All organisms depend on large networks of multicomponent complexes to carry out the biochemical processes of life. However, at its most fundamental, all biology is binary, dependent on the productive interaction of 2 molecules. A binds to B creating AB which then can assemble with C in what is essentially another binary event; and from these simple binary events, complex networks of interactions can arise that precisely regulate complex physiologic processes. Genomic and proteomic analyses support the importance of binary interactions, wherein binding partners coevolve, with “hot spots” displaying high levels of conservation in a chemically complementary manner.2,–4
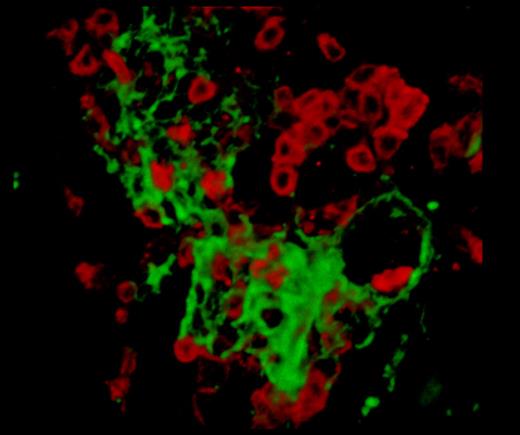
A single frame captured from supplemental Video 1 (Connolly et al1 ), showing the inflammatory lesions in PlauGFDhu/GFDhu mice. T lymphocytes (red, CD3 stain) surround a central core of fibrin (green).
A single frame captured from supplemental Video 1 (Connolly et al1 ), showing the inflammatory lesions in PlauGFDhu/GFDhu mice. T lymphocytes (red, CD3 stain) surround a central core of fibrin (green).
The study of protein interactions can appear to be deceptively simple. In the test tube the interaction of 2 proteins can be described kinetically and thermodynamically in great detail, or proteins can be expressed in cell lines, and their interacting partners can be captured and identified. Unfortunately, in the context of a whole organism, such descriptions do not always yield direct insight into physiologic function. Ultimately, a hypothesis for the physiologic role of any factor or interaction must be verified in the whole organism, and one of the most successful approaches for this involves the use of genetically modified animal models. Even in standard genetic models, where genes are deleted or overexpressed, discerning which effects are due to specific interactions, when a protein may interact with multiple factors, is difficult.
Connolly et al describe the creation of an animal model designed to investigate a specific binary interaction between a protease and its cell-surface receptor in the whole organism.1 The protease-receptor pair, uPA and uPAR, not only bind to each other, but both molecules can also associate with multiple other ligands. The profusion of interaction partners, particularly for uPAR, has been extensively studied both in vitro and in vivo.5,6 In this mouse model, 4 amino-acid substitutions have been introduced into the growth factor domain of the protease. These changes abolish the binding of uPA to its murine receptor, but the mutations were chosen to swap specificity such that the mutant uPA will bind to the human receptor with an affinity similar to that of human uPA. This demonstrates that the mutant protein retains its structural integrity and also permits selective reconstitution of the uPA:uPAR interaction by introduction of the human receptor. The gene encoding mutant uPA was homologously recombined into the uPA locus thus preserving the natural expression of uPA. This technique allowed the researchers to study the long-term effects of ablating a single property of uPA in an otherwise wild-type animal. Following the animals essentially throughout their lifespan enabled the authors to uncover a syndrome of chronic inflammation associated with multiorgan fibrin microdeposition, reminiscent of many chronic human inflammatory diseases (see figure).1 Interestingly as well, Connolly et al show that in several pathologic paradigms, the phenotype of the PlauGFDhu/GFDhu mice was less severe than the uPA-null mice phenotype, but identical to that of the uPAR-null mice. This suggests that at least in the disease models tested, the primary role of uPAR appears to be to localize uPA proteolytic activity. This is in contrast to several earlier studies where uPAR was suggested to have pleiotropic effects that were in part independent of uPA.5,6 The reasons for these discrepancies are not clear, but may be related to variations in the genetic backgrounds of the mice, a common confounding factor in many studies using genetic models.7 In the case of Connolly et al, the PlauGFDhu/GFDhu mice were created in the C57BL/6J background and all strains were backcrossed into this strain.
This novel strain not only represents a powerful new tool for the in vivo study of the uPA:uPAR interaction, but may also potentially be used to gain insight into fibrin-exacerbated chronic inflammatory diseases. Connolly et al have impressively shown that the ablation of a single binary interaction can lead to organism-wide pathology, demonstrating that complex biologic networks remain finely balanced in regard to their component fundamental interactions.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal