Abstract
Despite improvement in the treatment of advanced classical Hodgkin lymphoma, approximately 30% of patients relapse or die as result of the disease. Current predictive systems, determined by clinical and analytical parameters, fail to identify these high-risk patients accurately. We took a multistep approach to design a quantitative reverse-transcription polymerase chain reaction assay to be applied to routine formalin-fixed paraffin-embedded samples, integrating genes expressed by the tumor cells and their microenvironment. The significance of 30 genes chosen on the basis of previously published data was evaluated in 282 samples (divided into estimation and validation sets) to build a molecular risk score to predict failure. Adequate reverse-transcription polymerase chain reaction profiles were obtained from 262 of 282 cases (92.9%). Best predictor genes were integrated into an 11-gene model, including 4 functional pathways (cell cycle, apoptosis, macrophage activation, and interferon regulatory factor 4) able to identify low- and high-risk patients with different rates of 5-year failure-free survival: 74% versus 44.1% in the estimation set (P < .001) and 67.5% versus 45.0% in the validation set (P = .022). This model can be combined with stage IV into a final predictive model able to identify a group of patients with very bad outcome (5-year failure-free survival probability, 25.2%).
Introduction
Classical Hodgkin lymphoma (cHL) is assumed to be a curable tumor, but an important fraction of patients with advanced disease do not respond favorably to the current standard chemotherapy regimens whose base is adriamycin. The most widely used and reproducible prognostic score is the product of clinical and analytical parameters integrated in the International Prognostic Score (IPS), but it still fails to identify accurately, at the moment of diagnosis, a significant fraction of patients with very poor prognosis.1-3 Thus, the identification of biomarkers that, at diagnosis, may be consistently associated with nonresponse is essential for the recognition of patients at high risk of treatment failure to establish a more rational risk-adapted treatment strategy.
cHL represents a distinctive model of histologic complexity, with a minor population of the neoplastic Hodgkin and Reed-Sternberg (HRS) cells diluted in a reactive inflammatory background composed of nonneoplastic B and T cells, macrophages, eosinophils, neutrophils, and plasma cells. The complex relationship between the HRS cells and their microenvironment is only partially understood; however, important, if fragmentary, advances in our understanding are steadily being made.4 The clinical outcome of cHL has been found to be related to the expression of multiple biologic markers alone5-8 or in combination,9 expressed either by the tumor HRS cells, macrophages, regulatory T cells, or other nonneoplastic cell subpopulations.10-13
Some of these previous analyses rely on array-based gene expression analyses, which use frozen tissue and in most cases can only provide retrospective information. Investigators of other studies have used immunohistochemical staining, with some inherent limitations to the reproducibility of the data thus generated. It is now feasible to apply multigenic predictive molecular tests in advanced cHL patients in a routine setting by the use of a quantitative reverse-transcription polymerase chain reaction (RT-PCR) assay as we here describe, incorporating a selected number of genes that capture information from tumor and microenvironment cell components, designed for application to routine formalin-fixed paraffin-embedded (FFPE) samples and that can be used at the moment of the initial diagnosis.
Methods
Patients and samples
Previous studies allowed us to identify a group of genes whose expression was associated with the response of patients with advanced cHL to standard first-line treatment. Thus, the selection of genes to be analyzed was determined by results previously obtained in 2 independent series of 29 and 52 advanced cHL patients.10,11
The patients included in this study fulfilled stringent, previously described criteria (ie, age older than 16 years; advanced cHL; Ann Arbor stage IV, III, or IIB with bulky masses1 ; proven HIV-negative status) who have been treated with a first-line standard chemotherapy regimen that included adriamycin—ABVD (adriamycin, bleomycin, vinblastine, and dacarbazine) or ABVD variants—and for whom information was available about the achievement of complete remission (CR) and a follow-up of at least 12 months thereafter, which is a well-known and accepted surrogate indication of the course of the disease. With respect to the latter, patients were considered to have had a favorable course if they had achieved CR and maintained it for at least 12 months or with an unfavorable course if they had either not achieved CR or if they had once had it but had relapsed during the following 12 months. All tissue samples consisted of representative pretreatment lymph node biopsies collected after revision and approved by the institutional review board of the participating institutions of the Spanish Hodgkin Lymphoma Study Group. The study initially included 282 FFPE patients, who were randomly split and assigned to the training (194 cases) or validation sets (88 cases) on the basis of the minimum estimated sample size to derive the final model (Table 1).
Clinical characteristics of the cHL series
| Characteristic . | Estimation, n (%) . | Validation, n (%) . | Total, n (%) . | P . |
|---|---|---|---|---|
| Age, y | (n = 183) | (n = 79) | (n = 262) | |
| Younger than 45 | 133 (72.67) | 56 (70.88) | 189 (72.14) | .883 |
| 45 or older | 50 (27.32) | 23 (29.11) | 73 (27.86) | |
| Sex | (n = 183) | (n = 79) | (n = 262) | |
| Male | 99 (54.10) | 51 (64.55) | 150 (57.25) | .151 |
| Female | 84 (45.90) | 28 (35.44) | 112 (42.75) | |
| Stage IV | (n = 182) | (n = 79) | (n = 261) | |
| No | 132 (72.52) | 47 (59.49) | 179 | .053 |
| Yes | 50 (37.87) | 32 (40.50) | 82 | |
| IPS code | (n = 182) | (n = 79) | (n = 261) | |
| Less than 3 | 109 (59.89) | 41 (51.90) | 150 (57.47) | .288 |
| 3 or greater | 73 (40.11) | 38 (48.10) | 111 | |
| Outcome | (n = 183) | (n = 79) | (n = 262) | |
| F | 132 (72.14) | 57 (72.16) | 189 (72.14) | > .999 |
| U | 51 (27.86) | 22 (27.84) | 73 (27.86) |
| Characteristic . | Estimation, n (%) . | Validation, n (%) . | Total, n (%) . | P . |
|---|---|---|---|---|
| Age, y | (n = 183) | (n = 79) | (n = 262) | |
| Younger than 45 | 133 (72.67) | 56 (70.88) | 189 (72.14) | .883 |
| 45 or older | 50 (27.32) | 23 (29.11) | 73 (27.86) | |
| Sex | (n = 183) | (n = 79) | (n = 262) | |
| Male | 99 (54.10) | 51 (64.55) | 150 (57.25) | .151 |
| Female | 84 (45.90) | 28 (35.44) | 112 (42.75) | |
| Stage IV | (n = 182) | (n = 79) | (n = 261) | |
| No | 132 (72.52) | 47 (59.49) | 179 | .053 |
| Yes | 50 (37.87) | 32 (40.50) | 82 | |
| IPS code | (n = 182) | (n = 79) | (n = 261) | |
| Less than 3 | 109 (59.89) | 41 (51.90) | 150 (57.47) | .288 |
| 3 or greater | 73 (40.11) | 38 (48.10) | 111 | |
| Outcome | (n = 183) | (n = 79) | (n = 262) | |
| F | 132 (72.14) | 57 (72.16) | 189 (72.14) | > .999 |
| U | 51 (27.86) | 22 (27.84) | 73 (27.86) |
Clinical characteristics of patients with adequate RT-PCR profiles. Summary of the clinical characteristics of the patients in the estimation and validation sets that yielded suitable analyzable data (262 of 282; 92.90%). Differences in distribution of standard clinical parameters (age, sex, stage, IPS and outcome) between estimation and validation datasets tested by Pearson chi-square with Yates correction were not statistically significant (IPS values of 0-2 classified as low IPS; IPS values > 2 classified as high IPS).
cHL indicates classical Hodgkin lymphoma; F, favorable response; IPS, International Prognostic Score; RT-PCR, reverse transcription polymerase chain reaction; U, unfavorable response.
Additional exclusion criteria were insufficient RNA quality (purity ratio A260:A230 < 1.7) or a weak RT-PCR signal (average cycle threshold > 35) for the reference genes or in more than 10 genes of the assay. As a result, 20 patients were excluded, and the remaining 262 patients (183 in the training group and 79 in the validation group) meeting these criteria were included in the statistical analysis (Table 1).
Gene selection
The genes included in the assay initially were selected from 2 preliminary expression-profiling studies10,11 that rendered a list of genes expressed by HRS and microenvironment cell subpopulations identified in unfavorable cHL patients. Selected genes were primarily chosen on the basis of their prognostic ability and capacity to represent biologic functions identified as relevant in cHL pathogenesis.11 In addition, the strength and consistency of primer and probe performance also were taken into account.11 The initial selection consisted of 30 genes, including genes expressed by the neoplastic cells involved in the cell cycle (G2/M), apoptosis, histones, chaperones, drug metabolism, and mitogen-activated protein kinase signatures, and from microenvironment genes expressed by different cellular or functional populations of T cells, monocytes, macrophages, and dendritic cells. Details of the RT-PCR assays are available in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Analysis of gene expression
Gene expression was analyzed by the use of a customized TaqMan low-density array platform (Micro Fluidic Cards; Applied Biosystems) on FFPE as previously described.11,14 A preamplification step (PreAmp; Applied Biosystems) was used to improve the sensitivity of our assay for low-abundance target genes available from FFPE samples.15-17 Reactions were performed by use of the ABI PRISM 7900HT Sequence Detection system (Applied Biosystems), and we measured the expression of each gene in triplicate and then normalized it with a set of 2 reference genes (HMBS and GUSB) whose uniform expression in cHL tumor samples was tested in previous studies.11 Missing values were imputed using the K-nearest neighbor algorithm.18
Statistical analysis
Differences in the distributions of standard clinical parameters (age, sex, stage, IPS, the individual variables contained in IPS, and outcome) in the estimation and validation datasets were tested by the Pearson χ2 test (Table 1).
The first end point of this study was the response to standard first-line treatment considering favorable response (F) and unfavorable response (U), as mentioned previously. Data from second-line and salvage therapies and/or bone-marrow transplantation were not considered.
The selection of the best predictive genes and the logistic regression model was on the basis only of the data from the training group of 183 patients, without any previous survival analysis that used information from the validation group. Univariate regression analysis was performed with treatment response (F vs U) as the dependent variable to identify genes significantly associated (P < .05) with outcome. In addition, final gene selection analysis was performed by cross-validation with the use of 3 prediction algorithms (http://tnasas.bioinfo.cnio.es/): diagonal linear discriminant analysis,19 support vector machines,20 and K-nearest neighbor.21 Cross validation was used to test the classification ability of the initial set of significant genes to choose the strongest predictor genes, which were classified into functional groups on the basis of their known biologic relationship and their coregulated expression as estimated by the Pearson correlation coefficient. Individual genes from each functional group were weighted by the use of linear discriminant analysis.22 Finally, these functional gene clusters associated with cHL outcome were analyzed in a multivariate logistic regression model with response to therapy (F vs U) as a dependent variable. In this way, an algorithm was derived that combines these measurements into a quantitative “molecular risk score” (MRS), which can be used as a continuous variable to estimate the probability of treatment response. The MRS cut-off points were prespecified by the use of an area under the receiver operating characteristic curve (ROC) analysis to define different risk groups. (See the supplemental Appendix for details of the statistical analysis and methods.) Finally, performance of the logistic regression model was tested in the validation group of patients (n = 79).
For graphical representation, survival analyses were performed with the Kaplan-Meier method and long-rank test separately in the training and validation series and in the entire series. Because the primary objective of the study was to identify patients at high risk of treatment failure, we used failure-free survival (FFS) as the fundamental end point for survival analysis. FFS was defined as the time interval between treatment initiation and treatment failure or last follow-up. Failure was defined as either the failure to achieve CR or the occurrence of progressive disease, irrespective of whether there had been an initial CR. Overall survival (OS), an end point whose significance is imperfect because it is conditioned by the effect of subsequent eventual treatments and complication of treatment, was included as a secondary end point in the survival analyses, defined as the time interval between diagnosis and death caused by the lymphoma.
Finally, in the whole series, a multivariate Cox proportional hazards model, including the data at diagnosis, the IPS stratified as previously defined (0-2 vs ≥ 3),1 and its 7 individual variables (hemoglobin < 1.5 g/dL; albumin < 4 g/dL; leukocytosis ≥ 15 000/mm3; lymphopenia < 600/mm3; age ≥ 45 years; male sex; stage IV), was applied to test the independence of the MRS, including the remaining significant variables in a final integrative model.
All statistical analyses were 2-sided; values of P less than .05 were considered to be significant. These were performed with SPSS 15.0 (SPSS Inc). Survival curves were assessed by the Kaplan-Meier method, and risk groups were compared by the log-rank test. Plots were generated with the use of GraphPad Prism Version 5 (GraphPad Software, Inc).
Results
Gene selection and development of the predictor model
In training series, univariate regression analysis of the expression data for the 30 initially selected genes revealed 20 genes to significantly predict failure to first-line treatment (supplemental Table 3). When cross validation was applied, the genes most frequently found in prognostic models consisted of a panel of 11 genes that were included in the final model: BCL2, BCL2L1, CASP3, HMMR, CENPF, CCNA2, CCNE2, CDC2, LYZ, STAT1, and IRF4.
To derive the model, we took a 2-step approach, first combining individual gene-expression patterns into precise functional pathways and then subsequently correlating these functional groups with the clinical outcome by using multivariate logistic regression. Final selected genes were weighted by the use of linear discriminant analysis and clustered into their corresponding functional pathways defined as macrophage activation (LYZ, STAT1), cell cycle (HMMR, CENPF, CCNA2, CCNE2, CDC2), and apoptosis (BCL2, BCL2L1, CASP3; Figure 1A). The Pearson correlation coefficient was significant for the genes included in each of the signatures (P < .001) apart from IRF4, which was included as an independent predictive gene because there were neither distinct functional relationships nor statistically significant correlations with other genes or pathways (supplemental Table 4). These functional groups captured information about the tumoral HRS cells and their nontumoral microenvironment, in agreement with previous studies.10 The multivariate logistic regression analysis integrating these pathways showed that cell cycle and apoptosis terms were associated with an unfavorable outcome of patients, whereas macrophage activation and IRF4 signatures had protective effects.
Panel of 11 genes and the molecular risk algorithm. (A) The molecular risk algorithm is determined on the basis of the relative contributions of each of the 4 gene functional groups from the tumoral HRS and their reactive microenvironment as follows: MRS = exp (fx)/(1 + exp [fx]), where fx = (−0.913) + (0.401 × apoptosis) + (0.284 × cell cycle) + (−0.301 × monocyte) + (−0.143 × IRF4). Coefficients were derived from a multivariate analysis in which positive values indicate that a greater level of expression is correlated with a worse outcome, and negative coefficients indicate that a greater level of expression of the pathways is associated with a better outcome. (B) MRS as a continuous function was used to set a threshold for stratifying patients by ROC analysis. Patients were stratified according to the levels of the molecular risk score into low-risk (< 0.3) and high-risk (≥ 0.3) groups. (C-D) Survival estimates of FFS in patients from estimation (n = 183) and validation (n = 79) sets after classification into risk groups. Kaplan-Meier analysis and the log-rank test gave significant results in both estimation and validation sets, indicating the potential prognostic capacity of the algorithm developed here.
Panel of 11 genes and the molecular risk algorithm. (A) The molecular risk algorithm is determined on the basis of the relative contributions of each of the 4 gene functional groups from the tumoral HRS and their reactive microenvironment as follows: MRS = exp (fx)/(1 + exp [fx]), where fx = (−0.913) + (0.401 × apoptosis) + (0.284 × cell cycle) + (−0.301 × monocyte) + (−0.143 × IRF4). Coefficients were derived from a multivariate analysis in which positive values indicate that a greater level of expression is correlated with a worse outcome, and negative coefficients indicate that a greater level of expression of the pathways is associated with a better outcome. (B) MRS as a continuous function was used to set a threshold for stratifying patients by ROC analysis. Patients were stratified according to the levels of the molecular risk score into low-risk (< 0.3) and high-risk (≥ 0.3) groups. (C-D) Survival estimates of FFS in patients from estimation (n = 183) and validation (n = 79) sets after classification into risk groups. Kaplan-Meier analysis and the log-rank test gave significant results in both estimation and validation sets, indicating the potential prognostic capacity of the algorithm developed here.
Thus, the optimized final model was determined on the basis of the relative contributions of each of the 4 functional terms, as described in the following equation: constant (−0.913) + (0.401 × apoptosis) + (0.284 × cell cycle) + (−0.301 × macrophage activation) + (−0.143 × IRF4). The continuous probability function generated by the logistic regression was defined as the MRS to treatment failure and ranged from 0.06 to 0.813 (Figure 1B). ROC analysis was used to define a threshold for stratifying patients, and the largest area under the curve was obtained by the use of 0.3 as the threshold, thus dividing the series into high-risk (MRS ≥ 0.3) and low-risk (MRS < 0.3) cases (Figure 1B-C).
Validation of the MRS
The MRS differed significantly between the various outcome groups (supplemental Figure 2) and predicted treatment response with an accuracy of 68.9% in the estimation dataset and 70% in the validation dataset. Because treatment failure was the main end point used to derive the logistic regression function, FFS as previously defined, was used for validation of the model and Kaplan-Meier analysis of survival. Predicted probabilities also identified 2 risk groups associated with FFS in both the estimation and validation sets (Figure 1C; supplemental Figure 3). FFS probabilities at 5 years were 74.0% versus 44.1% (P < .001) in the training set and 67.5% versus 45.0% (P = .022) in the validation set.
In addition, the analyses of OS showed different risk groups identified by the MRS in the estimation group of patients. The differences were not significant in the validation series, probably because of the limited number of events (supplemental Figure 3).
There were no significant statistical differences in the MRS distributions of the 2 histologic subtypes, nodular sclerosis and mixed-cellularity cHL (P = .186, t test), and the differences in survival between risk groups identified by the MRS remained significant in both the estimation and validation sets stratified by histologic subtype (supplemental Figure 4).
Integrative model using MRS and clinical variables
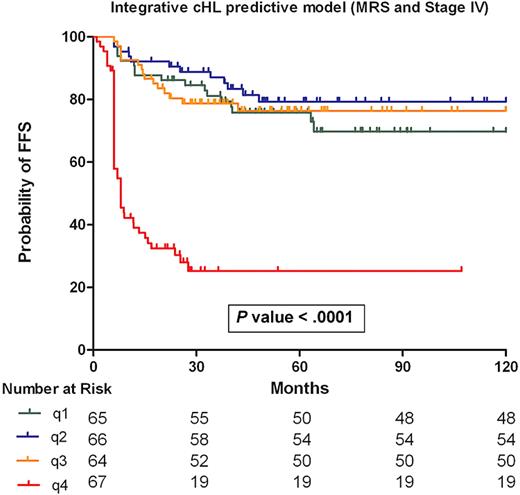
In the whole series, a multivariate Cox proportional hazards model with FFS as the dependent variable and including the MRS and the IPS, we found that only the MRS was significant (Table 2). No interaction was observed between the IPS or the individual IPS variables and the MRS low- and high-risk groups (supplemental Table 5). Thus, to compare the molecular risk algorithm and the individual IPS components, a backward stepwise selection Cox model was tested, with FFS as the dependent variable and including the MRS and the individual components of the IPS in the global series of samples. Only MRS and stage IV were statistically significant (Table 2) and so were retained in the final Cox regression model. Patient stratification into quartiles on the basis of the Cox model identified a subgroup of advanced cHL patients (fourth quartile) with a very poor outcome: 5-year FFS of 24.3% (P < .001; Figure 2).
MRS and the IPS variables
| . | P . | Hazard ratio (95% CI) . |
|---|---|---|
| IPS and MRS (n = 262)* | ||
| MRS | < .001§ | 24.362 (6.268-94.691) |
| IPS | .205 | 1.111 (0.944-1.309) |
| IPS variables and MRS (n = 262)† | ||
| MRS | < .001§ | 24.715 (6.804-89.768) |
| Hemoglobin < 10.5 g/dL | .352 | 1.243 (0.787-1.963) |
| Albumin < 4 g/dL | .504 | 1.157 (0.755-1.772) |
| Leucocytosis ≥ 15 000/mm3 | .256 | 1.312 (0.821-2.095) |
| Lymphopenia < 600/mm3 | .555 | 0.820 (0.425-1.583) |
| Age ≥ 45 y | .369 | 1.227 (0.785-1.916) |
| Stage IV | .041§ | 1.552 (1.018-2.360) |
| Male sex | .753 | 0.936 (0.618-1.416) |
| PT-BR Integrative Cox model (n = 262)‡ | ||
| MRS | < .001§ | 23.782 (6.041-94.340) |
| Stage IV | .025§ | 1.409 (1.044-1.900) |
| . | P . | Hazard ratio (95% CI) . |
|---|---|---|
| IPS and MRS (n = 262)* | ||
| MRS | < .001§ | 24.362 (6.268-94.691) |
| IPS | .205 | 1.111 (0.944-1.309) |
| IPS variables and MRS (n = 262)† | ||
| MRS | < .001§ | 24.715 (6.804-89.768) |
| Hemoglobin < 10.5 g/dL | .352 | 1.243 (0.787-1.963) |
| Albumin < 4 g/dL | .504 | 1.157 (0.755-1.772) |
| Leucocytosis ≥ 15 000/mm3 | .256 | 1.312 (0.821-2.095) |
| Lymphopenia < 600/mm3 | .555 | 0.820 (0.425-1.583) |
| Age ≥ 45 y | .369 | 1.227 (0.785-1.916) |
| Stage IV | .041§ | 1.552 (1.018-2.360) |
| Male sex | .753 | 0.936 (0.618-1.416) |
| PT-BR Integrative Cox model (n = 262)‡ | ||
| MRS | < .001§ | 23.782 (6.041-94.340) |
| Stage IV | .025§ | 1.409 (1.044-1.900) |
95% CI indicates 95% confidence interval; IPS, International Prognostic Score; and MRS, molecular risk score.
Multivariate Cox regression analysis considering MRS and the IPS.
Univariate Cox regression analyses considering individual IPS variables and MRS.
Cox regression analysis of the final variables included in the integrative model (molecular risk plus stage IV) obtained by backward stepwise selection.
Significant.
Integrative risk model of cHL. The final Cox model integrates the MRS and clinical variable stage IV. Patients in quartiles 1, 2, and 3 have comparable FFS rates at 5 years (76.4%, 79.3%, and 69.7%, respectively) whereas patients in quartile 4 show a 5-year FFS of 24.3% (P < .001).
Integrative risk model of cHL. The final Cox model integrates the MRS and clinical variable stage IV. Patients in quartiles 1, 2, and 3 have comparable FFS rates at 5 years (76.4%, 79.3%, and 69.7%, respectively) whereas patients in quartile 4 show a 5-year FFS of 24.3% (P < .001).
Discussion
Here we describe, in a series of patients with advanced cHL, a 4-cluster/11-gene model derived from an initial selection of 30 potentially predictive markers that can be detected by RT-PCR and integrated into a molecular risk algorithm that can identify patient subgroups with very different probabilities of treatment failure. This approach is determined on the basis of reliable quantitative RT-PCR techniques applicable to paraffin-embedded diagnostic samples and follows similar approaches taken in breast cancer and other tumor types.23,24 This benefits from previous expression profiling studies performed in cHL,10 and the improved knowledge about the role of the tumoral cell and the microenvironment in the pathogenesis and outcome of this disease.13,25,26 This MRS, calculated in an initial estimation set, was confirmed in an independent set. Genes included in the score were selected on the basis of previous gene expression profiling data generated in independent sets of patients11,27 and can be classified in 4 functional pathways.
The final 4-cluster/11-gene model can additionally incorporate one of the well-established clinical variables (stage IV), thus integrating the main molecular characteristics of the tumors related with treatment response and tumor burden estimation in a single scoring system. The multivariate Cox model indicates that most patients with stage IV cHL and with a high MRS (≥ 0.3) will have a very poor outcome, with 5-year FFS probability of 24.3% and OS probability of 76.3%. Therefore, this combination of stage IV and high MRS identifies a group of patients with very bad outcome who could have been initially missed by consideration of the IPS alone.1,3
It is of note that in the present series the IPS did not show any significant prognostic influence on FFS. From the individual components of the IPS, only stage IV disease remained significant in multivariate analyses. This finding is in agreement with recent studies in which the authors demonstrated that IPS is of limited utility in advanced HL cases treated in the modern era, where more accurate pathologic diagnosis, improved control of therapy, use of growth factors, and enhanced supportive care are yielding better outcomes compared with historic results.3 Thus, deviations from the standard therapy cannot be justified on the basis of the survival prediction capacity of the IPS, and identification of high-risk populations needs to be supplemented with molecular markers.
In the model, the expression of BCL2, BCL2L1, CASP3, HMMR, CENPF, CCNA2, CCNE2, and CDC2 included in the apoptosis and cell-cycle pathways, respectively, were correlated with short FFS. The expression of various antiapoptotic BCL2 family proteins has been repeatedly reported in HRS cells,7,28,29 thereby contributing to the survival of HRS cells. BCL2 and BCL2L1 (BCL-XL) both frequently are expressed by HRS cells in cHL, and their levels have been associated with inferior FFS in patients treated with ABVD or equivalent regimens,5 thus confirming the importance of this group of apoptotic regulators for cHL outcome prediction. The second signature, cell cycle, is mainly composed of genes coding for regulatory proteins of the S and G2/M phases of the cell cycle, thus directly related with cell proliferation. Again, expression of some of these markers has been previously described at the protein level in cHL,7,30,31 and a significant prognostic value for CDC2 (CDK1) and CCNA2 (cyclin A2) protein expression in non-Hodgkin lymphomas was also found for both disease-free survival and OS. Interestingly, this aberrant association between increased expressions of antiapoptotic proteins and growth fraction-associated proteins in HRS cells provides further evidence that cell cycle and apoptosis regulation are profoundly disturbed and closely related in the disease, further justifying the inclusion of these 2 pathways in the predictive model.
Moreover, pathways involved in cell cycle and apoptosis regulation are rational therapeutic targets. Indeed, inhibitors of Cyclin-Cdk complexes (including Cyclin E-Cdk2, Cyclin A-Cdk2, and Cyclin B-Cdk1 complexes) are currently under preclinical and clinical investigation in different cancer types,32-34 and these drugs could be considered for the treatment of advanced and refractory cHL patients. Likewise, the potential for targeting BCL2-related proteins in lymphoma is promising. Small molecule inhibitors of the Bcl-2 family have demonstrated high target affinity and an improved toxicity profile, and clinical trials of these agents are yielding interesting results.35
Confirming previous observations about the importance of the reactive microenvironment for cHL patient outcome,9,11,12 LYZ and STAT1 genes, expressed at high levels in a subset of tissue monocytes and activated macrophages, also are included in this model, and correlated with prolonged FFS and better outcome. The relevance of the cell composition of the reactive background in cHL has been reinforced by the data recently reported by Steidl et al.36 They used gene expression profiling to identify a gene signature of tumor-associated macrophages that is associated with treatment failure, in an approach methodologically similar to previous reports from our group and others.10,37 The discrepancy in the results concerning the role of macrophages may have arisen from technical differences (RT-PCR vs microarray gene-expression and immunohistochemistry) or the selection of markers such as LYZ and STAT1 in this study, reflecting a specific functional status of the monocyte-macrophages.38
In addition, IRF4 (MUM1) expression was associated with longer FFS. This gene is an interferon regulatory factor, lymphocyte specific, induced after nuclear factor-κB (NF-κB) activation, which controls B-cell proliferation and differentiation and has recently been shown to be up-modulated by CD40 engagement in HL cells.39 Interestingly, the lack of IRF4 protein has been previously associated with outcome in cHL,40 representing a potential adverse prognostic factor. In addition, both IRF4 and BCL2L1 represent well-known NF-κB target genes41,42 whose expression is induced after NF-κB pathway activation. Thus, our final model includes important subrogates from the NF-κB activation, which is thought to be an essential pathogenetic mechanism in this disease. It is of note that Bednarski et al43 recently described how the inhibition of the canonical NF-κB pathway enhances the proapoptotic effects of adriamycin, thus also identifying NF-κB inhibition as an interesting therapeutic approach.
In conclusion, we have developed a molecular risk algorithm, on the basis of feasible and reproducible molecular techniques, that is capable of stratifying at the moment of diagnosis advanced cHL patients with different outcome. Moreover, a combination of this algorithm with the presence of clinical stage IV disease can be used to identify a group of cHL patients with a very bad outcome who could benefit from more intensive therapeutic approaches.
These results are promising, but further validation in larger and independent series of patient is needed for the model to become established as part of the clinical routine. Also the predictive value of this model should also be tested in patients treated with modern intensive chemotherapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Nuria Malats, from the Genetic and Molecular Epidemiology Group at the CNIO, for her useful advice and supervision of the statistical analysis. We also thank Marién Castillo and Laura Cereceda, at the CNIO Tumor Bank, for collecting the human tumor samples and for their excellent assistance with data management. Members of the Spanish Hodgkin Lymphoma Study Group are cited in the supplemental Appendix.
This work was supported by grants from the Fondo de Investigaciones Sanitarias (PI08/1985, PI05/1623, PI05/2800, PI05/2327, RETIC RD06/0020/0107) and the Ministerio de Ciencia y Tecnología (SAF2008-03 871), Spain. B.S.-E. is supported by a grant from the Ministerio de Ciencia e Innovación (FIS), Spain.
Authorship
Contribution: B.S.E., C.M., M.A.P., and J.F.G. contributed to the conception and design of the study, the analysis and interpretation of the data, and the drafting of the article; B.S.E. and A.L. performed statistical analysis; M.M.M. managed tissue banking; J.M., P.S., C.R.M., A.L., R.R., J.R., A.C., C.C., M.C., J.A., R.A., A.A., A.S., S.S., A.B., J.M.M., P.S.-G., F.B., C.R., M.F.F., J.G.L., M.G.-C., C.S., J.L.L., M.L.I., M.M., J.G.C., A.M., J.F., R.G.C, and J.F.T. managed patient databases and contributed with tumor samples and clinical follow-up; and all the authors read and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Members of the Spanish Hodgkin Lymphoma Study Group are listed in the supplemental Appendix.
Correspondence: Miguel A Piris, MD, Lymphoma Group, Spanish National Cancer Centre (CNIO). E-28029 Madrid, Spain; e-mail: mapiris@cnio.es.
References
Author notes
B.S.-E. and C.M. contributed equally to this work.

![Figure 1. Panel of 11 genes and the molecular risk algorithm. (A) The molecular risk algorithm is determined on the basis of the relative contributions of each of the 4 gene functional groups from the tumoral HRS and their reactive microenvironment as follows: MRS = exp (fx)/(1 + exp [fx]), where fx = (−0.913) + (0.401 × apoptosis) + (0.284 × cell cycle) + (−0.301 × monocyte) + (−0.143 × IRF4). Coefficients were derived from a multivariate analysis in which positive values indicate that a greater level of expression is correlated with a worse outcome, and negative coefficients indicate that a greater level of expression of the pathways is associated with a better outcome. (B) MRS as a continuous function was used to set a threshold for stratifying patients by ROC analysis. Patients were stratified according to the levels of the molecular risk score into low-risk (< 0.3) and high-risk (≥ 0.3) groups. (C-D) Survival estimates of FFS in patients from estimation (n = 183) and validation (n = 79) sets after classification into risk groups. Kaplan-Meier analysis and the log-rank test gave significant results in both estimation and validation sets, indicating the potential prognostic capacity of the algorithm developed here.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/8/10.1182_blood-2010-02-270009/4/m_zh89991055940001.jpeg?Expires=1769325450&Signature=NOQr7LgwwHgU0faPyMFMCCs-RLtnJ-WcHCBwWrGEx1O0GpG~~vC5PPvpoUYuG6CMNuyvxl09Cj7GIeHQIUOiCb0mLvaqSE6Y3dEVbFWZpJEyGQ5BBGHIgLVTquz4cJqUyOA6OuiZ5bKNUDoAdSreXjXOA0juVvUGWzEGstAvcCoUgHzXpepo7q~-SW-kUL2T0RbHbYHCKcM5JFnhbznpqYSItTyRhRP8Pq0qvvYpg6BZ-YsvIAwJ9skvkBGHBHNRLJU7ghjd~qKs4sKM1Cipg5x5pmH7ncNlLvlAPTOOYZhZ3FZET8T7QHWbCjXayg5ZXg2fzoIRgfDrWZEzyC-deQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)