Abstract
Several common genomic loci, involving various immunity- and metabolism-related genes, have been associated with plasma fibrinogen in European Americans (EAs). The genetic determinants of fibrinogen in African Americans (AAs) are poorly characterized. Using a vascular gene-centric array in 23 634 EA and 6657 AA participants from 6 studies comprising the Candidate Gene Association Resource project, we examined the association of 47 539 common and lower frequency variants with fibrinogen concentration. We identified a rare Pro265Leu variant in FGB (rs6054) associated with lower fibrinogen. Common fibrinogen gene single nucleotide polymorphisms (FGB rs1800787 and FGG rs2066861) significantly associated with fibrinogen in EAs were prevalent in AAs and showed consistent associations. Several fibrinogen locus single nucleotide polymorphism associated with lower fibrinogen were exclusive to AAs; these include a newly reported association with FGA rs10050257. For IL6R, IL1RN, and NLRP3 inflammatory gene loci, associations with fibrinogen were concordant between EAs and AAs, but not at other loci (CPS1, PCCB, and SCL22A5-IRF1). The association of FGG rs2066861 with fibrinogen differed according to assay type used to measure fibrinogen. Further characterization of common and lower-frequency genetic variants that contribute to interpopulation differences in fibrinogen phenotype may help refine our understanding of the contribution of hemostasis and inflammation to atherothrombotic risk.
Introduction
Fibrinogen plays a central role in hemostasis and is also a marker of inflammation. Increased plasma fibrinogen concentration is a well-established risk factor for cardiovascular disease (CVD).1 Recent genome-wide scans have provided strong evidence that common polymorphisms of the fibrinogen structural genes on chromosome 4 (FGA, FGB, and FGG) and several other genomic loci (IL6R, CPS1, PCCB, NLRP3, IL1RN, CD300LF, IRF1-SCL22A5) influence fibrinogen in European-American (EA) populations.2-6 Nonetheless, common polymorphisms explain only a small portion of the heritable component of fibrinogen, which has estimates of approximately 20% to 50% in EAs.7-9 Thus, additional genetic variants with more subtle effects, gene-environment interactions, or lower frequency variants with large effects might account for additional inter-individual variation in fibrinogen.
Fibrinogen levels differ by race/ethnicity, with higher levels among African Americans (AAs) than EAs.10-14 Fibrinogen predicts CVD risk in AAs as well as EAs.15 Heterogeneity between EAs and AAs in the allelic patterns of fibrinogen phenotype association has been noted for the fibrinogen structural genes on chromosome 4.3,16 Little is known about other genomic loci that might account for inter-individual differences in fibrinogen between EAs and other U.S. minority populations.
The ITMAT-Broad-CARe (IBC) genotyping array is a custom, CVD gene-centric single nucleotide polymorphism (SNP) genotyping platform that contains greater SNP marker density and linkage disequilibrium (LD) coverage for more than 2000 CVD candidate regions than current genome-wide arrays. The IBC array is particularly informative for individuals of African descent, enabling analyses of common and lower frequency variants in diverse populations.17
We analyzed the association between the SNPs on the IBC candidate gene array and fibrinogen levels in a large number of EA and AA participants from 6 population-based cohorts from the Candidate Gene Association Resource (CARe) project. Our objectives were (1) to perform a detailed characterization and comparative analysis of the allelic patterns of association for the fibrinogen gene locus and other fibrinogen-associated candidate gene regions in EAs and AAs, and (2) to evaluate whether other common or less frequent variants at these or other candidate gene loci, or gene-environment interaction, explain any of the “missing heritability” of fibrinogen. Secondarily, we assessed the potential heterogeneity of fibrinogen SNP associations according to the type of assay used to measure fibrinogen (functional versus immunologic).
Methods
The CARe Consortium is a National Heart, Lung, and Blood Institute (NHLBI)-supported resource for analyses of the association of genotypes with heart, lung, and blood phenotypes. CARe cohort studies with fibrinogen measurements include the Atherosclerosis Risk in Communities (ARIC) study, the Coronary Artery Risk Development in Young Adults (CARDIA) study, the Cleveland Family Study (CFS), the Cardiovascular Health Study (CHS), the Framingham Heart Study (FHS), and the Multi-Ethnic Study of Atherosclerosis (MESA). All participating institutions and care sites gave institutional review board approval for this study. Further details of the participating CARe studies are given under supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Fibrinogen measurement
In ARIC, CFS, and CHS, the Clauss clotting rate method18 was used for measuring plasma fibrinogen. In MESA and CARDIA, fibrinogen was determined by an immunonephelometric method (Dade Behring Marburg GmbH) on a Behring Nephelometer II analyzer. The FHS used the Clauss method in the offspring and generation 3 participants, and a modified method of Ratnoff and Menzie19 in the original cohort subjects.
Genotyping
The IBC genotyping array17 enables efficient capture of genetic diversity across approximately 2100 genes related to cardiovascular, inflammatory, hemostasis/coagulation, and metabolic phenotypes and pathways. There are a total of 49 320 SNPs on the IBC array, including approximately 15 000 candidate gene SNPs not present in the HapMap. The tagging approach utilized on the IBC array was designed to capture maximal genetic information from the HapMap populations as well as EA and AA individuals for both common and lower frequency SNPs.17 Further details regarding the SNP selection and tagging approach are given under supplemental Methods.
After excluding study participants who did not provide informed consent for genetic testing, subjects with a mismatch between genotypic and phenotypic sex, and other individuals who were genetic outliers, there were a total of 23 634 EA and 6657 AA CARe study participants available for analysis (see supplemental Methods).
Statistical analysis
Sex-specific fibrinogen phenotype residuals were constructed within cohort and race strata, after accounting for age and field site/center. A normal quantile transformation was performed on the covariate-adjusted residuals to improve normality of model residuals and to preserve the rank order of measurements assayed in different laboratories and in some instances, using different fibrinogen assays. For FHS, because fibrinogen was measured at different times using different assays for the parent, offspring, and generation 3 cohorts, phenotype residuals were constructed separately within each FHS cohort.
Genotype-phenotype data analyses were performed separately for EA and AA subjects within each study. To further adjust for population stratification, principal components were incorporated as covariates in analyses (see supplemental Methods). For studies involving unrelated individuals, linear regression was used on quantile transformed standardized fibrinogen residuals, assuming an additive genetic model. Association analyses were performed in PLINK V 1.0.7.20 To allow for the presence of related individuals in CFS and FHS, linear mixed effects models were used.21 Results from each study were combined using fixed and random effects meta-analysis based on inverse-variance weighting. To assess heterogeneity of SNP-fibrinogen association results between cohort studies, we used the I2 inconsistency metric.
To correct for multiple testing, we first determined the effective number of independent SNPs present on the IBC array is 26 482 for AAs and 20 544 for EAs. To maintain an overall type 1 error rate of 5%, a statistical threshold of α = 2 × 10−6 was used to declare array-wide significance. To test for gene-environment interaction, we estimated the effect of the top SNPs within strata of sex, obesity (body mass index ≥ 30 kg/m2 vs < 30), and current smoking status by introducing an interaction term into the linear model.
To assess the number of independent SNPs associated with fibrinogen phenotype within a candidate region, association signals were initially characterized by grouping significant SNPs according to LD, using a pair-wise r2 threshold of greater than 0.50 to define a smaller number of clusters of correlated SNPs. To further assess the presence of multiple independent SNPs in a candidate region, regression analyses were repeated, conditional on the mostly strongly associated SNP in the region. Finally, for fine-mapping of candidate regions on chromosomes 4q32 and 5q31, additional SNP genotypes were imputed using phase 2 HapMap genotype data. Further details regarding imputation and haplotype estimation can be found in the supplemental Methods.
Results
The sample size and participant characteristics from each study are shown in Table 1. Mean fibrinogen levels were higher in AAs than EAs. A total of 32 SNPs spanning 11 candidate gene regions were significantly associated with fibrinogen in EAs (P value range 10−6-10−39, supplemental Table 1 and supplemental Figure 1). Results were similar when the analyses were restricted to the approximately 3867 CARe EA participants from CARDIA, MESA, and CFS, who were not included as part of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium fibrinogen genome-wide association studies (GWAS; data not shown). In the smaller AA sample, a total of 6 SNPs were significantly associated with fibrinogen (P value range 10−6-10−11, supplemental Table 2 and supplemental Figure 1), all located in the chromosome 4 fibrinogen structural gene region.
Characteristics of the study participants*
| . | ARIC . | CARDIA . | CFS . | CHS . | FHS† . | MESA . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA . | EA . | AA . | EA . | AA . | EA . | AA . | EA . | EA . | AA . | EA . | ||
| N | 2853 | 9406 | 1178 | 1370 | 341 | 244 | 719 | 3859 | 6502 | 1566 | 2253 | |
| Female, % | 63 | 54 | 60 | 52 | 58 | 50 | 63 | 56 | 53 | 54 | 52 | |
| Current smoker, % | 29 | 24 | 32 | 21 | 18 | 8 | 16 | 11 | 14 | 18 | 12 | |
| Diabetes, % | 13 | 5 | 2 | 1 | 12 | 5 | 16 | 7 | 11 | 15 | 4 | |
| Age, y | 53 ± 6 | 54 ± 6 | 25 ± 4 | 26 ± 3 | 41 ± 19 | 45 ± 20 | 73 ± 6 | 73 ± 6 | 49 ± 14 | 62 ± 10 | 63 ± 10 | |
| Body mass index, kg/m2 | 30.6 ± 6.3 | 28.3 ± 5.3 | 25.8 ± 5.9 | 23.7 ± 4.0 | 33.1 ± 9.8 | 31.9 ± 9.3 | 28.5 ± 5.6 | 26.4 ± 4.5 | 27.4 ± 5.5 | 30.2 ± 5.9 | 27.8 ± 5.1 | |
| Systolic blood pressure, mm Hg | 128 ± 21 | 118 ± 17 | 112 ± 11 | 109 ± 11 | 123 ± 18 | 122 ± 15 | 142 ± 29 | 135 ± 21 | 121 ± 17 | 132 ± 22 | 123 ± 1 | |
| Diastolic blood pressure, mm Hg | 80 ± 12 | 72 ± 10 | 70 ± 11 | 69 ± 9 | 74 ± 11 | 73 ± 8 | 75 ± 11 | 70 ± 11 | 75 ± 10 | 75 ± 10 | 70 ± 10 | |
| Total cholesterol, mg/dL | 215 ± 45 | 215 ± 41 | 179 ± 33 | 176 ± 33 | 184 ± 42 | 192 ± 42 | 209 ± 39 | 212 ± 39 | 194 ± 36 | 189 ± 36 | 196 ± 36 | |
| HDL cholesterol, mg/dL | 55 ± 17 | 51 ± 17 | 54 ± 13 | 52 ± 13 | 45 ± 13 | 43 ± 12 | 58 ± 16 | 54 ± 16 | 54 ± 16 | 52 ± 15 | 52 ± 16 | |
| Fibrinogen, mg/dL | 320 ± 71 | 297 ± 61 | 363 ± 89 | 320 ± 69 | 323 ± 85 | 328 ± 70 | 346 ± 75 | 320 ± 64 | 357 ± 75 | 362 ± 80 | 336 ± 70 | |
| . | ARIC . | CARDIA . | CFS . | CHS . | FHS† . | MESA . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA . | EA . | AA . | EA . | AA . | EA . | AA . | EA . | EA . | AA . | EA . | ||
| N | 2853 | 9406 | 1178 | 1370 | 341 | 244 | 719 | 3859 | 6502 | 1566 | 2253 | |
| Female, % | 63 | 54 | 60 | 52 | 58 | 50 | 63 | 56 | 53 | 54 | 52 | |
| Current smoker, % | 29 | 24 | 32 | 21 | 18 | 8 | 16 | 11 | 14 | 18 | 12 | |
| Diabetes, % | 13 | 5 | 2 | 1 | 12 | 5 | 16 | 7 | 11 | 15 | 4 | |
| Age, y | 53 ± 6 | 54 ± 6 | 25 ± 4 | 26 ± 3 | 41 ± 19 | 45 ± 20 | 73 ± 6 | 73 ± 6 | 49 ± 14 | 62 ± 10 | 63 ± 10 | |
| Body mass index, kg/m2 | 30.6 ± 6.3 | 28.3 ± 5.3 | 25.8 ± 5.9 | 23.7 ± 4.0 | 33.1 ± 9.8 | 31.9 ± 9.3 | 28.5 ± 5.6 | 26.4 ± 4.5 | 27.4 ± 5.5 | 30.2 ± 5.9 | 27.8 ± 5.1 | |
| Systolic blood pressure, mm Hg | 128 ± 21 | 118 ± 17 | 112 ± 11 | 109 ± 11 | 123 ± 18 | 122 ± 15 | 142 ± 29 | 135 ± 21 | 121 ± 17 | 132 ± 22 | 123 ± 1 | |
| Diastolic blood pressure, mm Hg | 80 ± 12 | 72 ± 10 | 70 ± 11 | 69 ± 9 | 74 ± 11 | 73 ± 8 | 75 ± 11 | 70 ± 11 | 75 ± 10 | 75 ± 10 | 70 ± 10 | |
| Total cholesterol, mg/dL | 215 ± 45 | 215 ± 41 | 179 ± 33 | 176 ± 33 | 184 ± 42 | 192 ± 42 | 209 ± 39 | 212 ± 39 | 194 ± 36 | 189 ± 36 | 196 ± 36 | |
| HDL cholesterol, mg/dL | 55 ± 17 | 51 ± 17 | 54 ± 13 | 52 ± 13 | 45 ± 13 | 43 ± 12 | 58 ± 16 | 54 ± 16 | 54 ± 16 | 52 ± 15 | 52 ± 16 | |
| Fibrinogen, mg/dL | 320 ± 71 | 297 ± 61 | 363 ± 89 | 320 ± 69 | 323 ± 85 | 328 ± 70 | 346 ± 75 | 320 ± 64 | 357 ± 75 | 362 ± 80 | 336 ± 70 | |
Mean ± SD for continuous variables, % for categorical variables.
Combines parent, offspring, and generation 3 cohort.
Common fibrinogen-associated SNPs in EAs
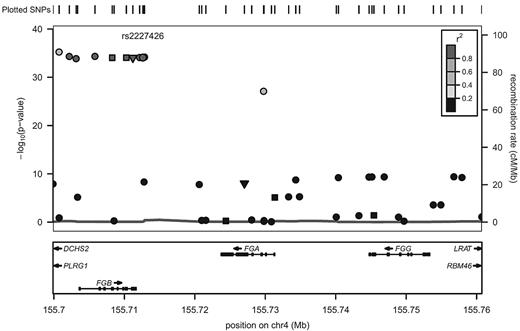
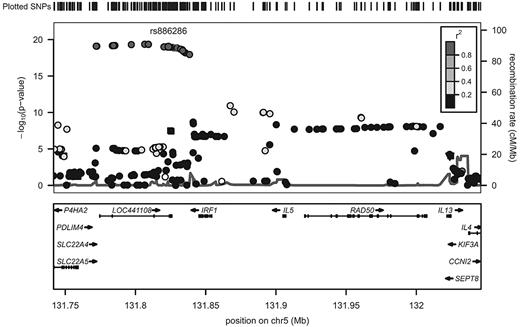
The majority of SNPs significantly associated with fibrinogen in EAs were common, with minor allele frequencies > 20%. Of the 32 SNPs that reached the threshold of experiment-wide significance, 14 SNPs could be considered to represent potential independent association signals. SNPs previously associated with fibrinogen in GWAS of EAs that are replicated in the current report, and those SNPs newly associated with fibrinogen in the current study, are indicated in Table 2. Based on assessment of regional LD patterns and conditional regression analyses in the overall CARe EA sample (n = 23 634), there was most likely a single association signal present within each of the IL6R, CPS1, PCCB, and HDLBP genes (tagged by rs7529229, rs7422339, rs3821445, and rs6752050, respectively). In 4 genes, we found evidence of more than one independent fibrinogen-associated SNP. Within NLRP3, the minor allele of rs4925659 associated with higher fibrinogen and the minor allele of rs12239046 associated with lower levels. The minor allele of IL1RN rs315921 was associated with lower fibrinogen levels. At a slightly less restrictive significance threshold, (P < 10−5) IL1RN rs4251961 was associated with higher fibrinogen levels. In the fibrinogen structural gene region, 2 discrete clusters of SNPs associated with fibrinogen phenotype could be distinguished: 1 associated with higher fibrinogen levels derived from the FGB region (tagged by rs1800787), and the other associated with lower fibrinogen levels derived primarily from the FGG region (tagged by rs2066861); (Table 2 and Figure 1). The strongest fibrinogen association signal in the cytokine gene cluster region on chromosome 5 was derived from a block of highly correlated SNPs, tagged by rs6874639, that span the genes encoding SLC22A5 and IRF1 (meta-analysis P values in the range of 10−19 to 10−20; Table 2 and Figure 2). This LD bin includes rs1016988 and rs2522056, which were previously reported to be associated with circulating fibrinogen.5,6 In a conditional regression analysis of this region (adjusting for rs6874639), only rs6873426 remained associated with fibrinogen (P = 1.2 × 10−5).
Nonredundant candidate gene SNPs associated with fibrinogen in EAs
| Chromosome . | Gene . | dbSNP reference number . | Minor allele frequency . | Meta-analysis regression coefficient (SE) . | Meta-analysis P . | Comment . |
|---|---|---|---|---|---|---|
| 1q21 | IL6R | rs7529229 (T > C) | 0.41 | −0.050 (0.009) | 8.3 × 10−8 | Confirmation of previous GWAS result |
| 1q44 | NLRP3 | rs4925659 (G > A) | 0.38 | 0.052 (0.010) | 5.3 × 10−8 | Confirmation of previous GWAS result |
| 1q44 | NLRP3 | rs12239046 (C > T) | 0.39 | −0.049 (0.010) | 3.3 × 10−7 | Fine-mapping of previous GWAS result |
| 2q13 | IL1RN | rs315921 (G > A) | 0.18 | −0.057 (0.012) | 2.6 × 10−6 | Confirmation of previous GWAS result |
| 2q13 | IL1RN | rs4251961 (T > C) | 0.38 | 0.040 (0.0102) | 2.1 × 10−5 | Fine-mapping of previous GWAS result |
| 2q34 | CPS1 | rs7422339 (C > A) | 0.32 | −0.060 (0.010) | 1.9 × 10−9 | Confirmation of previous GWAS result |
| 2q37 | HDLBP | rs6752050 (T > C) | 0.15 | −0.063 (0.013) | 1.2 × 10−6 | New association in current study |
| 3q22 | PCCB | rs3821445 (A > G) | 0.20 | 0.056 (0.012) | 1.2 × 10−6 | Confirmation of previous GWAS result |
| 4q32 | FGB | rs1800787 (C > T) | 0.21 | 0.151 (0.012) | 1.4 × 10−9 | Confirmation of previous GWAS result |
| 4q32 | FGB | rs6054 (C > T) | 0.004 | −0.580 (0.068) | 1.2 × 10−17 | New association in current study |
| 4q32 | FGG | rs2066861 (C > T) | 0.23 | −0.068 (0.011) | 4.2 × 10−10* | Fine-mapping of previous GWAS result |
| 5q31 | SLC22A5-IRF1 | rs6874639† (A > G) | 0.19 | −0.128 (0.014) | 4.3 × 10−20 | Confirmation of previous GWAS result |
| 5q31 | IRF1 | rs6873426† (G > T) | 0.31 | 0.089 (0.014) | 2.1 × 10−10 | Fine-mapping of previous GWAS result |
| 20q13 | HNF4A | rs1800961 (C > T) | 0.03 | −0.124 (0.027) | 4.5 × 10−6 | New association in current study |
| Chromosome . | Gene . | dbSNP reference number . | Minor allele frequency . | Meta-analysis regression coefficient (SE) . | Meta-analysis P . | Comment . |
|---|---|---|---|---|---|---|
| 1q21 | IL6R | rs7529229 (T > C) | 0.41 | −0.050 (0.009) | 8.3 × 10−8 | Confirmation of previous GWAS result |
| 1q44 | NLRP3 | rs4925659 (G > A) | 0.38 | 0.052 (0.010) | 5.3 × 10−8 | Confirmation of previous GWAS result |
| 1q44 | NLRP3 | rs12239046 (C > T) | 0.39 | −0.049 (0.010) | 3.3 × 10−7 | Fine-mapping of previous GWAS result |
| 2q13 | IL1RN | rs315921 (G > A) | 0.18 | −0.057 (0.012) | 2.6 × 10−6 | Confirmation of previous GWAS result |
| 2q13 | IL1RN | rs4251961 (T > C) | 0.38 | 0.040 (0.0102) | 2.1 × 10−5 | Fine-mapping of previous GWAS result |
| 2q34 | CPS1 | rs7422339 (C > A) | 0.32 | −0.060 (0.010) | 1.9 × 10−9 | Confirmation of previous GWAS result |
| 2q37 | HDLBP | rs6752050 (T > C) | 0.15 | −0.063 (0.013) | 1.2 × 10−6 | New association in current study |
| 3q22 | PCCB | rs3821445 (A > G) | 0.20 | 0.056 (0.012) | 1.2 × 10−6 | Confirmation of previous GWAS result |
| 4q32 | FGB | rs1800787 (C > T) | 0.21 | 0.151 (0.012) | 1.4 × 10−9 | Confirmation of previous GWAS result |
| 4q32 | FGB | rs6054 (C > T) | 0.004 | −0.580 (0.068) | 1.2 × 10−17 | New association in current study |
| 4q32 | FGG | rs2066861 (C > T) | 0.23 | −0.068 (0.011) | 4.2 × 10−10* | Fine-mapping of previous GWAS result |
| 5q31 | SLC22A5-IRF1 | rs6874639† (A > G) | 0.19 | −0.128 (0.014) | 4.3 × 10−20 | Confirmation of previous GWAS result |
| 5q31 | IRF1 | rs6873426† (G > T) | 0.31 | 0.089 (0.014) | 2.1 × 10−10 | Fine-mapping of previous GWAS result |
| 20q13 | HNF4A | rs1800961 (C > T) | 0.03 | −0.124 (0.027) | 4.5 × 10−6 | New association in current study |
dbSNP indicates National Center for Biotechnology Information (NCBI) Single Nucleotide Polymorphism Database; and SE, standard error of regression coefficient.
Indicates meta-analysis P value for heterogeneity < .01.
SNP genotype imputed from HapMap CEU (EA) genotype data.
Imputed regional association results for chromosome 4 SNPs associated with fibrinogen levels. The left y-axis displays the -log(P value), the right y-axis the recombination rate, and the x-axis the SNP position on the chromosome. The degree of LD (r2) is shown by various colors (legend in top right had corner). The symbol shapes represent the type of SNP: inverted triangles represent nonsynonymous coding variants, squares represent synonymous coding variants, and circles represent intron and other variant types. LD is in reference to rs2227426, which is in the same LD bin as the most strongly associated SNP, rs1800787.
Imputed regional association results for chromosome 4 SNPs associated with fibrinogen levels. The left y-axis displays the -log(P value), the right y-axis the recombination rate, and the x-axis the SNP position on the chromosome. The degree of LD (r2) is shown by various colors (legend in top right had corner). The symbol shapes represent the type of SNP: inverted triangles represent nonsynonymous coding variants, squares represent synonymous coding variants, and circles represent intron and other variant types. LD is in reference to rs2227426, which is in the same LD bin as the most strongly associated SNP, rs1800787.
Imputed regional association results for chromosome 5 SNPs associated with fibrinogen levels. The left y-axis displays the -log(P value), the right y-axis the recombination rate, and the x-axis the SNP position on the chromosome. The degree of LD (r2) is shown by various colors (legend in top right had corner). The symbol shapes represent the type of SNP: inverted triangles represent nonsynonymous coding variants, squares represent synonymous coding variants, and circles represent intron and other variant types. LD is in reference to rs886286, which is in the same LD bin as the most strongly associated SNP, rs6874639.
Imputed regional association results for chromosome 5 SNPs associated with fibrinogen levels. The left y-axis displays the -log(P value), the right y-axis the recombination rate, and the x-axis the SNP position on the chromosome. The degree of LD (r2) is shown by various colors (legend in top right had corner). The symbol shapes represent the type of SNP: inverted triangles represent nonsynonymous coding variants, squares represent synonymous coding variants, and circles represent intron and other variant types. LD is in reference to rs886286, which is in the same LD bin as the most strongly associated SNP, rs6874639.
Low-frequency fibrinogen-associated variants
Two SNPs with minor allele frequencies of < 5% showed evidence of association with fibrinogen in EAs (Table 2). A low-frequency nonsynonymous Pro265Leu variant of the fibrinogen β-chain encoded by FGB rs6054 (MAF = 0.4% in EAs and 0.2% in AAs) was significantly associated with fibrinogen concentration, even after adjustment for the top common FGB variant rs1800787 (P = 1 × 10−18). Haplotype analysis across the FGB region confirmed that the rs6054 rare variant allele likely arose on a common ancestral haplotype (allele frequency approximately 30% in EAs) that is distinct from the high-fibrinogen FGB haplotype tagged by rs1800787. A nonsynonymous coding variant of HNF4A with a MAF of 3% in EAs (rs1800961 encoding an Ile139Thr missense substitution) was associated with lower fibrinogen, although it did not reach the threshold of statistical significance (P = 5 × 10−6).
Fibrinogen-associated loci in AAs and comparison with EA results
The 6 SNPs significantly associated with fibrinogen in the AA sample (Table 3) had minor allele frequencies that ranged from approximately 2% to 12%; these SNPs represent 5 independent association signals within the FGB-FGA-FGG gene cluster. The association of fibrinogen with rs10050257, located 5 kb upstream of the FGA transcription start site, has not been previously reported. Two additional independent signals, tagged by rs10034922 and rs4463047, were present among the imputed HapMap SNPs in this region.
Nonredundant candidate gene tagSNPs associated with fibrinogen in AAs
| Chromosome . | Gene . | dbSNP reference number . | Minor allele frequency . | Meta-analysis regression coefficient (SE) . | Meta-analysis P . | Comment . |
|---|---|---|---|---|---|---|
| 4q32 | FGG | rs2066874 (T > C) | 0.03 | −0.333 (0.050) | 2.9 × 10−11 | Replication of previous candidate gene study |
| 4q32 | FGA | rs2070017 (C > T) | 0.11 | −0.163 (0.028) | 4.8 × 10−9 | Replication of previous candidate gene study |
| 4q32 | FGA | rs10050257 (T > G) | 0.06 | −0.213 (0.037) | 7.7 × 10−9 | New association in current study |
| 4q32 | FGB | rs1800787 (C > T) | 0.10 | 0.146 (0.029) | 4.7 × 10−7 | Replication of previous candidate gene study |
| 4q32 | FGB | rs6058 (G > T) | 0.07 | −0.170 (0.036) | 9.5 × 10−7 | Replication of previous candidate gene study |
| 4q32 | FGA-FGG | rs10034922* (C > T) | 0.19 | −0.121 (0.024) | 3.9 × 10−7 | New association in current study |
| 4q32 | FGB | rs4463047* (C > T) | 0.13 | −0.179 (0.036) | 7.3 × 10−7 | New association in current study |
| Chromosome . | Gene . | dbSNP reference number . | Minor allele frequency . | Meta-analysis regression coefficient (SE) . | Meta-analysis P . | Comment . |
|---|---|---|---|---|---|---|
| 4q32 | FGG | rs2066874 (T > C) | 0.03 | −0.333 (0.050) | 2.9 × 10−11 | Replication of previous candidate gene study |
| 4q32 | FGA | rs2070017 (C > T) | 0.11 | −0.163 (0.028) | 4.8 × 10−9 | Replication of previous candidate gene study |
| 4q32 | FGA | rs10050257 (T > G) | 0.06 | −0.213 (0.037) | 7.7 × 10−9 | New association in current study |
| 4q32 | FGB | rs1800787 (C > T) | 0.10 | 0.146 (0.029) | 4.7 × 10−7 | Replication of previous candidate gene study |
| 4q32 | FGB | rs6058 (G > T) | 0.07 | −0.170 (0.036) | 9.5 × 10−7 | Replication of previous candidate gene study |
| 4q32 | FGA-FGG | rs10034922* (C > T) | 0.19 | −0.121 (0.024) | 3.9 × 10−7 | New association in current study |
| 4q32 | FGB | rs4463047* (C > T) | 0.13 | −0.179 (0.036) | 7.3 × 10−7 | New association in current study |
dbSNP indicates NCBI Single Nucleotide Polymorphism Database; and SE, standard error of regression coefficient.
SNP genotype imputed from HapMap EA (CEU) plus Yoruban (YRI) genotype data.
Table 4 compares the top SNP genotype effects (change in fibrinogen per minor allele, adjusted for age, sex, and clinic site) and minor allele frequencies for EAs and AAs, for each region significantly associated with fibrinogen phenotype in either population. At the FGB-FGA-FGG fibrinogen gene locus, FGB rs1800787 and FGG rs2066861 were less common in AAs, but showed consistent direction and magnitude of phenotypic effects in AAs. In contrast, several fibrinogen-associated SNPs were exclusive to AAs (rs2066874, rs10050257, rs2070017, rs2066877, and rs6058); all had minor allele frequencies of < 0.1% in EAs.
Comparison of SNPs associated with circulating levels of fibrinogen in EAs and AAs
| Chromosome . | Coordinate . | Gene . | SNP . | Allele 1 . | Allele 2 . | Variant type . | EAs . | AAs . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAF* . | β . | P . | CAF* . | β . | P . | |||||||
| 1 | 152687402 | IL6R | rs7529229 | t | c | intron | 0.41 | −0.050 | 8.3 × 10−8 | 0.64 | −0.012 | .51 |
| 1 | 152685503 | IL6R | rs8192284 | a | c | coding-nonsynon | 0.40 | −0.046 | 9.3 × 10−7 | 0.14 | −0.060 | .02 |
| 1 | 245670086 | NLRP3 | rs4925659 | g | a | intron | 0.38 | 0.052 | 5.3 × 10−8 | 0.17 | 0.018 | .44 |
| 1 | 245668218 | NLRP3 | rs12239046 | c | t | intron | 0.39 | −0.049 | 3.3 × 10−7 | 0.49 | −0.039 | .02 |
| 2 | 113588522 | IL1RN | rs315921 | g | a | 5′ upstream | 0.18 | −0.057 | 2.6 × 10−6 | 0.03 | 0.030 | .54 |
| 2 | 113590938 | IL1RN | rs4251961 | t | c | 5′ upstream | 0.38 | 0.040 | 2.1 × 10−5 | 0.18 | 0.097 | 1.8 × 10−5 |
| 2 | 211248752 | CPS1 | rs7422339 | c | a | coding-nonsynon | 0.32 | −0.060 | 1.9 × 10−9 | 0.37 | −0.021 | .25 |
| 2 | 241877453 | HDLBP | rs6752050 | t | c | untranslated | 0.15 | −0.063 | 1.2 × 10−6 | 0.03 | −0.033 | .51 |
| 3 | 137485499 | PCCB | rs3821445 | a | g | intron | 0.20 | 0.056 | 1.2 × 10−6 | 0.11 | −0.005 | .85 |
| 4 | 155703465 | FGB | rs1800787 | c | t | 5′ upstream | 0.21 | 0.151 | 1.4 × 10−39 | 0.10 | 0.146 | 4.7 × 10−7 |
| 4 | 155709058 | FGB | rs6054 | c | t | coding-nonsynon | 0.004 | −0.580 | 1.2 × 10−17 | 0.001 | −0.343 | .14 |
| 4 | 155746886 | FGG | rs2066861 | c | t | 5′ upstream | 0.23 | −0.068 | 4.2 × 10−10 | 0.29 | −0.073 | 6.8 × 10−5 |
| 4 | 155749031 | FGG | rs2066874 | t | c | intron | 8 × 10−5 | −0.289 | .62 | 0.03 | −0.333 | 2.9 × 10−11 |
| 4 | 155734845 | FGA | rs10050257 | t | g | 5′ upstream | 0.0009 | −0.288 | .10 | 0.06 | −0.213 | 7.7 × 10−9 |
| 4 | 155729726 | FGA | rs2070017 | c | t | intron | 0.002 | −0.303 | .03 | 0.11 | −0.163 | 4.8 × 10−9 |
| 4 | 155709794 | FGB | rs6058 | g | t | coding-synon | 0.0001 | −0.676 | .02 | 0.07 | −0.170 | 9.5 × 10−7 |
| 4 | 155740078 | FGA-FGG | rs10034922† | c | t | intergenic | 0.01 | 0.115 | .09 | 0.19 | −0.121 | 3.9 × 10−7 |
| 5 | 131677085 | SLC22A4 | rs270607† | a | g | intron | 0.30 | −0.063 | 2.0 × 10−8 | 0.24 | −0.065 | .005 |
| 5 | 131806615 | SLC22A5-IRF1 | rs6874639† | a | g | intergenic | 0.19 | −0.128 | 4.3 × 10−20 | 0.32 | −0.0273 | .29 |
| 20 | 42475778 | HNF4A | rs1800961 | c | t | coding-nonsynon | 0.03 | −0.124 | 4.5 × 10−6 | 0.006 | −0.098 | .38 |
| Chromosome . | Coordinate . | Gene . | SNP . | Allele 1 . | Allele 2 . | Variant type . | EAs . | AAs . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAF* . | β . | P . | CAF* . | β . | P . | |||||||
| 1 | 152687402 | IL6R | rs7529229 | t | c | intron | 0.41 | −0.050 | 8.3 × 10−8 | 0.64 | −0.012 | .51 |
| 1 | 152685503 | IL6R | rs8192284 | a | c | coding-nonsynon | 0.40 | −0.046 | 9.3 × 10−7 | 0.14 | −0.060 | .02 |
| 1 | 245670086 | NLRP3 | rs4925659 | g | a | intron | 0.38 | 0.052 | 5.3 × 10−8 | 0.17 | 0.018 | .44 |
| 1 | 245668218 | NLRP3 | rs12239046 | c | t | intron | 0.39 | −0.049 | 3.3 × 10−7 | 0.49 | −0.039 | .02 |
| 2 | 113588522 | IL1RN | rs315921 | g | a | 5′ upstream | 0.18 | −0.057 | 2.6 × 10−6 | 0.03 | 0.030 | .54 |
| 2 | 113590938 | IL1RN | rs4251961 | t | c | 5′ upstream | 0.38 | 0.040 | 2.1 × 10−5 | 0.18 | 0.097 | 1.8 × 10−5 |
| 2 | 211248752 | CPS1 | rs7422339 | c | a | coding-nonsynon | 0.32 | −0.060 | 1.9 × 10−9 | 0.37 | −0.021 | .25 |
| 2 | 241877453 | HDLBP | rs6752050 | t | c | untranslated | 0.15 | −0.063 | 1.2 × 10−6 | 0.03 | −0.033 | .51 |
| 3 | 137485499 | PCCB | rs3821445 | a | g | intron | 0.20 | 0.056 | 1.2 × 10−6 | 0.11 | −0.005 | .85 |
| 4 | 155703465 | FGB | rs1800787 | c | t | 5′ upstream | 0.21 | 0.151 | 1.4 × 10−39 | 0.10 | 0.146 | 4.7 × 10−7 |
| 4 | 155709058 | FGB | rs6054 | c | t | coding-nonsynon | 0.004 | −0.580 | 1.2 × 10−17 | 0.001 | −0.343 | .14 |
| 4 | 155746886 | FGG | rs2066861 | c | t | 5′ upstream | 0.23 | −0.068 | 4.2 × 10−10 | 0.29 | −0.073 | 6.8 × 10−5 |
| 4 | 155749031 | FGG | rs2066874 | t | c | intron | 8 × 10−5 | −0.289 | .62 | 0.03 | −0.333 | 2.9 × 10−11 |
| 4 | 155734845 | FGA | rs10050257 | t | g | 5′ upstream | 0.0009 | −0.288 | .10 | 0.06 | −0.213 | 7.7 × 10−9 |
| 4 | 155729726 | FGA | rs2070017 | c | t | intron | 0.002 | −0.303 | .03 | 0.11 | −0.163 | 4.8 × 10−9 |
| 4 | 155709794 | FGB | rs6058 | g | t | coding-synon | 0.0001 | −0.676 | .02 | 0.07 | −0.170 | 9.5 × 10−7 |
| 4 | 155740078 | FGA-FGG | rs10034922† | c | t | intergenic | 0.01 | 0.115 | .09 | 0.19 | −0.121 | 3.9 × 10−7 |
| 5 | 131677085 | SLC22A4 | rs270607† | a | g | intron | 0.30 | −0.063 | 2.0 × 10−8 | 0.24 | −0.065 | .005 |
| 5 | 131806615 | SLC22A5-IRF1 | rs6874639† | a | g | intergenic | 0.19 | −0.128 | 4.3 × 10−20 | 0.32 | −0.0273 | .29 |
| 20 | 42475778 | HNF4A | rs1800961 | c | t | coding-nonsynon | 0.03 | −0.124 | 4.5 × 10−6 | 0.006 | −0.098 | .38 |
Coded allele frequency of allele 2.
SNP genotype imputed from HapMap CEU or CEU plus YRI.
For several additional loci (IL6R, IL1RN rs4251961, NLRP3 rs12239046, and SLC22A4 rs270607), the EA fibrinogen phenotype association results were concordant (similar direction of effect and nominal P < .05) in AAs. In contrast, there was no evidence of fibrinogen phenotype association in AAs for NLRP3 rs4925659, CPS1, PCCB, and SLC22A5-IRF1. There was also no evidence of association for IL1RN rs315921 or HDLBP rs6752050, although the frequencies of the respective minor alleles were considerably lower among AAs than EAs.
By comparing the SNP phenotype association results between EAs and AAs, in some instances the regional LD patterns suggested further localization of the putative causal variant(s). These findings are described in further detail under supplemental Results.
Between-study heterogeneity and gene-environment interaction
In the combined meta-analysis, the FGG SNP cluster tagged by rs2066861 showed strong inter-cohort differences in association with fibrinogen (P value for heterogeneity = 1 × 10−9). Table 5 shows the individual study association results for the 3 fibrinogen structural gene region nonredundant SNPs associated with fibrinogen phenotype in EAs. While the SNP regression coefficients for rs1800787 and rs6054 are consistent across studies, rs2066861 is strongly associated with lower fibrinogen only in ARIC and CHS, the 2 largest studies that used the Clauss functional fibrinogen assay. By testing several study characteristics and summary measures, the type of fibrinogen assay explained almost all (96.3%) of the between-study variance associated with rs2066861 in FGG. Similar between-cohort differences for rs2066861 were observed among the AA cohorts (P value for heterogeneity = 7 × 10−5). Within FHS, there was no evidence for heterogeneity of results between the parent, offspring, and generation 3 cohorts. There was no significant interaction between the significant SNPs and sex, current smoking, or obesity in either EAs or AAs (data not shown).
Cohort-specific association results for fibrinogen gene SNPs in EAs
| SNP . | Gene . | ARIC N = 9554 . | CARDIA N = 1311 . | CFS N = 252 . | CHS N = 3918 . | FHS N = 6733 . | MESA N = 2288 . | P for between-study heterogeneity . |
|---|---|---|---|---|---|---|---|---|
| rs1800787 | FGB | 0.132 ± 0.018 P = 1 × 10−13 | 0.164 ± 0.04 P = .0004 | NA | 0.150 ± 0.027 P = 3 × 10−8 | 0.178 ± 0.023 P = 2 × 10−14 | 0.159 ± 0.037 P = 2 × 10−5 | .62 |
| rs6054 | FGB | −0.588 ± 0.112 P = 1 × 10−7 | −0.904 ± 0.350 P = .01 | NA | −0.648 ± 0.192 P = .0007 | −0.526 ± 0.111 P=2 × 10−6 | −0.545 ± 0.0218 P = .013 | .87 |
| rs2066861 | FGG | −0.116 ± 0.017 P = 8 × 10−12 | −0.0588 ± 0.045 P = .20 | 0.0224 ± 0.115 P = .85 | −0.159 ± 0.02 P=1 × 10−9 | 0.0216 ± 0.02 P = .33 | 0.057 ± 0.03 P = .11 | 1.48 × 10−9 |
| SNP . | Gene . | ARIC N = 9554 . | CARDIA N = 1311 . | CFS N = 252 . | CHS N = 3918 . | FHS N = 6733 . | MESA N = 2288 . | P for between-study heterogeneity . |
|---|---|---|---|---|---|---|---|---|
| rs1800787 | FGB | 0.132 ± 0.018 P = 1 × 10−13 | 0.164 ± 0.04 P = .0004 | NA | 0.150 ± 0.027 P = 3 × 10−8 | 0.178 ± 0.023 P = 2 × 10−14 | 0.159 ± 0.037 P = 2 × 10−5 | .62 |
| rs6054 | FGB | −0.588 ± 0.112 P = 1 × 10−7 | −0.904 ± 0.350 P = .01 | NA | −0.648 ± 0.192 P = .0007 | −0.526 ± 0.111 P=2 × 10−6 | −0.545 ± 0.0218 P = .013 | .87 |
| rs2066861 | FGG | −0.116 ± 0.017 P = 8 × 10−12 | −0.0588 ± 0.045 P = .20 | 0.0224 ± 0.115 P = .85 | −0.159 ± 0.02 P=1 × 10−9 | 0.0216 ± 0.02 P = .33 | 0.057 ± 0.03 P = .11 | 1.48 × 10−9 |
For each study, the number in the top row indicate regression β coefficient for standardized fibrinogen ± standard error, based on an additive genetic model. The last column indicates the P value for heterogeneity based on the combined meta-analysis.
Discussion
In this large population-based study, we were able to simultaneously confirm and more precisely characterize the genetic variants associated with fibrinogen in EAs5,6 and also compare and contrast results of the association findings within an under-represented minority population of AAs. In EAs, we identified multiple SNPs independently associated with fibrinogen at FGB-FGA-FGG, NLRP3, IL1RN, and IRF1-SCL22A5. We note concordance of EA and AA fibrinogen association results at 2 common fibrinogen loci (FGB rs1800787 and FGG rs2066861), as well as at several additional loci within inflammatory genes (IL6R, IL1RN, NLRP3) for which there is strong prior evidence of association with fibrinogen based on EA GWAS or CARe IBC results. We also identified a new low-frequency nonsynonymous FGB variant (rs6054 encoding β-fibrinogen Pro265Leu) associated with lower fibrinogen levels.
The current study includes by far the largest AA sample (n = 6657) evaluated for genetic factors associated with plasma fibrinogen. Our results confirm the association of several SNPs (rs2066874, rs2070017, and rs6058) previously identified through candidate gene analyses in single cohort studies of CARDIA and CHS AAs.3,16 We also report a novel association with rs10050257, located approximately 5 kb upstream of the FGA transcription start site. We used a genotyping array that has denser LD and broader population coverage of previously identified candidate gene regions for AAs than current GWAS platforms. Currently, our ability to perform replicate the novel rs10050257 association is limited by (1) the unavailability of additional AA samples genotyped using the IBC genotyping array and (2) the relatively low allele frequency of these AA SNPs, which are not present on current GWAS genotyping platforms.
While some loci were consistently associated with fibrinogen between EAs and AAs, others were not. Several fibrinogen-associated SNPs in FGA, FGB, and FGG were exclusive to AAs. Other loci were associated with fibrinogen phenotype only among EAs. The latter could possibly be due to the smaller AA size compared with the EA sample or to different LD patterns between these populations. Based on population-specific effect sizes and/or allele frequency differences, it is possible that some of these loci might account in part for the higher fibrinogen levels in AAs compared with EAs.
Despite the consistent heritability of fibrinogen,7-9 none of the previously or newly identified significant SNPs in our study explain a large portion of the variation in fibrinogen. Together, the common and low-frequency SNPs explained 2.2%-15% of the variation in circulating fibrinogen among EA cohorts and 1.8%-3.8% among AA cohorts. For the newly identified lower frequency variants, the percent explained by rs6054 and rs1800961 together ranged from 0.3%-1.0%. We also found no evidence that gene by environment interactions are important in determining fibrinogen levels in either racial/ethnic group, although the number of environmental factors examined was limited.
Characterization of the cis-acting and coding variants within the fibrinogen structural genes have contributed to our understanding of fibrinogen synthesis and the potential role of fibrinogen and inflammation in atherothrombotic disease. Fibrinogen is composed of 3 chains of amino acids, α, β, and γ, and each chain is encoded by a distinct gene (FGA, FGB, and FGG), all of which reside in a cluster on chromosome 4. Higher fibrinogen levels have been observed with either the FGB −455G > A (rs1800790) or −148C > T polymorphism (rs1800787).22 Another SNP in perfect LD with rs1800787 (rs4508864), has been previously associated with FGB expression in liver tissue.23 Interleukin-6 (IL-6) is the primary mediator of fibrinogen synthesis in response to inflammation, and sequences responsive to IL-6 are present in the promoter regions of the 3 genes encoding fibrinogen. The FGB rs1800787 polymorphism is located in between a hepatocyte nuclear factor-3 (HNF-3) site and a CCAAT box/enhancer-binding protein (C/EBP) site within the FGB promoter that comprise a an IL-6-response element. The rs1800787-148 C/T polymorphism influences IL-6–induced FGB promoter activity by interfering with the cooperation between adjacent HNF-3 and C/EBP promoter binding sites.24 Interestingly, we additionally note that rs10050257, the newly identified FGA promoter region SNP associated with fibrinogen in AAs is also located within C/EBP and HNF-3 consensus transcription factor binding sequences. Additional in vitro transcriptional analysis of the FGA promoter is required to confirm the mechanism of association for rs10050257.
While evidence for association of common FGA, FGB, and FGG variants with thrombotic risk has been inconsistent,25-29 familial dysfibrinogenemias due to rare fibrinogen gene mutations have been associated with either thrombosis or hemorrhage.30 Mutations in the fibrinogen genes leading to amyloidosis have also been described;31 and recent evidence suggests that fibrinogen may be involved in the pathogenesis of Alzheimer disease.32 The β-fibrinogen Pro265Leu polymorphism (rs6054) is located within an evolutionarily conserved intra-chain disulfide loop that appears to affect fibrinogen secretion and assembly;33 this SNP is predicted to alter normal function.34 The role of the rare Pro265Leu variant in clinical atherothrombotic disease remains to be determined.
Altered splicing regulation may be another mechanism by which fibrinogen gene variants contribute to phenotypic differences in fibrinogen. Both rs6054 and rs6058 (a synonymous SNP located near the splice site of exon 6 of FGB) are predicted to alter splicing enhancer elements.34 SNPs rs2066861 and rs2066865 (FGG 10034C > T) tag a common FGG haplotype and are located in a region of alternative pre-mRNA processing that results in the formation of the fibrinogen γ chain isoform, fibrinogen γ′ that appears to be protective against deep vein thrombosis.35 FGG 10034C > T results in a decrease in the ratio of fibrinogen γ′ to γA and therefore increases risk of deep vein thrombosis36 and possibly stroke.29 These FGG SNPs are also in LD with the Thr312Ala polymorphism (rs6050) of FGA, which is located within the αC domain of fibrinogen, and is important for lateral aggregation and factor XIII-induced cross-linking of fibrin. The Ala312 fibrinogen-α chain variant has been associated with venous thrombosis37 or post-stroke mortality in subjects with atrial fibrillation.38 The Ala312 variant has also been associated with altered clot structure, and therefore may predispose to clot embolization.39
Fibrinogen can be measured using either a functional Clauss method or an antigen method. In most individuals, the results of the 2 assays are highly correlated, but discrepancies can occur because the immunoassay and functional assay measure different properties of fibrinogen.40,41 In the current study, we tested for between-study differences in genotype-fibrinogen association results according to type of fibrinogen assay at the stage of meta-analysis. The use of the fibrinogen immunoassay in 2 of the 6 CARe studies allowed us to confirm that the common FGG haplotype associated with the fibrinogen γ′ chain isoform is differentially associated with decreased fibrinogen levels as measured by functional assay, but not by immunoassay. The same FGG haplotype was recently associated with higher D-dimer levels. Together, these findings have potentially important implications for thrombotic risk assessment.42
Several of the genes harboring polymorphisms associated with fibrinogen are implicated in pathways that link the sensing and regulation of cellular injury, inflammation and metabolic stress, and autoimmunity. Hepatic fibrinogen synthesis is regulated by cytokines IL-1 and IL-6,43 and IL6R and IL1RN encode important regulatory proteins for each of these cytokines. One of our significant SNPs, rs4537545, influences IL6R expression in lymphocytes,44 and is in strong LD with rs8192244, which encodes a functional Ala358Asp substitution at the site where the IL-6 receptor is cleaved to form soluble IL-6 receptor. IL-1 receptor antagonist, the protein encoded by IL1RN mediates a variety of IL-1 related inflammatory responses. The NLRP3 inflammasome complex functions as an upstream activator of IL-1, IL-18, and nuclear factor-κB signaling. Mutations in either NLRP3 or IL1RN result in dysregulated IL-1β production and autoinflammatory disease.45 Chromosome 5q31 contains a cluster of coordinately regulated genes involved in the immune response, including a 250-kb risk haplotype associated with Crohn disease susceptibility.46 The protein encoded by CPS1 is an enzyme that catalyzes the initial step of the hepatic urea cycle. The nonsynonymous CPS1 variant rs7422339 (Thr1405Asn) has been associated with vascular and inflammatory disease,47 and was recently associated with homocysteine.48
Several other fibrinogen phenotype-associated genes have functional associations with lipid metabolism, another important determinant of vascular risk. A variant of PCCB, which encodes the β subunit of the amino acid catabolic enzyme propionyl-CoA carboxylase, was recently associated with small high-density lipoprotein (HDL) particle concentration.49 Previous studies have consistently found that HDL cholesterol is inversely associated with fibrinogen.50 High-density lipoprotein-binding protein (HDLBP) may function in the removal of excess cellular cholesterol and has been colocalized with apolipoprotein E in cholesterol-loaded plaque macrophages.51 The Thr130Ile variant of HNF4A (rs1800961) is located in the DNA binding domain of the transcription factor HNF-4 and has been associated with decreased transcriptional activity in vitro, increased risk of T2D52 and lower HDL cholesterol levels.53
In summary, both common and less frequent sequence variants of inflammatory and hemostatic genes are associated with fibrinogen. It appears that at least some loci contributing to fibrinogen levels differ between AAs and EAs. Additional studies are needed to better understand the relative contribution of genetic and environmental factors to the chronic systemic inflammatory burden that may underlie interpopulation differences in fibrinogen and potentially contribute to CVD risk.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the investigators, staff, and participants of ARIC, CARDIA, CHS, CFS, FHS, and MESA for their valuable contributions. The authors additionally thank Taylor Young and Deborah Farlow (The Broad Institute, Cambridge, MA), and Guillaume Lettre (Montreal Heart Institute/Universite de Montreal, Montreal, QC) for their significant efforts and contributions to the CARe consortium.
A.P.R., C.L.W., L.A.L., and E.M.L. are supported by R01 HL71862-06, “Thrombosis Genetics, MI, and Stroke in Older Adults.” N.L.S. is supported by NHLBI grant nos. HL073410 and HL095080. CARe is supported by contract no. HHSN268200625226C and from the National Institutes of Health/NHLBI, and subcontract no. 5215810-55000000041 to C.L.W. A full listing of the grants and contracts that have supported CARe is provided at http://public.nhlbi.nih.gov/GeneticsGenomics/home/care.aspx.
National Institutes of Health
Authorship
Contribution: C.L.W., L.A.L., K.C.T., E.M.L., A.D.J., L.A.H., C.P., Y.L., Q.Y., A.P.R., and C.O.J. contributed to analysis and interpretation of the data; B.J.K. took part in designing the IBC candidate gene array; C.L.W. and A.P.R. drafted the manuscript; D.G., N.L.S., C.O.J., D.R.J., A.R.F., A.D.J., B.J.K., J.A.D., J.G.W., G.T., R.P.T., and W.T. contributed to substantial revision of the manuscript; and senior investigators participating in data collection for cohorts represented in CARe are C.J.O., S.R., H.A.T., J.G.W., A.R.F., D.R.J., and A.P.R.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alexander P. Reiner, Box 357236, Department of Epidemiology, University of Washington, Seattle, WA 98195; e-mail: apreiner@u.washington.edu.
References
Author notes
C.L.W., C.J.O., and A.P.R. contributed equally to this manuscript.