4B). The antibodies to IRF8 and PU.1 were also tested as potential supershift reagents. As shown in supplemental Figure 4C, the DNA-protein complex was indeed supershifted with anti-IRF8 antibodies, and was inhibited with antibodies against PU.1. No change was seen with the control antibody (anti-GST). These data show that IRF8 and its partner PU.1 interact with the IFN-β promoter.
IRF8 functionally synergizes with IRF3 for IFN-β transcription
The occupancy site of IRF8 is located at -GAAA- (−70 to −67 bp) (Figure 4E), which is close to one of IRF3 binding sites on the IFN-β gene38 (Figure 5A). To determine whether IRF3, similarly to IRF8, binds within the IFN-β promoter, EMSA using the antibody against IRF3 was performed. The heterocomplex band, which was supershifted by antibodies specific for IRF8 (Figure 2E), was similarly inhibited by the antibody against IRF3 (supplemental Figure 5A), indicating that the DNA-protein complex contained IRF3 in addition to IRF8. This raised the possibility of an interaction between IRF8 and IRF3. We used a series of approaches to detect any interaction of IRF8 with IRF3. We first examined whether IRF3 interacts with IRF8 using coimmunoprecipitation analysis. We found that immunoprecipitation of IRF3 coimmunoprecipitates IRF8 (Figure 5B). Therefore, IRF3 interacts with IRF8. Subsequently, siRNA-mediated knockdown experiments were conducted in monocytes to assess the functional importance of IRF3 and IRF8. Knockdown of either IRF3 (supplemental Figure 5B) or IRF8 markedly reduced the transcription of IFN-β, but knockdown of both IRF3 and IRF8 had an additive effect, and completely abolished the presence of IFN-β transcripts (Figure 5C). Next, we transfected 32Dcl3 cells with IFN-β promoter luciferase construct and then additionally overexpressed IRF8 and IRF3. When cells were transfected with IRF3 or IRF8 alone, a modest transcriptional activation of the IFN-β-driven luciferase reporter construct was observed (Figure 5D). However, cotransfection of IRF3 and IRF8 expression vectors induced a strong synergistic transactivation (Figure 5D). These findings demonstrate that IRF3 and IRF8 interact functionally to regulate IFN-β induction.
IRF8 cooperates with IRF3 to regulate IFN-β transcription. (A) The genomic sequence of human IFN-β from −79 bp to −58 bp. The IRF3- and IRF8-binding sites are labeled in blue and red, respectively. (B) THP-1 cells were induced with LPS for 2 hours before total lysates were harvested and immunoprecipitation was performed. Western blotting of immunoprecipitated proteins is shown. Similar results were obtained in 3 independent experiments. (C) Combined knockdown of IRF8 and IRF3 using siRNA results in a pronounced reduction of IFN-β transcription in primary monocytes. Monocytes were transfected with control siRNA, IRF8 siRNA, IRF3 siRNA, or both IRF8 and IRF3 siRNAs. mRNA levels of IFN-β in LPS simulated-monocytes were determined by real-time PCR. Similar results were obtained in 3 independent experiments with monocytes from 3 different donors, one of which is shown. (D) 32Dcl3 cells were transfected with control, IRF3 plasmid, IRF8 plasmid, or both IRF8 and IRF3 plasmids, and then cells were stimulated with Sendai virus for 2 hours to activate expression of an IFN-β promoter luciferase. Values are mean plus or minus SD from 3 independent experiments. (E) EMSA was performed with siRNA-transfected THP-1 nuclear extract. Lane 1 indicates free probe; lanes 2 and 3, nuclear extract from LPS-stimulated THP-1 cells transfected with siRNA (lane 2 indicates control siRNA; and lane 3, IRF8 siRNA). (F) EMSA was performed with LPS stimulated THP-1 nuclear extracts. WT(−83 to −55) probe and M-70(−83 to −55) probes were used. Similar results were obtained in 3 independent experiments, one of which is shown. (G) IRF8–2HA mutants overexpressed in HEK293 cells with IRF3–5D-2FLAG or IRF3–5D were immunoprecipitated with anti-FLAG antibody. Western blot was performed with anti-HA antibody. (H) IRF3–5D-2FLAG and IRF3–5D mutants were overexpressed together with IRF8–2HA in HEK293 cells and immunoprecipitated with anti-FLAG antibody or antimouse IgG. Western blot was performed with anti-HA antibody. (E-H) Similar results were obtained in 3 independent experiments, one of which is shown.
IRF8 cooperates with IRF3 to regulate IFN-β transcription. (A) The genomic sequence of human IFN-β from −79 bp to −58 bp. The IRF3- and IRF8-binding sites are labeled in blue and red, respectively. (B) THP-1 cells were induced with LPS for 2 hours before total lysates were harvested and immunoprecipitation was performed. Western blotting of immunoprecipitated proteins is shown. Similar results were obtained in 3 independent experiments. (C) Combined knockdown of IRF8 and IRF3 using siRNA results in a pronounced reduction of IFN-β transcription in primary monocytes. Monocytes were transfected with control siRNA, IRF8 siRNA, IRF3 siRNA, or both IRF8 and IRF3 siRNAs. mRNA levels of IFN-β in LPS simulated-monocytes were determined by real-time PCR. Similar results were obtained in 3 independent experiments with monocytes from 3 different donors, one of which is shown. (D) 32Dcl3 cells were transfected with control, IRF3 plasmid, IRF8 plasmid, or both IRF8 and IRF3 plasmids, and then cells were stimulated with Sendai virus for 2 hours to activate expression of an IFN-β promoter luciferase. Values are mean plus or minus SD from 3 independent experiments. (E) EMSA was performed with siRNA-transfected THP-1 nuclear extract. Lane 1 indicates free probe; lanes 2 and 3, nuclear extract from LPS-stimulated THP-1 cells transfected with siRNA (lane 2 indicates control siRNA; and lane 3, IRF8 siRNA). (F) EMSA was performed with LPS stimulated THP-1 nuclear extracts. WT(−83 to −55) probe and M-70(−83 to −55) probes were used. Similar results were obtained in 3 independent experiments, one of which is shown. (G) IRF8–2HA mutants overexpressed in HEK293 cells with IRF3–5D-2FLAG or IRF3–5D were immunoprecipitated with anti-FLAG antibody. Western blot was performed with anti-HA antibody. (H) IRF3–5D-2FLAG and IRF3–5D mutants were overexpressed together with IRF8–2HA in HEK293 cells and immunoprecipitated with anti-FLAG antibody or antimouse IgG. Western blot was performed with anti-HA antibody. (E-H) Similar results were obtained in 3 independent experiments, one of which is shown.
We have determined that IRF3 and IRF8 interact with each other and IRF8 binds constitutively to the promoter region of the IFN-β before induction. To determine whether the binding of IRF8 promotes the recruitment of IRF3 to IFN-β DNA, we depleted IRF8 and examined the occupancy of IRF3 at IFN-β promoter by EMSA. Interestingly, on depletion of IRF8, we observed the reduction in DNA-protein complex formation (Figure 5E), which previously contained IRF3 (supplemental Figure 5A). Mutations within the consensus IRF8 binding motif (−70 -GAAA- −67) similarly led to the disruption of DNA-protein complex formation (Figure 5F). These data suggest that the binding of IRF8 stabilizes DNA-protein complex formation and facilitates the occupancy of IRF3 to the IFN-β promoter.
We then investigated the physical interaction between IRF8 and IRF3 by immunoprecipitation. Using the lysates of cells transfected with IRF3 and IRF8, we showed that the 2 proteins were able to interact (supplemental Figure 5C). IRF8 mutant clones were also made and then assessed for interactions with IRF3 (Figure 5G; supplemental Figure 5D). WT-IRF8 and IRF8(K79E) interacted with IRF3–5D-2FLAG (a constitutively active form of IRF339 ), suggesting that both proteins can interact independently of the IRF8 DBD to form a strong complex (Figure 5G lanes 1 and 3). The point mutation R291E in the IAD domain of IRF8 also retained the interaction with IRF3–5D-FLAG (Figure 5G lane 5). Similarly, IRF8 mutants with a major deletion in the IAD, designated IRF8(1-390), IRF8(1-305), and IRF8(1-253) (Figure 5G lanes 7, 9, and 11), as well as the DBD-deletion mutant IRF8(Δ1-122), were able to interact with IRF3 (Figure 5G lane 15). All the mutations that most greatly impaired the interaction between IRF8 and IRF3 occurred in the region between the DBD and the IAD of IRF8 (Figure 5G lane 13). The observation that interactions between IRF8 and IRF3 were preserved, even in the various DBD and IAD mutants, raises the possibility that the IRF8 DBD and IAD subdomains may not be involved in protein-protein interactions between IRF8 and IRF3. To confirm this, another mutant IRF8(100-270) was tested with WT-IRF3. Figure 5G lane 17 shows that IRF8(100-270) is able to bind the constitutively active form of IRF3. Thus, residues 100 to 270 of IRF8 are necessary for its interaction with IRF3. Because this truncated mutant lacks the DBD and IAD domains, it cannot be sufficient to lead to IFN-β transcription (supplemental Figure 5E).
To complete our understanding of the nature of the interaction between IRF8 and IRF3, we proceeded to map the corresponding domains of IRF3 responsible for its association with IRF8. First, a series of deletions were made at the carboxyl and amino termini of the molecule (supplemental Figure 5H). Deletions from position 253 to the C-terminal end of IRF3 had no effect on its ability to associate with IRF8 (Figure 5H lane 9). In contrast, IRF3(1-131) and IRF3(1-120) with additional deletions toward the N-terminus were unable to form complexes with IRF8 (Figure 5H lanes 11 and 13). This suggests that the carboxyl end of the association domain is around residue 253. To further refine this estimation, a series of IRF3 mutants were constructed. When the first 74 amino acids of the DBD were deleted, the mutant, IRF3(75-253) was still able to associate with IRF8 (Figure 5H lane 15). Therefore, the interaction between IRF3 and IRF8 depends on intact residues in the 75 to 253 amino acid region of IRF3.
Discussion
IFN-β is a type I interferon produced by many cell types after viral infection. It is one of the most important mammalian cytokines in innate and adaptive immunity. As IFN-β serves as part of the early host defense against pathogens and subsequently activates cellular defense systems, it is evident that its rapid initiation is necessary. This notion is corroborated by our observations that monocytes, but not other cell types, exhibited remarkably faster IFN-β transcription after challenge with viral or bacterial Toll-like receptor ligands (Figure 1A-E). In this study, we dissected the molecular mechanisms underlying this unique phenomenon of rapid transcription of the IFN-β gene in monocytes after pathogenic stimulation.
Our experiments identified IRF8 as the critical regulator of rapid IFN-β transcription in monocytes. The siRNA-mediated knockdown of IRF8 in LPS-stimulated monocytes profoundly reduced IFN-β levels (Figure 2A), whereas the reintroduction of IRF8 into murine 32Dcl3 (IRF8−/−) cells restored their ability to transcribe IFN-β (Figure 3B). These data indicate that IRF8 plays a major role in regulating the transcription of IFN-β in monocytes, thereby contributing to the innate immune response against infection. Moreover, given the finding that IFN-β can also play a role in the development of other immune cell types, such as CD4+ and CD8+ T cells,9,10 it is possible that IRF8 also has a vital role in broadly influencing the adaptive immune response. Investigation by other groups demonstrated that IRF8-deficient mice were extremely susceptible to bacterial or viral infections and exhibited a defect in Th1 CD4+ T-cell responses.40 This is consistent with our current work establishing the active involvement of IRF8 in IFN-β induction, which plays a pivotal role in the activation of innate cellular responses, such as the maturation of dendritic cells and the activation of natural killer cells,5,41 as well as for the development of CD4+ T cell-mediated immunity.9 Thus, we suggest that impaired IFN-β production by IRF8−/− macrophages may partly account for the observed phenotype of IRF8-deficient mice.
Our investigations reveal that an IRF8-binding site exists within the IFN-β promoter region defined by the sequence 5′-GAAA-3′ from −70 to −67 bp (Figures 2D, 4E). Our ChIP assay confirmed that the endogenous IRF8 constitutively binds to the IFN-β promoter (supplemental Figure 2B). Moreover, mutations within this sequence adversely affect the DNA-binding affinity of IRF8 and inhibit formation of the regulatory protein complex consisting of both IRF8 and its interacting partner, PU.1. This site appears essential for the binding of IRF8 to the IFN-β promoter, further affirming that IRF8 induces IFN-β transcription by direct regulation rather than indirect action.
We found that PU.1 functions as a critical partner of IRF8 to regulate IFN-β transcription in monocytes. Paralleling the results with IRF8, knockdown of PU.1 in LPS-stimulated human monocytes drastically reduced IFN-β transcript levels (Figure 4B). In addition, a DNA-protein complex containing PU.1 as an additional DNA-binding factor at the IFN-β promoter region was demonstrated (Figure 4C). As IRF8, PU.1 was also constitutively bound to the IFN-β promoter. Moreover, it is interesting to note that, although PU.1 has been shown to bind to its cognate site in other immune genes, such as IgGλL, independently of IRF8,27 the presence of both IRF8 and PU.1 was necessary for their binding and activation of IFN-β gene expression in a cooperative synergistic fashion.
Our data show that IRF8 also interacts directly with IRF3 (Figure 5B,G-H), a ubiquitously expressed transcription factor, required for the induction of IFN-β transcription in macrophages.39 Based on our observations, we anticipated that IRF8 would interact with IRF3 to induce the transcription of IFN-β. This was indeed the case, and using immunoprecipitation, we defined the critical domains required for this interaction, namely, residues 100 to 270 of IRF8 and residues 75 to 253 of IRF3. To our knowledge, this is the first demonstration of a direct association between IRF8 and IRF3. Therefore, our findings provide evidence that these 2 transcription factors play a nonredundant role in regulating the IFN-β gene. The constitutive binding of IRF8 to the promoter region of the IFN-β gene could contribute to rapid IFN-β transcription in monocytes through facilitating IRF3 recruitment.
In conclusion, we showed that IRF8 is responsible for the prompt IFN-β transcription, and we have identified its binding site within the IFN-β locus. We revealed that these effects were achieved in concert with PU.1 as a partner of IRF8, and uncovered a previously unknown IFN-β regulatory complex consisting of IRF8 and IRF3. In light of our results, we propose a novel hypothetical model of fast IFN-β transcription in pathogen-stimulated monocytes (supplemental Figure 5G). In this model, the constitutive binding of IRF8 and PU.1 to the IFN-β promoter constitutes a preformed activation complex that is responsible for the rapid transcription of IFN-β seen exclusively in monocytes. In response to pathogenic stimulation, the complex facilitates swift recruitment of activated IRF3 to the IFN-β promoter to enable the prompt expression of IFN-β. In contrast, in nonmyeloid cells, the absence of a prebound IRF8 and PU.1 priming complex results in delayed IFN-β production. This unique rapid IFN-β transcription in monocytes is probably a vital signal for the initiation of innate and adaptive immunity and, in particular, a Th1 type immune response.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Lucy Robinson, Dr Jo Keeble, and Mr En Jun Yang for their critical review and help in manuscript preparation; Dr Tom Maniatis and Dr John Hiscott for providing plasmids for this study; and Health Sciences Authority of Singapore for preparing the buffy coat for this work.
This work was supported by the Singapore Immunology Network (grant 06-001; K.-C.C.).
National Institutes of Health
Authorship
Contribution: P.L. performed research, analyzed data, and wrote the paper; J.J.-Y.W. performed research; C.S. and W.-X.S. contributed to experimental design and produced critical reagents; K.-Q.N. and M.B.C.K. prepared for the blood sample; and K.-C.C. directed the study, designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Keh-Chuang Chin, Laboratory of Gene Regulation and Inflammation, Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Immunos #04-00, 8A Biomedical Grove, Singapore 138648; e-mail: kehchuang_chin@immunol.a-star.edu.sg.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal