Abstract
Chromosomal translocations in hematological malignancies often result in novel fusion chimeric genes. We report a case of acute myeloid leukemia with a clonal translocation t(11;12)(p15;q13) displaying morphologic and immunophenotypic features resembling the classical hypergranular subtype of acute promyelocytic leukemia. The gene fused to NUP98 (nucleoporin 98) was detected by comparative genomic hybridization array as the retinoid acid receptor gamma gene (RARG). The involvement of RARG in a chimeric fusion transcript has not been reported previously in human leukemia.
Introduction
Chromosomal translocations in hematological malignancies often result in the generation of novel chimeric genes. The nucleoporin 98 gene (NUP98) located at chromosome 11p15 is recurrently involved in a variety of rearrangements in both myeloid and lymphoid malignancies.1,2 After the first NUP98 rearrangement, Homeobox A9 gene (NUP98/HOXA9), was discovered, more than 20 different fusion partners were reported.1 Among these partners, other homeobox and non-homeobox genes have been identified as fusion partners of NUP98. We report here a novel NUP98 rearrangement in a patient with acute myeloid leukemia displaying morphologic and immunophenotypic features resembling the classical hypergranular subtype of acute promyelocytic leukemia. This case harbored a novel fusion gene as a result of a chromosomal break point at 12q13 detected in a new 11p15 rearrangement. The gene fused to NUP98 was identified by comparative genomic hybridization (CGH) array as the retinoid acid receptor gamma gene (RARG). RARG is a member of the nuclear receptor superfamily and shares high homology (90%) with RARA and RARB, the other retinoic acid receptors that are involved in retinoid signaling. While an artificial construct has been reported in which RARG is fused to the promyelocytic leukemia gene product (PML), resulting in an oncogenic protein,3 no cases of human leukemia–bearing RARG fusions have been described.
Methods
Case reports
This study was approved by the institutional review board of the Hospital Universitario La Fe. A 35-year-old man was referred to our department with asthenia, mucosal bleeding, spontaneous ecchymoses, and fever. Blood tests showed a hemoglobin level of 6 g/dL, a platelet count of 8 × 109/L, and a white blood cell count of 12 × 109/L with 82% blasts and atypical promyelocytes. Although increased levels of D-dimer were present, the patient did not fulfill the criteria for coagulopathy.4 Morphologic examination of bone marrow (BM) smears disclosed a monomorphic infiltration by blast cells, 80% hypergranular (atypical promyelocytes, which frequently displayed Auer rods) resembling the hypergranular subtype of acute promyelocytic leukemia (M3) of the French-American-British classification (Figure 1A-B). The immunophenotype of peripheral blood blasts was positive for CD13, CD33, CD45, CD117, and cMPO, weakly positive for CD34, and negative for HLA-DR and B-cell and T-cell markers. The immunofluorescence staining with the anti-PML monoclonal antibody (PG-M3)5 was negative in both BM and peripheral blood samples. Moreover, reverse transcription polymerase chain reaction (RT-PCR) to amplify the PML/RARA fusion gene and conventional karyotyping to search for the t(15;17) were also negative. Due to the clinical suspicion of acute promyelocytic leukemia, the patient was started on all-trans retinoic acid (ATRA) treatment. One day after ATRA initiation, due to the absence of a PML/RARA rearrangement, ATRA was discontinued and treatment was switched to a standard 7 + 3 schedule with cytarabine and idarubicin as induction therapy. After the achievement of complete remission, the patient underwent consolidation chemotherapy followed by autologous peripheral blood stem-cell transplant. Eight months after transplantation, he remains alive and in complete hematologic remission.
Cytogenetic analysis and FISH
A BM sample was processed after short-term culture (24 hours) following standard procedures. The chromosomes were stained by G-banding and the karyotype reported according to International System for Human Cytogenetic Nomenclature recommendations.6 Fluorescent in situ hybridization (FISH) was performed on 200 interphase cells using the dual-color translocation probes Vysis LSI PML/RARA and the Vysis LSI RARA dual color break apart (both from Abbott Laboratories) to discard variant translocations of RARA.
RT-PCR
Molecular analysis by reverse transcription polymerase chain reaction (RT-PCR) for different PML/RARA isoforms was performed as described elsewhere.7 The presence of NUP98 rearrangements with the genes described in 12q13, HOXC11 and HOXC13, was also determined using previously reported assays.8,9 The NUP98/RARG mRNA was reverse-transcribed into cDNA using random hexamers, and PCR was performed using the following primers: NUP98 exon 10 NUP98F: 5′-GGG CTT GGT GCA GGA TTT GG-3′, and RARG exon 7 RARGR: 5′-TGG GTC CGG TTC AGG GTC AGC-3′. These primers were also used to amplify the fusion transcript break points (GenBank accession number: HQ229990).
CGH array
Genomic DNA was quantified and its purity confirmed by spectrometry (ND1000, NanoDrop Technologies). Non-amplification labeling of DNA (direct method) was obtained following the Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis protocol (Version 5.0; Agilent Technologies). DNA (2000 ng) from patient leukemic cells was fragmented in a restriction-digestion step. Digestion was confirmed and evaluated by the DNA 7500 Bioanalyzer assay. Cyanine 3-dUTP and cyanine 5-dUTP were used for fluorescent labeling of, respectively, the BM sample and the patient DNA control sample for the patient digested gDNA using a genomic DNA enzymatic labeling kit according to the manufacturer's instructions. Labeled DNA was hybridized with Human Genome CGH Microarray 244K containing 236,000+ coding and noncoding human sequences represented. Arrays were scanned in a microarray scanner (Agilent) according to the manufacturer's protocol, and data were extracted using feature extraction software Version 9.5.3.1; Agilent). All microarray data are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-2928.
Results and discussion
G-banding karyotype analysis in BM cells revealed a clonal translocation t(11;12)(p15;q13) in 16 of 20 metaphases analyzed (Figure 1C). FISH analysis using the PML/RARA and RARA probes hybridized to the normal copies of chromosomes 15 and 17, indicating the absence of cryptic PML/RARA fusion or RARA rearrangements. Two previous rearrangements in apparently similar cases bearing t(11;12) aberrations have been described,8,9 and these involved the NUP98 gene in 11p15 and HOXC11 or HOXC13 genes in 12q13. These fusion transcripts were ruled out by specific RT-PCR analyses.
Morphologic features and results of CGH array of an acute myeloid leukemia patient with a clonal translocation t(11;12)(p15;q13) resembling the classical hypergranular subtype of acute promyelocytic leukemia. May-Grümwald-Giemsa–stained bone marrow smear (1000×, Nikon Eclipse 80i microscope and Nikon DS-Fi 1 camera) showing (A) promyelocytes with hypergranulated cytoplasm; several nuclei are invaginated and (B) promyelocyte with bundles of Auer rods (faggot cell). (C) Representative karyotype of BM cells with t(11;12)(p15;q13). (D) Results of CGH array. Top, microdeletions in chromosome 11p15 involving the NUP98 gene. Bottom, microdeletions in chromosome 12q13 involving the RARG gene.
Morphologic features and results of CGH array of an acute myeloid leukemia patient with a clonal translocation t(11;12)(p15;q13) resembling the classical hypergranular subtype of acute promyelocytic leukemia. May-Grümwald-Giemsa–stained bone marrow smear (1000×, Nikon Eclipse 80i microscope and Nikon DS-Fi 1 camera) showing (A) promyelocytes with hypergranulated cytoplasm; several nuclei are invaginated and (B) promyelocyte with bundles of Auer rods (faggot cell). (C) Representative karyotype of BM cells with t(11;12)(p15;q13). (D) Results of CGH array. Top, microdeletions in chromosome 11p15 involving the NUP98 gene. Bottom, microdeletions in chromosome 12q13 involving the RARG gene.
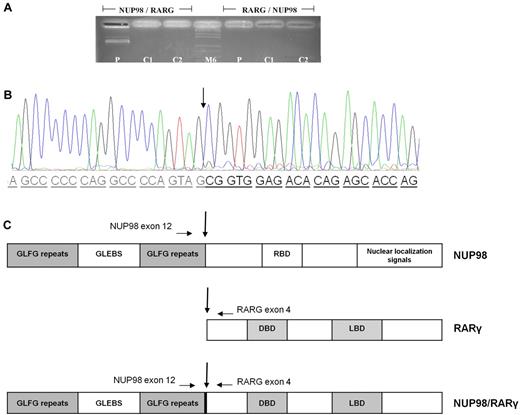
A CGH array was performed to better map the rearrangement region and to search for minor or cryptic genomic imbalances flanking the break-point translocation. A 1.0-Mb microdeletion in 11p15 and a 2.5-Mb microdeletion in 12q13, involving the NUP98 and RARG genes, respectively, were identified. No other copy number variations were found (Figure 1D). To confirm a NUP98/RARG fusion, an 881-bp product was specifically amplified from the patient's cDNA but not from the control (Figure 2A). The transcript fusion product was coamplified in the same reaction and sequenced. Sequence analysis of the NUP98/RARG fusion transcript revealed that NUP98 exon 12 was fused in-frame to RARG exon 4 (Figure 2B). The reciprocal RARG/NUP98 fusion product was not detected with either single-step or nested PCR (Figure 2A). This latter finding might be related to the demonstrated microdeletions in both genes. The NUP98/RARG fusion mRNA (Figure 2C) is predicted to encode an 862–amino acid protein. The NUP98 5′-region encoding the GLFG-repeat and the GLEBS-like motifs were fused to the 3′-region of RARG, which included the DNA- and ligand-binding domains of the gene. We hypothesize that, as was observed with RARA fusion genes in acute promyelocytic leukemia, this newly described chimeric fusion can disrupt normal retinoid signaling by acting as an aberrant RARG receptor (Figure 2C).
Electrophoresis agarose gel and chromatogram showing the NUP98/RARG rearrangement, and schematic representation of the NUP98/RARG fusion protein. (A) NUP98/RARG fusion. The reciprocal RARG/NUP98 fusion product was not detected. (B) Nucleotide and protein sequences surrounding the NUP98/RARG fusion region. The fusion was in-frame, so that the open reading frames of both genes in the fusion transcript were retained. The arrows indicate the break-point and fusion sites of the NUP98/RARG gene. (C) Schematic representation of the NUP98/RARG fusion proteins and its wild-type counterparts. The locations of the primers used for RT-PCR are indicated by horizontal arrows. GLFG repeats have been shown to be docking sites for transport receptors. P indicates patient cDNA (881bp); C1 and C2, PML/RARA patient's cDNA; M6, molecular weight; GLEBS, motif of nuclear export of poly(A)+ RNA and nuclear pore complex (NPC) structure; RBD, RNA-binding domain; DBD, In the abbreviations for Figure 2, “DBD, DNA-binding domain and distribution and arrows the joining site” has been rephrased as “DBD, DNA-binding domain. Arrows indicate the joining sites.” Please confirm or correct. DNA-binding domain. Arrows indicate the joining sites.
Electrophoresis agarose gel and chromatogram showing the NUP98/RARG rearrangement, and schematic representation of the NUP98/RARG fusion protein. (A) NUP98/RARG fusion. The reciprocal RARG/NUP98 fusion product was not detected. (B) Nucleotide and protein sequences surrounding the NUP98/RARG fusion region. The fusion was in-frame, so that the open reading frames of both genes in the fusion transcript were retained. The arrows indicate the break-point and fusion sites of the NUP98/RARG gene. (C) Schematic representation of the NUP98/RARG fusion proteins and its wild-type counterparts. The locations of the primers used for RT-PCR are indicated by horizontal arrows. GLFG repeats have been shown to be docking sites for transport receptors. P indicates patient cDNA (881bp); C1 and C2, PML/RARA patient's cDNA; M6, molecular weight; GLEBS, motif of nuclear export of poly(A)+ RNA and nuclear pore complex (NPC) structure; RBD, RNA-binding domain; DBD, In the abbreviations for Figure 2, “DBD, DNA-binding domain and distribution and arrows the joining site” has been rephrased as “DBD, DNA-binding domain. Arrows indicate the joining sites.” Please confirm or correct. DNA-binding domain. Arrows indicate the joining sites.
The NUP98 gene encodes a protein component of the nuclear pore complex that regulates the nucleocytoplasmic transport of proteins and mRNAs. NUP98 contains a domain with a GLFG repeat that has been shown to activate transcription, providing docking sites for nuclear transport to conduct RNA and protein traffic between the nucleus and cytoplasm. Chimeric transcripts formed by the NUP98 N-terminal GLFG repeats fused to the C-terminus of the partner proteins are expressed in all NUP98 fusions reported, suggesting that the NUP98 N-terminus may be important for leukemogenesis.10,11 Here we identified RARG as a new NUP98 partner.
RARG is a member of the retinoid acid receptor (RAR) family, along with RARA, which is known to be involved in the PML/RARA fusion of acute promyelocytic leukemia, and RARB. RARs are nuclear hormone receptors functioning as ligand-activated transcription factors that interact specifically to modulate transcription of DNA elements.12 They function as heterodimers with retinoid X receptors. Specifically, heterodimers formed by RXRA-RARG are necessary for growth arrest and visceral and primitive endodermal differentiation.3,12
RARG rearrangements in human leukemia have not been previously described. Recently, La Starza et al1 reported a case of an adult patient with acute myeloid leukemia of the M2 subtype carrying a t(11;12)(p15;q13) translocation that involved a NUP98 rearrangement with 1 of the following 3 putative candidate genes localized at the translocation break point: RARG, MFSD5, or ESPL1. Although, in the above case the partner gene of NUP98 was not identified, the authors speculated that RARG was probably not implicated because it was incorrectly oriented. This hypothesis is reinforced by the fact that, unlike our case, the patient reported by La Starza et al1 did not share any morphologic features characteristic of acute promyelocytic leukemia. It should be noted that an artificial PML/RARG fusion protein is able to produce the same biological effects mediated in the cells by PML/RARA and also to confer responsiveness to differentiation treatment with retinoid acid.3 In addition, inoculated transduced cells expressing PML/RARG were able to trigger leukemia in vivo in a murine model, displaying morphologic features resembling those obtained with transduction of PML/RARA.12
In conclusion, we describe here a novel NUP98/RARG gene rearrangement that conferred to leukemic cells acute promyelocytic leukemia–like morphologic and immunophenotypic features. The favorable outcome with a standard 7 + 3 chemotherapy approach, followed by consolidation and intensification with autologous stem-cell transplantation, does not allow us ascertain the sensitivity to ATRA, due to the early discontinuation of the drug in this patient. Further studies are needed to better investigate the biological properties, in particular sensitivity to retinoids, of leukemias bearing this new NUP98/RARG fusion protein.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Federico Gomis for the morphological photographs, Beatriz Costán for her daily support in performing molecular and cytogenetic studies, and the Microarray Analysis Service of the Centro de Investigación Principe Felipe of Valencia for CGH arrays. We also thank Dr Francesco Lo Coco for his invaluable help and critical review of the manuscript.
This work was supported in part by grant R06/0020/0031 from Red Temática de Investigación Cooperativa en Cancer, grant BES2008-008053 from the Ministerio de Ciencia e Innovación, and grants CA08/00141, CM09/00038, PI06/0657, and 09/01828 from Instituto de Salud Carlos III.
Authorship
Contribution: E.S. designed molecular studies and wrote the paper; M.L.P.-S. and L.S. performed morphologic analysis; E.S. and A.V. performed cytogenetic analysis and FISH experiments; A.S. performed PG-M3 immunofluorescence study; M.I. and O.F. performed molecular studies; E.B. supervised molecular studies; J.C. supervised clinical and experimental findings; J.M. and G.M.-A. were involved in the management of the patient; I.L. provided clinical data; and M.A.S. was responsible for the conception of the study and final approval of the draft. All authors reviewed the manuscript and contributed to the final draft.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Miguel A. Sanz, Hospital Universitario La Fe, Avenida Campanar 21, 46009 Valencia, Spain; e-mail: msanz@uv.es.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal