Abstract
Human natural killer (NK) cells comprise 2 main subsets, CD56bright and CD56dim cells, that differ in function, phenotype, and tissue localization. To further dissect the heterogeneity of CD56dim cells, we have performed transcriptome analysis and functional ex vivo characterization of human NK-cell subsets according to the expression of markers related to differentiation, migration or competence. Here, we show for the first time that the ability to respond to cytokines or to activating receptors is mutually exclusive in almost all NK cells with the exception of CD56dim CD62L+ cells. Indeed, only these cells combine the ability to produce interferon-γ after cytokines and proliferate in vivo during viral infection with the capacity to kill and produce cytokines upon engagement of activating receptors. Therefore, CD56dim CD62L+ cells represent a unique subset of polyfunctional NK cells. Ex vivo analysis of their function, phenotype, telomere length, frequencies during ageing as well as transfer experiments of NK-cell subsets into immunodeficient mice suggest that CD56dim CD62L+ cells represent an intermediate stage of NK-cell maturation, which after restimulation can accomplish multiple tasks and further develop into terminally differentiated effectors.
Introduction
In the course of the immune response against microbes, naive T cells proliferate and generate varied classes of effector as well as memory cells, with distinct properties and functions. This program appears to be essential to achieve protection and immunologic memory. In recent years, it became clear that natural killer (NK) cells do not represent a homogenous population of cells ready to kill, but they also undergo a differentiation program, which includes major histocompatibility complex (MHC)–dependent education,1-3 priming,4 and even generation of memory during recall responses.5,6 This complexity implies the existence of distinct stages of NK-cell differentiation, which can guarantee an efficient division of labor, as it has been shown for T cells. However, in contrast to T cells, understanding NK cells increasing complexity is hampered by the lack of adequate markers that would enable us to distinguish defined steps of NK-cell differentiation history. In humans, 2 NK-cell subsets have been characterized.7,8 While human CD56bright NK cells can strongly proliferate9 and produce interferon-γ (IFN-γ) in response to cytokine stimulation,10-12 CD56dim NK cells are highly cytotoxic.10,11,13 Many studies have shown that the CD56dim NK-cell subset may be derived directly from the CD56bright NK-cell subset,14-17 supporting Lanier et al's original proposal.7 However, CD56dim NK cells represent a heterogeneous population concerning the expression of several markers, namely Killer immunoglobulin-like receptors (KIR), NKG2A or CD94, CD27, and CD62L. Some of these markers have been associated with different NK-cell functions.3,18-21 Thus, the expression of self MHC-specific inhibitory receptors such as KIR and NKG2A correlates with higher ability to kill and to produce IFN-γ in response to stimulation via activating receptors1-3,18 or even cytokines.3,22 Moreover, others and we have shown that KIR can be acquired by CD56bright as well as by CD56dim KIR− NK cells after cytokine stimulation in vitro and in vivo14,17,23 and that KIR expression correlates to a lower proliferative capacity.14,22 Altogether, these data suggest that KIR expression among CD56dim cells may define a terminally differentiated subset characterized by low proliferative ability and high effector potential. On the other hand, it is also reported that CD27 expression influences NK-cell functions as it correlates with high ability to proliferate and to produce IFN-γ and with low cytotoxic potential, at least in humans.20,21 Interestingly, although expression of CD62L within a subset of CD56dim NK cells was previously reported,24 functional analysis according to CD62L expression has never been performed. Because the expression of KIR, NKG2A, CD27, or CD62L is not mutually exclusive but CD56dim NK cells partially co-express these markers, the aim of this study was to dissect how the expression of CD62L and of other markers correlates to different NK-cell function and phenotype, to better define the differentiation history of human NK cells. Here we show that during maturation from CD56bright to CD56dim, NK cells lose the ability to respond to cytokines and gain the ability to respond to activating receptor stimulation. Only the CD56dim CD62L+ subset combines the high potential of CD56bright cells to produce IFN-γ after cytokine stimulation and proliferate with the capacity to kill and produce cytokines upon activating receptor stimulation, typical of CD56dim cells. Moreover, the analysis of transcriptome signatures, telomere length, frequencies during ageing and transfer experiments into immunodeficient mice suggest that CD56bright, CD56dim CD62L+ and CD56dim CD62L− may correspond to sequential steps of NK-cell maturation.
Methods
Cells and culture conditions
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll Hypaque density gradient centrifugation (Amersham Pharmacia Biotech) from leukocyte concentrates obtained from healthy donors (DRK) after donor informed consent was in accordance with the Declaration of Helsinki and approval by the local ethics committees on human studies (Charité Berlin, Germany). NK-cell subsets were separated by magnetic cell sorting (MACS) to enrich CD56+ cells (Miltenyi Biotec) and subsequently sorted using a FACSAria (BD Biosciences). All sorted subsets displayed purity above 98%. NK cells (5 × 105 or 106/mL) were cultured in 96-well round-bottom plates (Greiner Bio-One) in RPMI-1640 (Gibco BRL) supplemented with 10% human AB serum (Lonza), 100 U/mL penicillin, and 0.1 mg/mL streptomycin. For NK-cell stimulation, the following cytokines were used: interleukin-2 (IL-2; R&D Systems), IL-15, IL-12, and IL-18 (Miltenyi Biotec). All microarray data are available in the Gene Expression Omnibus (GEO) under accession number GSE21774. Further information is available in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Analysis of cytokine production
NK cells (106/mL) were stimulated in the presence of IL-12 and IL-18 or IL-12 and IL-15 or in the presence of myeloid dendritic cells (mDCs) plus R848 and lipopolysaccharide (see supplemental Methods). Stimulation via activating receptors was performed as follows: goat F(ab)2 anti–mouse immunoglobulin G (Beckman Coulter) was coated to plastic wells (96-well, U-Bottom; Greiner Bio-One) for 2 hours in phosphate-buffered saline (PBS) at 37°C at 20 μg/mL. After washes, purified mouse anti–human NKG2D (BAT221; provided by D. Pende, INRC, University of Genoa, Genoa, Italy), NKp30 (AZ20), NKp46 (BAB281), 2B4 (PP35), all provided by A. Moretta (University of Genova, Genova, Italy) and CD2 (PPA-2.10; BD Biosciences) monoclonal antibodies (mAbs) were incubated for 30 minutes at 4°C at 10 μg/mL in PBS. For CD16 stimulation, purified mouse anti–human CD16 (3G8; BD Biosciences) was coated directly to plastic wells for 2 hours in PBS at 37°C at 10 μg/mL. After washes, 2 × 105 cells per well were plated in each well and stimulated for 8 hours. Cytokine production was measured in cell supernatants by enzyme-linked immunosorbent assay according to manufacturer's instructions (eBioscience) or intracellularly by flow cytometry. mAbs used are described in supplemental Methods. For intracellular detection of the cytokines, 10 μg/mL BrefeldinA (Sigma-Aldrich) was added for the final 8 hours of stimulation. Cells were fixed with 1.8% formaldehyde solution and permeabilized with Perm2 Solution (BD Biosciences). IFN-γ mRNA was measured by quantitative reverse-transcribed polymerase chain reaction (see supplemental Methods).
Analysis of cytotoxic potential
NK-cell cytotoxicity was analyzed in all experiments by co-cultivating the indicated NK-cell subset with the MHC-class I negative target cell line K562 (ATTC) in an effector-to-target ratio of 5:1. To measure degranulation we performed the CD107a Mobilization Assay using an anti–CD107a-fluorescein isothiocyanate mAb (BD Biosciences), as previously described.25 In some experiments, cytotoxicity was directly assessed using a flow cytometric assay for NK-cell killing developed by McGinnes et al with slight modifications.26 Briefly, target cells were loaded with 5μM carboxyfluorescein succinimidyl ester (CFSE) and incubated with NK cells at 37°C for 6 hours. After wash, propidium iodide was added. Live target cells were identified as CFSE+ PI− whereas dead target sells (Td) were CFSE+ PI+. Specific lysis was calculated as Td (cultured with effector cells) − Td (cultured without effector cells).
In vivo experimental procedures
BALB/c nonobese diabetic/severe combined immune deficient (NOD/SCID) and BALB/c NOD/SCID/γc−/− mice (The Jackson Laboratory) were maintained under defined flora conditions. For NK-cell transfer, 106 human CD56bright CD62L+, CD56dim CD62L+, or CD56dim CD62L− NK cells were sorted (98% purity) and injected into the tail veins of 6-week-old 137Cs source 350cGy irradiated mice. NOD/SCID/γc−/− mice were injected intraperitoneally with 2.5 μg of hIL-15 (Miltenyi Biotec) plus 7.5 μg of hIL15Rα-Fc (R&D Systems) preincubated in 200 μL of PBS for 30 minutes at 37°C, as previously described.17 Mice were killed at day 7 after transfer and experiments were repeated 3 times for each mouse strain.
Statistical analysis
For Gaussian distributed variables, the paired Student t test was used for statistical analysis. Gaussian distribution was tested by Shapiro Wilk normality test and D'Agostino & Pearson omnibus normality test. If variables were not Gaussian distributed, Wilcoxon signed rank test was employed for statistical analysis. Linear correlation was analyzed using the Pearson correlation coefficient.
Results
CD56bright and CD56dim CD62L+ NK cells display high proliferative ability in vitro and in vivo during a defined antiviral immune response
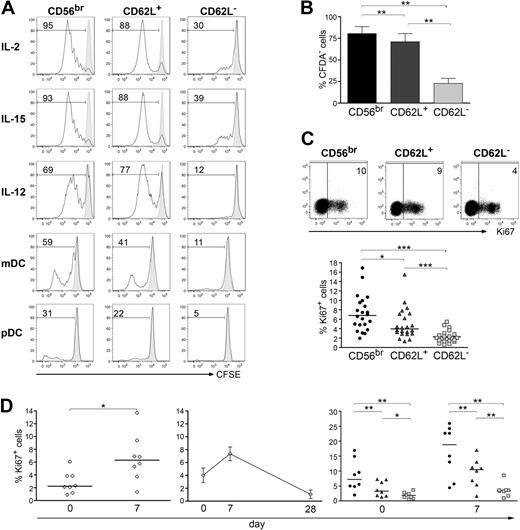
CD56bright NK cells exhibit a much higher capacity to proliferate after in vitro stimulation with cytokines or DCs than CD56dim NK cells.8 Because CD56dim NK cells are not a homogenous population concerning the expression of several surface markers including CD62L, we compared the proliferative ability of CD56bright, CD56dim CD62L+, and CD56dim CD62L− subsets after stimulation with cytokines or peripheral blood (PB)–derived DCs, either myeloid or plasmacytoid. Activation with IL-2 (Figure 1A-B), IL-15, IL-12, or DCs (Figure 1A) resulted in higher proliferation of CD56bright and CD56dim CD62L+ cells compared with CD56dim CD62L− ones. Cell survival after 5 days was consistently comparable in all 3 NK-cell subsets (supplemental Figure 1A-B). CD56bright NK cells proliferated slightly more than CD56dim CD62L+ ones at high concentrations of IL-2 (Figure 1B), while the difference in proliferation efficiency between the 2 subsets became more striking only at very low concentrations of IL-2 (supplemental Figure 1C). To determine whether the ability to proliferate in vitro actually mirrors what happens in vivo, we analyzed PBMCs ex vivo for the expression of Ki67, which is present in G1, G2, S, and M phases, but not in G0 phase of the cell cycle.27 Ki67 was expressed by a small fraction of CD3+ T cells and by a significantly higher proportion of NK cells (supplemental Figure 1D), thus showing that a consistent number of NK cells was undergoing homeostatic proliferation in vivo or has been recently activated due to an ongoing immune response. Interestingly, a higher percentage of in vivo proliferating cells could be detected among CD56bright and CD56dim CD62L+ cells compared with CD56dim CD62L− ones (Figure 1C). To investigate the proliferative response of NK cells in the course of a defined immune response, we explored NK-cell in vivo proliferation after vaccination with live yellow fever virus (YFV)–17D. Immunization with YFV-17D vaccine results in a self-limiting acute viral infection with viral replication,28 and therefore represents an ideal setting to assess NK-cell proliferation during an immune response. The frequency of Ki67+ NK cells was significantly increased at day 7 after YFV-17D vaccination compared with day 0 (Figure 1D left) and returned to basal levels by day 28 (Figure 1D middle), clearly indicating that a consistent proportion of NK cells was proliferating in vivo in response to viral infection. Importantly, cells that were proliferating at day 7 after YFV-17D vaccination were again comprised mostly within CD56bright and CD56dim CD62L+ NK-cell subsets (Figure 1D right), demonstrating that the high proliferative ability displayed in vitro by these 2 subsets actually reflects their in vivo behavior during a viral infection.
Proliferative ability of NK-cell subsets in vitro and in vivo after YFVvaccination. (A-B) Analysis of in vitro proliferation of natural killer (NK)–cell subsets (CD56bright, CD56dim CD62L+, and CD56dim CD62L− cells) was measured by CFSE dilution after 5 days of stimulation with either 50 ng/mL interleukin-2 (IL-2), IL-15, or IL-12, with myeloid dendritic cells (mDCs) in the presence of 100 ng/mL lipopolysaccharide and 10 μg/mL R848 or with plasmacytoid DC plus 10 μg/mL R848, 10 μg/mL CpG-A, and 10 ng/mL IL-3. (A) Representative data of 10 independent experiments is shown. Stimulated (open histograms) and unstimulated (gray-filled histograms) cells as well as the percentage of proliferating cells are depicted for each condition. (B) Mean percentage ± SEM of proliferating cells after high dose of IL-2 stimulation of 10 independent experiments is shown. P was calculated by Wilcoxon test. (C-D) In vivo proliferation of NK-cell subsets was measured by Ki67 staining of peripheral blood mononuclear cells (PBMCs) derived from healthy donors. (C) One representative donor (top panel) and percentage of Ki67+ cells plus corresponding medians of 22 donors analyzed ex vivo (bottom panel) are shown. (D) Ki67 expression was analyzed directly before (day 0), or after yellow fever virus (YFV) vaccination (days 7 and 28). Analysis was performed after gating on total CD3− CD56+ NK cells (left and middle) or on CD56bright (●), CD56dim CD62L+ ( ) and CD56dim CD62L− (
) and CD56dim CD62L− ( , right). Percentage of Ki67+ cells of 8 donors and corresponding median values (left and right graphs) or mean percentage ± SEM of Ki67+ cells of 3 donors (middle) are depicted. *P < .05; **P < .01; ***P < .0001 as calculated by Wilcoxon test.
, right). Percentage of Ki67+ cells of 8 donors and corresponding median values (left and right graphs) or mean percentage ± SEM of Ki67+ cells of 3 donors (middle) are depicted. *P < .05; **P < .01; ***P < .0001 as calculated by Wilcoxon test.
Proliferative ability of NK-cell subsets in vitro and in vivo after YFVvaccination. (A-B) Analysis of in vitro proliferation of natural killer (NK)–cell subsets (CD56bright, CD56dim CD62L+, and CD56dim CD62L− cells) was measured by CFSE dilution after 5 days of stimulation with either 50 ng/mL interleukin-2 (IL-2), IL-15, or IL-12, with myeloid dendritic cells (mDCs) in the presence of 100 ng/mL lipopolysaccharide and 10 μg/mL R848 or with plasmacytoid DC plus 10 μg/mL R848, 10 μg/mL CpG-A, and 10 ng/mL IL-3. (A) Representative data of 10 independent experiments is shown. Stimulated (open histograms) and unstimulated (gray-filled histograms) cells as well as the percentage of proliferating cells are depicted for each condition. (B) Mean percentage ± SEM of proliferating cells after high dose of IL-2 stimulation of 10 independent experiments is shown. P was calculated by Wilcoxon test. (C-D) In vivo proliferation of NK-cell subsets was measured by Ki67 staining of peripheral blood mononuclear cells (PBMCs) derived from healthy donors. (C) One representative donor (top panel) and percentage of Ki67+ cells plus corresponding medians of 22 donors analyzed ex vivo (bottom panel) are shown. (D) Ki67 expression was analyzed directly before (day 0), or after yellow fever virus (YFV) vaccination (days 7 and 28). Analysis was performed after gating on total CD3− CD56+ NK cells (left and middle) or on CD56bright (●), CD56dim CD62L+ ( ) and CD56dim CD62L− (
) and CD56dim CD62L− ( , right). Percentage of Ki67+ cells of 8 donors and corresponding median values (left and right graphs) or mean percentage ± SEM of Ki67+ cells of 3 donors (middle) are depicted. *P < .05; **P < .01; ***P < .0001 as calculated by Wilcoxon test.
, right). Percentage of Ki67+ cells of 8 donors and corresponding median values (left and right graphs) or mean percentage ± SEM of Ki67+ cells of 3 donors (middle) are depicted. *P < .05; **P < .01; ***P < .0001 as calculated by Wilcoxon test.
Signaling downstream of the common γ chain accounts for the better responsiveness of CD56dim CD62L+ NK cells to IL-2 and IL-15 stimulation
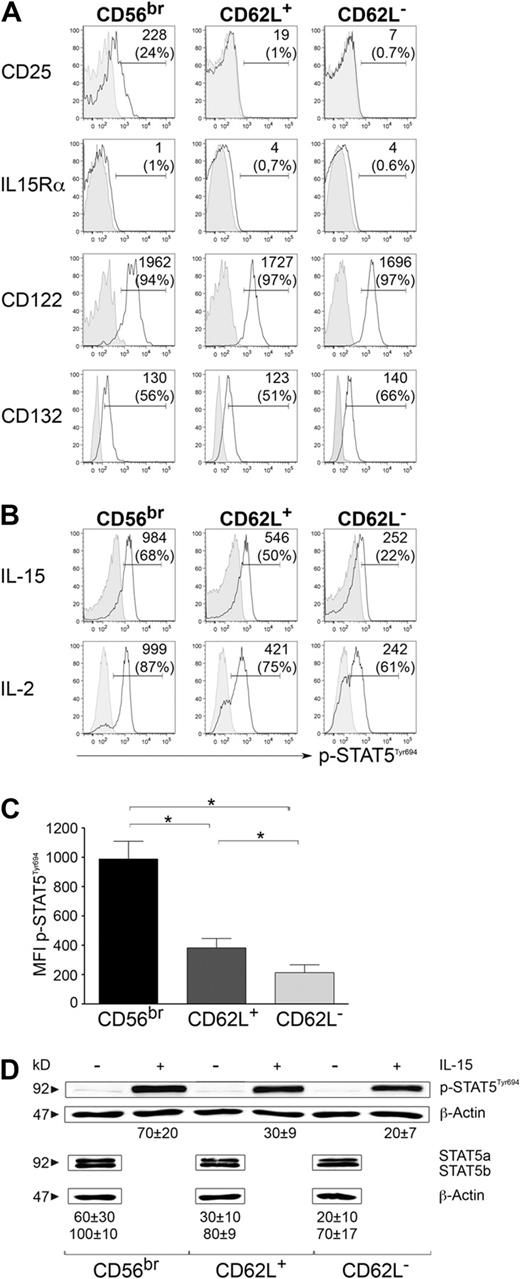
We investigated whether differences in cytokine receptor expression could account for the superior proliferative ability of CD56dim CD62L+ compared with CD62L− NK cells. The expression of IL2R/IL15Rβ chain (CD122), the common γ chain (CD132), and IL15Rα were comparable in the 3 subsets. IL2Rα chain (CD25) was exclusively expressed ex vivo on CD56bright cells, in line with previous observations,9 but not on CD56dim CD62L+ or CD62L− cells (Figure 2A). Therefore, while expression of CD25 on resting CD56bright cells might at least partially explain their higher proliferative ability, differences in cytokine receptor expression could not account for the distinct proliferative behavior of CD56dim CD62L+ and CD62L− cells. For that reason, we analyzed phosphorylation of signal transducer and activator of transcription 5 (STAT5), which is acting downstream of the common γ chain receptor. IL-2 or IL-15 stimulation resulted in extremely higher STAT5 phosphorylation in CD56bright cells compared with both CD56dim CD62L+ and CD62L− NK cells (Figure 2B-D). However, CD56dim CD62L+ cells displayed slightly but significantly higher p-STAT5 levels compared with CD56dim CD62L− cells (Figure 2B-D). Similar tendencies were also observed for total STAT5a and STAT5b levels in unstimulated NK-cell subsets (Figure 2D). These data imply that differences in signaling downstream of the common γ chain, rather than in cytokine receptor expression, account for the better responsiveness of CD56dim CD62L+ NK cells to IL-2 and IL-15 stimulation. However, other mechanisms in addition to p-STAT5 levels must exist to explain their behavior.
Responsiveness to γ-chain cytokines by different NK-cell subsets. (A) Ex vivo staining of CD25 (IL2Rα chain), IL15Rα chain, CD122 (IL15/IL2Rβ chain), and CD132 (common γ chain; open histograms) and corresponding isotype control staining (gray-filled histograms) on CD56bright, CD56dim CD62L+, and CD56dim CD62L− NK cells. Percentages of positive cells and median fluorescence intensity (MFI) values of specific staining minus MFI of isotype staining of 1 representative donor of 3 are depicted. (B-D) Levels of p-STAT5 after stimulation of sorted NK-cell subsets with 100 ng/mL IL-15 or 100 ng/mL IL-2 for 15 minutes. (B) One representative experiment of 6 for IL-15 and of 4 for IL-2 is shown. Stimulated cells (open histograms) and unstimulated controls (gray-filled histograms) are depicted. MFI values of stimulated minus MFI of unstimulated controls and frequencies of p-STAT5+ cells are shown in each plot. (C) Mean value of p-STAT5 MFI ± SEM after IL-15 stimulation of 6 independent experiments is shown. *P < .05 as calculated by Wilcoxon test. (D) Western blot analysis of p-STAT5T694 levels after stimulation of sorted NK-cell subsets with or without 100 ng/mL IL-15 for 15 minutes (top panel) and ex vivo levels of total STAT5a and STAT5b (bottom panel) are shown. One representative experiment of 4 is shown. Numbers under the β-actin bands represent signal intensity displayed by each band normalized to β-actin (mean value ± SEM of 4 independent experiments).
Responsiveness to γ-chain cytokines by different NK-cell subsets. (A) Ex vivo staining of CD25 (IL2Rα chain), IL15Rα chain, CD122 (IL15/IL2Rβ chain), and CD132 (common γ chain; open histograms) and corresponding isotype control staining (gray-filled histograms) on CD56bright, CD56dim CD62L+, and CD56dim CD62L− NK cells. Percentages of positive cells and median fluorescence intensity (MFI) values of specific staining minus MFI of isotype staining of 1 representative donor of 3 are depicted. (B-D) Levels of p-STAT5 after stimulation of sorted NK-cell subsets with 100 ng/mL IL-15 or 100 ng/mL IL-2 for 15 minutes. (B) One representative experiment of 6 for IL-15 and of 4 for IL-2 is shown. Stimulated cells (open histograms) and unstimulated controls (gray-filled histograms) are depicted. MFI values of stimulated minus MFI of unstimulated controls and frequencies of p-STAT5+ cells are shown in each plot. (C) Mean value of p-STAT5 MFI ± SEM after IL-15 stimulation of 6 independent experiments is shown. *P < .05 as calculated by Wilcoxon test. (D) Western blot analysis of p-STAT5T694 levels after stimulation of sorted NK-cell subsets with or without 100 ng/mL IL-15 for 15 minutes (top panel) and ex vivo levels of total STAT5a and STAT5b (bottom panel) are shown. One representative experiment of 4 is shown. Numbers under the β-actin bands represent signal intensity displayed by each band normalized to β-actin (mean value ± SEM of 4 independent experiments).
CD56bright and CD56dim CD62L+ NK cells display similar ability to produce IFN-γ after stimulation with cytokines or DCs
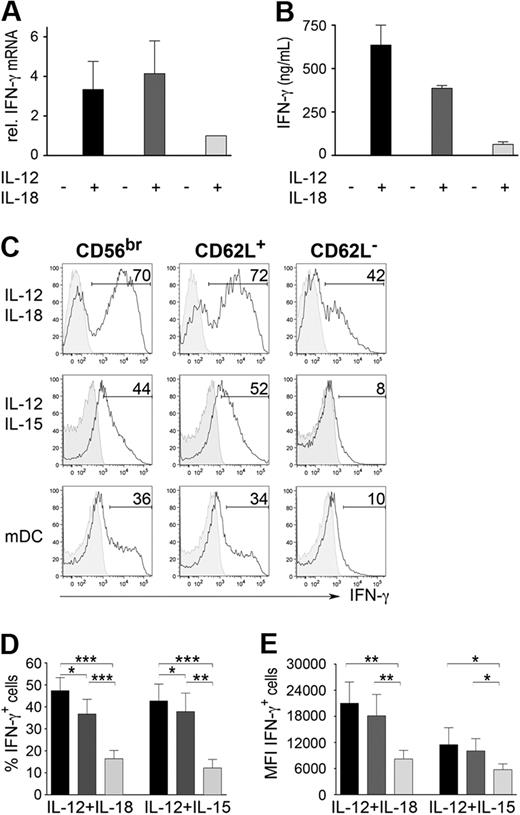
In addition to their high proliferative capacity, CD56bright NK cells are characterized by the higher ability to produce IFN-γ compared with total CD56dim NK cells after stimulation with cytokines or DCs.8 Here, we analyzed whether dissection of CD56dim NK cells into CD62L+ and CD62L− would result in differences concerning IFN-γ production. CD56bright and CD56dim CD62L+ NK cells expressed higher levels of IFN-γ transcripts (Figure 3A) and protein (Figure B-E) compared with CD56dim CD62L− NK cells after cytokine stimulation. Similar higher levels of IFN-γ protein could be observed also after coculture with PB-derived mDCs (Figure 3C). Analysis of intracellular levels of IFN-γ showed that CD56dim CD62L+ NK cells comprised intermediate percentages of IFN-γ secreting cells between CD56bright and CD56dim CD62L− NK cells (Figure 3C-D). Moreover, the amount of IFN-γ produced per cell, as measured by median fluorescence intensity (MFI) of the IFN-γ+ cells, was significantly higher in CD56bright and in CD56dim CD62L+ cells compared with CD62L− NK cells (Figure 3E), thus showing that only the first 2 subsets are skilled IFN-γ producers after cytokine stimulation.
IFN-γ production by NK-cell subsets after cytokine stimulation. Sorted CD56bright (black), CD56dim CD62L+ (dark gray) and CD56dim CD62L− cells (light gray; 106/mL) were stimulated with 50 ng/mL IL12+50 ng/mL IL-18, 50 ng/mL IL-12+50ng/mL IL-15 (A-E) or with PB-derived mDCs in the presence of 100 ng/mL lipopolysaccharide and 10 μg/mL R848 (C). Induction of IFN-γ transcripts by real-time quantitative reverse-transcribed polymerase chain reaction (A) and IFN-γ protein expression by enzyme-linked immunosorbent assay (B) was measured after 16 hours of stimulation. Intracellular staining of IFN-γ was analyzed by FACS (C-E) after 24 hours of stimulation in the presence of BrefeldinA for the final 8 hours. (A) Mean values ± SEM of IFN-γ mRNA fold induction in stimulated CD56bright and CD56dim CD62L+ compared with CD56dim CD62L− cells of 3 independent experiments is shown. (B) Mean levels of IFN-γ protein ± SEM of 3 donors. (C) One representative experiment of 16 (IL-12 + IL-18), 7 (IL-12 + IL-15) or 5 (DCs) is shown. Stimulated (open histograms) and unstimulated (gray-filled histograms) cells as well as percentage of IFN-γ producing cells are depicted for each condition. Mean percentage (D) and mean MFI (E) values ± SEM of IFN-γ expressing cells are shown. *P < .05, **P < .01; ***P < .0001 was calculated by the paired t test.
IFN-γ production by NK-cell subsets after cytokine stimulation. Sorted CD56bright (black), CD56dim CD62L+ (dark gray) and CD56dim CD62L− cells (light gray; 106/mL) were stimulated with 50 ng/mL IL12+50 ng/mL IL-18, 50 ng/mL IL-12+50ng/mL IL-15 (A-E) or with PB-derived mDCs in the presence of 100 ng/mL lipopolysaccharide and 10 μg/mL R848 (C). Induction of IFN-γ transcripts by real-time quantitative reverse-transcribed polymerase chain reaction (A) and IFN-γ protein expression by enzyme-linked immunosorbent assay (B) was measured after 16 hours of stimulation. Intracellular staining of IFN-γ was analyzed by FACS (C-E) after 24 hours of stimulation in the presence of BrefeldinA for the final 8 hours. (A) Mean values ± SEM of IFN-γ mRNA fold induction in stimulated CD56bright and CD56dim CD62L+ compared with CD56dim CD62L− cells of 3 independent experiments is shown. (B) Mean levels of IFN-γ protein ± SEM of 3 donors. (C) One representative experiment of 16 (IL-12 + IL-18), 7 (IL-12 + IL-15) or 5 (DCs) is shown. Stimulated (open histograms) and unstimulated (gray-filled histograms) cells as well as percentage of IFN-γ producing cells are depicted for each condition. Mean percentage (D) and mean MFI (E) values ± SEM of IFN-γ expressing cells are shown. *P < .05, **P < .01; ***P < .0001 was calculated by the paired t test.
Analysis of the impact of CD62L expression versus KIR, NKG2A or CD27 on proliferation and IFN-γ production
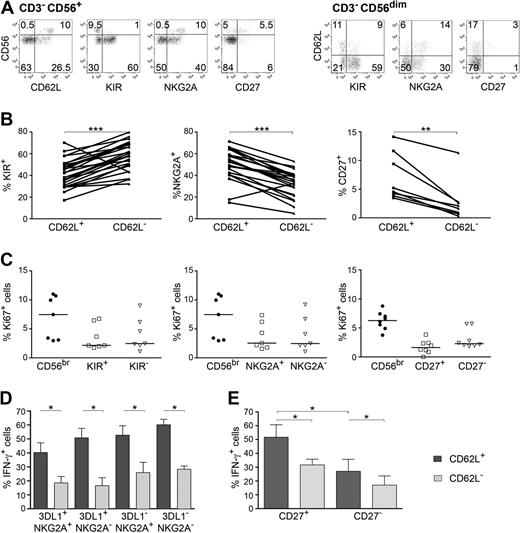
Previous studies have shown that expression of KIR, NKG2A, or CD27 can influence NK-cell proliferative ability as well as their capacity to produce IFN-γ.3,14,20-22 To interpret these apparently confusing results, we first analyzed co-expression of CD62L with KIR, NKG2A, and CD27 within NK cells. Thus, while CD56bright NK cells are mostly CD62L+, KIR−, NKG2A+, and CD27dim, the majority of CD56dim NK cells are CD62L−, KIR+, NKG2A−, and CD27− (Figure 4A). In particular, although CD62L and KIR expression was not mutually exclusive among CD56dim NK cells, the CD56dim CD62L+ subset contained higher percentages of KIR− as well as of NKG2A+ or CD27+ cells in all donors analyzed (Figure 4A-B). For that reason, we tested which of these markers were directly associated to the ability to proliferate in vivo and to efficiently produce IFN-γ after cytokine stimulation. While in vivo proliferating cells were preferentially found within CD56bright and CD56dim CD62L+ subsets (Figure 1C), no significant correlation of Ki67 expression with CD27, KIR, or NKG2A could be detected (Figure 4C). These data suggest that CD62L and not KIR, NKG2A, or CD27 expression identifies NK cells proliferating more extensively in vivo. Next, we evaluated whether the capacity to produce IFN-γ after stimulation with cytokines would also correlate exclusively to CD62L expression, or rather to self MHC-specific inhibitory receptors such as KIR or NKG2A. To this aim, CD56dim NK cells derived from HLA-Bw4 subjects were sorted for the expression of CD62L and NKG2A, stimulated with IL-12+IL-18 and stained for different KIR. IFN-γ expression was analyzed in all sorted subsets after gating on cells single positive for the self MHC (HLA-Bw4)–specific inhibitory receptor KIR3DL1, or cells that were negative for all KIR. Similar to proliferative ability, significantly more IFN-γ producing cells could be detected within CD56dim CD62L+ than within CD62L− cells independently of KIR or NKG2A expression (Figure 4D), thus clearly demonstrating that IFN-γ production in response to cytokines is not influenced by the presence of self MHC-specific inhibitory receptors or NKG2A but rather correlates to CD62L expression. Once we ruled out the possible influence of KIR and NKG2A, we investigated the impact of CD27 expression. Again, we detected a higher enrichment of IFN-γ–secreting cells within the CD56dim CD62L+ subset compared with the CD62L− one, both in CD56dim CD27+ as well as in CD27− cells. Remarkably, the CD56dim CD27+ CD62L+ subset comprises more cells able to produce IFN-γ than the CD27− CD62L+ one, demonstrating that CD56dim CD27+ CD62L+ cells, although representing a very minor population of NK cells (1.8% ± 0.3%), are the most potent IFN-γ producers within CD56dim NK cells (Figure 4E).
Analysis of CD62L, CD27, KIR or NKG2A expression in relation to NK-cell proliferation and IFN-γ production. (A) FACS analysis of PBMCs stained for the indicated markers and gated on CD3− CD56+ NK cells (left) or on CD3− CD56dim NK cells (right); 1 representative donor of 11 is shown. (B) Percentage of 2 + 3D KIR+ (indicated as KIR), NKG2A+ or CD27+ cells within CD56dim CD62L+ or CD56dim CD62L− subsets; 26 (KIR), 22 (NKG2A), and 10 (CD27) different healthy donors are shown. **P < .01; ***P < .0001 as calculated by Wilcoxon test. (C) Ki67 expression in PB-NK cell subsets after staining for CD62L, KIR (2 + 3D KIR), NKG2A and CD27 and gating on CD56bright or the indicated CD56dim subsets; 7 (KIR and NKG2A) or 8 (CD27) donors plus corresponding medians are shown. (D) CD56dim NK cells derived from HLA-Bw4 donors were sorted for NKG2A− CD62L+ or NKG2A− CD62L− cells and stimulated with 50 ng/mL IL-12 + 50 ng/mL IL-18 for 24 hours, in the presence of BrefeldinA for the final 8 hours. After staining for 2D KIR and KIR3DL1 (indicated as 3DL1), IFN-γ expression was analyzed within competent 2D KIR− KIR3DL1+ or hyporesponsive 2D KIR− KIR3DL1− cells. Mean ± SEM of 6 independent experiments is shown. (E) CD56dim NK cells were sorted according to CD27 and CD62L expression and IFN-γ was analyzed after stimulation with IL-12+IL-18. Mean ± SEM of 6 independent experiments is shown. *P < .05 as calculated by Wilcoxon test.
Analysis of CD62L, CD27, KIR or NKG2A expression in relation to NK-cell proliferation and IFN-γ production. (A) FACS analysis of PBMCs stained for the indicated markers and gated on CD3− CD56+ NK cells (left) or on CD3− CD56dim NK cells (right); 1 representative donor of 11 is shown. (B) Percentage of 2 + 3D KIR+ (indicated as KIR), NKG2A+ or CD27+ cells within CD56dim CD62L+ or CD56dim CD62L− subsets; 26 (KIR), 22 (NKG2A), and 10 (CD27) different healthy donors are shown. **P < .01; ***P < .0001 as calculated by Wilcoxon test. (C) Ki67 expression in PB-NK cell subsets after staining for CD62L, KIR (2 + 3D KIR), NKG2A and CD27 and gating on CD56bright or the indicated CD56dim subsets; 7 (KIR and NKG2A) or 8 (CD27) donors plus corresponding medians are shown. (D) CD56dim NK cells derived from HLA-Bw4 donors were sorted for NKG2A− CD62L+ or NKG2A− CD62L− cells and stimulated with 50 ng/mL IL-12 + 50 ng/mL IL-18 for 24 hours, in the presence of BrefeldinA for the final 8 hours. After staining for 2D KIR and KIR3DL1 (indicated as 3DL1), IFN-γ expression was analyzed within competent 2D KIR− KIR3DL1+ or hyporesponsive 2D KIR− KIR3DL1− cells. Mean ± SEM of 6 independent experiments is shown. (E) CD56dim NK cells were sorted according to CD27 and CD62L expression and IFN-γ was analyzed after stimulation with IL-12+IL-18. Mean ± SEM of 6 independent experiments is shown. *P < .05 as calculated by Wilcoxon test.
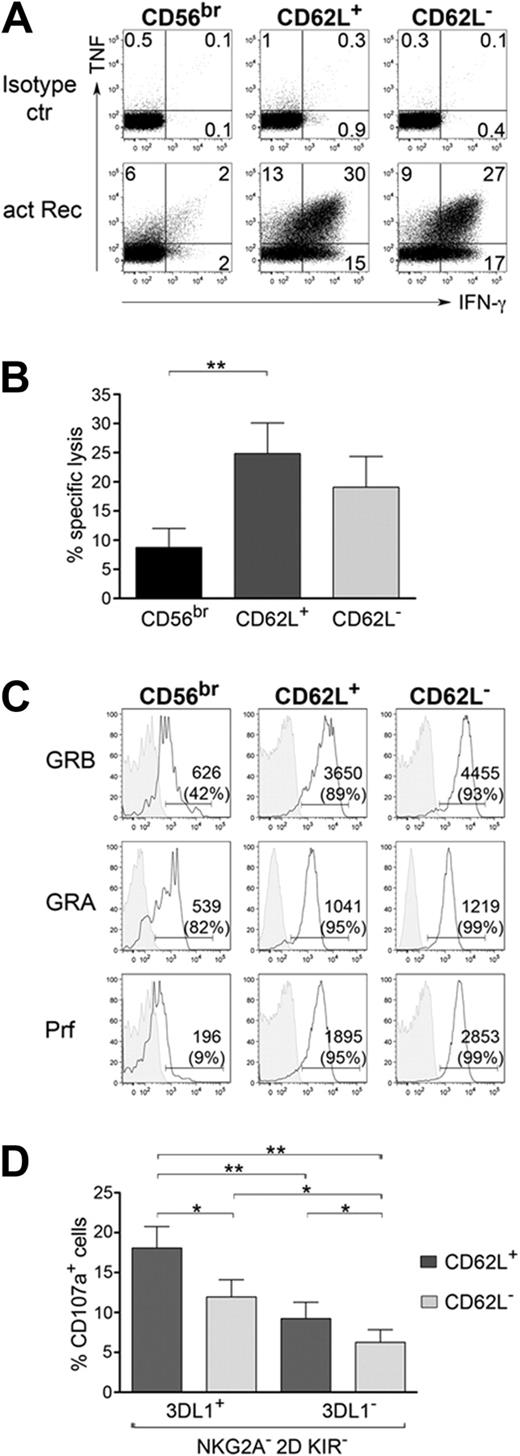
CD56dim CD62L+ and CD56dim CD62L− but not CD56bright NK cells are able to respond to activating receptor stimulation
IFN-γ expression can be induced in NK cells not only by cytokines but also by activating ligands expressed on target cells. Therefore, we analyzed IFN-γ and tumor necrosis factor (TNF) expression after cross-linking of multiple activating receptors in CD56bright, CD56dim CD62L+, and CD62L− cells. Surprisingly, although CD56bright cells are generally considered the most potent cytokine producers, they did not express significant amounts of either IFN-γ or TNF after activating receptor stimulation (Figure 5A). In contrast, both CD56dim cell subsets, CD62L+ and CD62L− cells, were proficient IFN-γ and TNF producers under the same stimuli (Figure 5A). Similarly, cross-linking of FcγRIII (CD16; supplemental Figure 2A) resulted in efficient IFN-γ production only in CD56dim CD62L+ and CD62L− subsets but not in CD56bright NK cells. These data show that CD56bright are not always the cytokine producers and that, during differentiation from CD56bright to CD56dim, NK cells lose the ability to produce cytokines after cytokine stimulation but acquire the capacity to express them after activating receptor stimulation. Comparative analysis of cytotoxicity revealed that CD56dim CD62L+ and CD62L− cells were both able to kill MHC class I negative tumor target cells K562, while CD56bright cells displayed much lower cytotoxic ability (Figure 5B), in line with previous observations.13 Indeed, although CD56dim CD62L+ NK cells display many functional and phenotypic features in common with CD56bright, cytotoxic ability after K562 (Figure 5B) or CD16 cross-linking (supplemental Figure 2A), as well as ex vivo expression of perforin, granzymeA, granzymeB (Figure 5C), and CD16 (supplemental Figure 2B) were high in both CD56dim CD62L+ and CD62L− NK-cell subsets. Because cytotoxic competence correlates to the presence of self MHC-specific inhibitory receptors, we investigated in more detail whether CD62L expression can influence NK-cell cytotoxicity within CD56dim KIR3DL1+ (competent) or CD56dim KIR− NKG2A− (hyporesponsive) cells in HLA-Bw4 donors. In line with previous data,3,18 expression of self MHC-specific KIR such as KIR3DL1 results in the most striking correlation with cytotoxic competence (Figure 5D). However, expression of CD62L confers a slightly higher cytotoxic potential to both, competent (KIR3DL1+) as well as hyporesponsive (KIR− NKG2A−) NK cells, which may reflect recent in vivo activation of CD56dim CD62L+ NK cells by cytokines. We observed the same slight difference in cytotoxic competence between CD62L+ and CD62L− cells, also when analyzing hyporesponsive KIR3DL1+ cells in HLA-Bw6 donors (supplemental Figure 2C). This small difference in cytotoxic competence between CD56dim CD62L+ and CD62L− NK-cell subsets was actually comparable or a little higher than the one between NKG2A+ and NKG2A− cells and lower than the effect associated to expression of self MHC specific KIR (supplemental Figure 2D). Interestingly, within hyporesponsive NK cells (KIR− NKG2A−: Figure 5D, or KIR3DL1+ cells in HLA-Bw6 donors: supplemental Figure 2C), potentially autoreactive ones are almost exclusively included within the CD62L+ subset. It has been previously shown that CD56dim KIR− (hyporesponsive) NK cells can up-regulate KIR after cytokine stimulation14,23 and that only those cells that de novo express self MHC-specific KIR are licensed to kill.29 Percentage of KIR up-regulation on proliferating cells is comparable between KIR− CD62L+ and KIR− CD62L− NK cells (supplemental Figure 2E). However, due to their extensive proliferation, a consistently higher proportion of KIR+ cells were generated from CD56dim CD62L+ compared with CD62L− NK cells, suggesting that cells endowed with strong proliferative ability might have a higher chance to be licensed.
Analysis of cytokine production and cytotoxicity in different NK-cell subsets after activating receptor stimulation. (A) Analysis of intracellular IFN-γ and TNF expression in CD56bright, CD56dim CD62L+, and CD56dim CD62L− NK cells after stimulation with a combination of plate-bound mAbs against NKp30, NKp46, NKG2D, 2B4, and CD2 or isotype control mAb; 1 representative experiment of 6 is shown. (B) Analysis of NK-cell subset cytotoxicity after K562 stimulation; mean ± SEM of 9 independent experiments is shown. (C) Ex vivo staining of granzymeA (GRA), granzymeB (GRB), perforin (Prf; open histograms), and of corresponding isotype control (gray-filled histograms) in CD56bright, CD56dim CD62L+, and CD56dim CD62L− NK cells; 1 representative donor of 3 is shown. MFI and percentage of positive cells is depicted. (D) CD56dim NK cells derived from HLA-Bw4 donors were sorted for NKG2A− CD62L+ and NKG2A− CD62L− cells and stimulated for 6 hours with K562. CD107a expression was analyzed within competent KIR3DL1+ 2D KIR− NKG2A− and hyporesponsive KIR3DL1− 2D KIR− NKG2A− NK-cell subsets after staining for KIR3DL1 (indicated as 3DL1) and 2D KIR and gating on the indicated subsets. Mean ± SEM of 8 independent experiments is shown. *P < .05; **P < .01; ***P < .0001 as calculated by Wilcoxon test.
Analysis of cytokine production and cytotoxicity in different NK-cell subsets after activating receptor stimulation. (A) Analysis of intracellular IFN-γ and TNF expression in CD56bright, CD56dim CD62L+, and CD56dim CD62L− NK cells after stimulation with a combination of plate-bound mAbs against NKp30, NKp46, NKG2D, 2B4, and CD2 or isotype control mAb; 1 representative experiment of 6 is shown. (B) Analysis of NK-cell subset cytotoxicity after K562 stimulation; mean ± SEM of 9 independent experiments is shown. (C) Ex vivo staining of granzymeA (GRA), granzymeB (GRB), perforin (Prf; open histograms), and of corresponding isotype control (gray-filled histograms) in CD56bright, CD56dim CD62L+, and CD56dim CD62L− NK cells; 1 representative donor of 3 is shown. MFI and percentage of positive cells is depicted. (D) CD56dim NK cells derived from HLA-Bw4 donors were sorted for NKG2A− CD62L+ and NKG2A− CD62L− cells and stimulated for 6 hours with K562. CD107a expression was analyzed within competent KIR3DL1+ 2D KIR− NKG2A− and hyporesponsive KIR3DL1− 2D KIR− NKG2A− NK-cell subsets after staining for KIR3DL1 (indicated as 3DL1) and 2D KIR and gating on the indicated subsets. Mean ± SEM of 8 independent experiments is shown. *P < .05; **P < .01; ***P < .0001 as calculated by Wilcoxon test.
Analysis of lymph node NK cells and chemokine receptor expression
CD56bright and CD56dim NK cells differ also for their main location, being CD56bright mainly found in secondary lymphoid organs. Indeed, CD56bright NK cells are CD62L+ CCR7+ while CD56dim cells are mostly CCR7−, but express CXCR1, which enables them to migrate into inflamed tissues.30 A small fraction of CD56dim CD62L+ cells were CCR7+ CXCR1− and a consistent proportion of them coexpressed CD27 (supplemental Figure 3A). Short IL-18 stimulation is able to induce CCR7 expression in CD56dim cells.31 CCR7 up-regulation after IL-18 stimulation occurred almost exclusively within CD56dim CD62L+ cells (supplemental Figure 3B). Analysis of lymph node (LN) showed that CD56dim cells, as defined by expression of CD16, could be found only in inflamed but not in nonreactive LN, which are conversely enriched in CD56bright NK cells, as previously shown.12,14,32 In inflamed LN, both CD56dim CD62L+ and CD62L− cells could be found (supplemental Figure 3C), although it is hard to conclude whether these cells were recruited or actually differentiating in situ from resident CD56bright NK cells in the course of the inflammatory response.
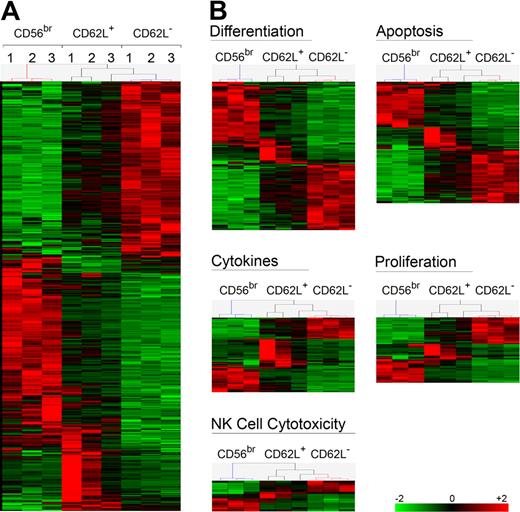
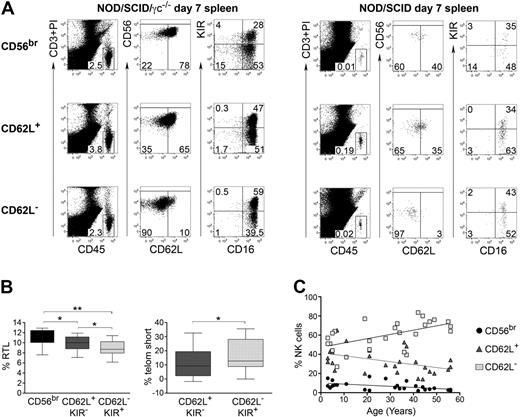
CD56dim CD62L+ NK cells have an intermediate phenotype between CD56bright and CD56dim CD62L− NK cells
CD56dim CD62L+ NK cells exhibit intermediate functions between CD56bright and CD56dim CD62L− cells. To have a global overview, we have performed transcriptome analysis of the 3 NK-cell subsets. As shown in Figure 6A, CD56dim CD62L+ NK cells displayed an intermediate signature between CD56bright and CD56dim CD62L− cells in all donors analyzed. This signature remained consistent when transcripts were grouped in different function-related clusters, such as cytotoxicity, proliferation, apoptosis, or cytokine related genes (Figure 6B). Based on their transcriptome signature as well as functional properties, we speculate that CD56dim CD62L+ NK cells could represent an intermediate step of NK-cell differentiation between CD56bright and CD56dim CD62L− cells. To test this hypothesis, we sorted and either transferred CD56bright, CD56dim CD62L+, or CD56dim CD62L− NK cells into NOD/SCID or NOD/SCID/γc−/− immunodeficient mice. NOD/SCID/γc−/− mice were treated with IL-15Rα+IL-15 to increase human NK-cell recovery, as previously published.17 Human CD45+ CD56+ CD3− NK cells could be found 7 days after transfer in the spleen of NOD/SCID and in higher amounts in NOD/SCID/γc−/− mice treated with IL-15Rα+IL-15 (Figure 7A). In line with previous publications,15,17 CD56bright acquired CD16 and KIR expression, thus confirming the proposed CD56bright differentiation into CD56dim NK cells in vivo. Importantly, a proportion of transferred CD56bright as well as of CD62L+ CD56dim NK cells down-regulated CD62L in vivo (38% ± 13% and 35% ± 9%, respectively), while CD62L up-regulation occurred only on a minority of transferred CD56dim CD62L− (5% ± 1.6%). To further support our hypothesis, we measured telomere length in different NK-cell subsets. Comparative analysis of CD62L+ and CD62L− or of KIR+ and KIR− CD56dim NK cells revealed no significant difference in telomere length (K.J., B.M., G.F., C.R., unpublished data, 2010). However, telomere length analysis of cells sorted according to CD62L and KIR revealed that CD56dim CD62L+ KIR− displayed intermediate telomere length between CD56bright and CD56dim CD62L− KIR+ NK cells (Figure 7B). It has been demonstrated that NK cells obtained from elderly people display increased frequencies of CD56dim and decreased frequencies of CD56bright cells,33 suggesting that terminally differentiated NK cells accumulate with aging. When we analyzed the percentage of CD56bright, CD56dim CD62L+ and CD56dim CD62L− NK cells in correlation to age, we found that frequencies of CD56bright as well as of CD56dim CD62L+ NK cells progressively decreased with aging, while CD56dim CD62L− cells increased (Figure 7C). Altogether, these data suggest that CD56dim CD62L+ NK cells might be enriched in cells displaying an intermediate step of maturation between CD56bright and CD56dim CD62L− NK cells.
Gene profile analysis of different NK cell subsets. Gene expression profiling of sorted NK-cell subsets (n = 3 for each of 3 subsets) identified 8003 probe sets in total, using High Performance Chip Data Analysis with Bioretis database as described in supplemental Methods. Hierarchical clustering was performed with Genes@Work with Pearson correlation and center of mass. (A) All 1816 differentially expressed probe sets between CD56dim CD62L+ and CD62L− are shown. (B) Five functional groups of probe sets were generated with Database for Annotation, Visualization and Integrated Discovery (DAVID) from significantly differentially regulated probe sets between CD56dim CD62L+ and CD62L− NK-cell subsets.
Gene profile analysis of different NK cell subsets. Gene expression profiling of sorted NK-cell subsets (n = 3 for each of 3 subsets) identified 8003 probe sets in total, using High Performance Chip Data Analysis with Bioretis database as described in supplemental Methods. Hierarchical clustering was performed with Genes@Work with Pearson correlation and center of mass. (A) All 1816 differentially expressed probe sets between CD56dim CD62L+ and CD62L− are shown. (B) Five functional groups of probe sets were generated with Database for Annotation, Visualization and Integrated Discovery (DAVID) from significantly differentially regulated probe sets between CD56dim CD62L+ and CD62L− NK-cell subsets.
CD56dim CD62L+ (KIR−) NK cells partially lose CD62L expression in vivo, have intermediate telomere length and decrease with aging. (A) FACS analysis of cells recovered from the spleen of immunodeficient mice (NOD/SCID/γc−/− mice, left and NOD/SCID, right), 7 days after transfer of human CD56bright CD62L+, CD56dim CD62L+ or CD56dim CD62L− cells, which have been sorted with high purity. NOD/SCIDγc−/− mice were treated with IL-15+IL-15RαFc. Human NK cells were identified as hCD45+ CD3− CD56+. Percentage of expression of CD56, CD62L, 2 + 3D KIR (indicated as KIR) and CD16 on recovered NK cells is shown in each quadrant. One representative experiment performed with both mouse strains of 3 independent experiments is shown. (B) Telomere length analysis of CD56bright, CD56dim CD62L+ KIR− and CD56dim CD62L− KIR+ NK-cell subsets; box plots with median percentage of relative telomere length (RTL, see supplemental Methods) plus interquartile range of 9 different donors is shown in the left graph; median percentage of telomere shortening (telom short) of CD56dim CD62L+ KIR− or of CD56dim CD62L− KIR+ cells relative to CD56bright ones is shown on the right. *P < .05; ** P < .01 as calculated by the paired Student t test. (C) Analysis of percentage of CD56bright (r = −0.56, P = .003), CD56dim CD62L+ (r = −0.44, P = .02), and CD56dim CD62L− (r = 0.56, P = .003) in correlation to age; Pearson correlation coefficient of 26 donors has been calculated.
CD56dim CD62L+ (KIR−) NK cells partially lose CD62L expression in vivo, have intermediate telomere length and decrease with aging. (A) FACS analysis of cells recovered from the spleen of immunodeficient mice (NOD/SCID/γc−/− mice, left and NOD/SCID, right), 7 days after transfer of human CD56bright CD62L+, CD56dim CD62L+ or CD56dim CD62L− cells, which have been sorted with high purity. NOD/SCIDγc−/− mice were treated with IL-15+IL-15RαFc. Human NK cells were identified as hCD45+ CD3− CD56+. Percentage of expression of CD56, CD62L, 2 + 3D KIR (indicated as KIR) and CD16 on recovered NK cells is shown in each quadrant. One representative experiment performed with both mouse strains of 3 independent experiments is shown. (B) Telomere length analysis of CD56bright, CD56dim CD62L+ KIR− and CD56dim CD62L− KIR+ NK-cell subsets; box plots with median percentage of relative telomere length (RTL, see supplemental Methods) plus interquartile range of 9 different donors is shown in the left graph; median percentage of telomere shortening (telom short) of CD56dim CD62L+ KIR− or of CD56dim CD62L− KIR+ cells relative to CD56bright ones is shown on the right. *P < .05; ** P < .01 as calculated by the paired Student t test. (C) Analysis of percentage of CD56bright (r = −0.56, P = .003), CD56dim CD62L+ (r = −0.44, P = .02), and CD56dim CD62L− (r = 0.56, P = .003) in correlation to age; Pearson correlation coefficient of 26 donors has been calculated.
Discussion
In our study we characterized unexplored ex vivo and in vivo properties of CD56bright and CD56dim NK cells and we identified a subset of polyfunctional NK cells according to the expression of the LN homing marker CD62L. In fact, we showed that CD56dim CD62L+ NK cells represent the only human NK-cell subset endowed with full effector functions after stimulation with cytokines or via activating receptors as well as with high self renewable ability. We show for the first time that not only CD56bright, but also a subset of CD56dim cells, proficiently respond to cytokine stimulation. The analysis of NK-cell proliferation and IFN-γ production in response to cytokine stimulation, dissecting for CD62L, KIR, NKG2A and CD27 expression, clearly showed that, in contrast to cytotoxic competence, cytokine responsiveness does not correlates with self MHC inhibitory receptors, but rather with CD62L expression. Previous data showing a correlation of KIR and/or NKG2A or CD94 expression with the ability to proliferate or produce IFN-γ after cytokine stimulation3,14,19,22 might be a consequence of enrichment of CD62L+ NK cells in these populations. Moreover, because even short-time cytokine stimulation modulates NKG2A and CD62L expression, cells should be sorted to clearly identify markers correlating with different properties ex vivo. Although CD56bright have been long considered the most potent cytokine producers, these cells were surprisingly almost unable to express IFN-γ and TNF after activating receptor stimulation, while CD56dim CD62L+ as well as CD62L− cells were conversely very efficient. This inability of CD56bright to respond to activating receptor stimulation did not correlate to the surface expression of such receptors. Indeed, most of NK-cell activating receptors cross-linked in our experimental setting are even higher expressed on CD56bright than on CD56dim NK cells.7,8 The molecular mechanisms underlying this phenomenon are currently under investigation in our laboratory. In line with cytokine expression after activating receptor stimulation, cytotoxicity against MHC class I− tumor target cells was very poor in CD56bright but comparably high in both CD62L+ and CD62L− CD56dim cells. Altogether, these data suggest that to acquire the ability to respond to activating ligands, NK cells need to be primed, as it has been proposed in mice.4 Conversely, the ability to proliferate and display effector functions after cytokine stimulation seems to be a property of NK cells at early stage of differentiation. Priming of CD56bright NK cells (probably by DC-derived cytokines, such as IL-15) would induce their differentiation into CD56dim cells, and this step could be critical to acquire the ability to respond to activating ligands, as well as the expression of cytotoxic effector molecules such as perforin, granzymes and CD16. KIR acquisition represents a crucial step during NK-cell differentiation from CD56bright to CD56dim cells, because the ability to respond to activating ligands correlated most strikingly with the expression of self MHC-specific KIR, in line with previous results.3,18 Conversely, CD62L or NKG2A expression had just minor enhancing impact, probably reflecting recent in vivo priming by cytokines. The influence of CD27 expression on cytotoxic capacity of NK cells is controversially debated both in humans and in mice.20,34,35 Data are difficult to interpret because these studies did not take into account the influence of self MHC-specific inhibitory receptors.20,34 To assess this issue, analysis of cytotoxicity should be performed on CD27+ and CD27− cells expressing or not self MHC-specific inhibitory receptors. However, due to the extremely low number of CD27+ KIR+ cells, this experiment is rather challenging, at least in humans. KLRG1 has been associated to a mature NK-cell phenotype in mouse.36 Although, we could confirm that KLRG1 is not expressed on CD56bright NK cells, as previously shown,37 we could not observe any correlation of KLRG1 with CD62L expression (supplemental Figure 4). Based on the results obtained in our study, it seems that the ability to proliferate and to produce IFN-γ in response to cytokines is clearly dissociated from the ability to kill and to produce IFN-γ after activating receptor stimulation in most NK cells with the exception of CD56dim CD62L+ cells, which therefore represent a unique polyfunctional subset. Recent studies suggested that NK cells primed during viral infections or by cytokines may exhibit memory-like properties during a recall viral response or ex vivo restimulation.5,6 Due to their peculiar ex vivo properties, CD56dim CD62L+ subset might be able to respond and expand rapidly after restimulation in a memory like fashion and to develop further into terminally differentiated effectors. Although in the human system it is difficult to provide data sustaining this hypothesis, transfer of human NK-cell subsets into immunodeficient mice as well as analysis of their telomere length at least supported the idea that CD56dim CD62L+ KIR− NK cells may represent an intermediate step of maturation between CD56bright and CD56dim CD62L− KIR+ ones. This concept would be in line with the intermediate phenotype observed in the transcriptome as well as in functional analysis. Our in vivo experiments showed that CD56bright acquired CD16 and KIR in line with previous data, proposing a model of linear NK-cell differentiation: CD56bright →CD56dim KIR− →CD56dim KIR− cells.17 Importantly, together with KIR acquisition, a fraction of CD56bright as well as of CD56dim CD62L+ cells lost CD62L expression. Because loss of CD62L and acquisition of KIR expression appear to be independent events, CD56dim CD62L+ KIR+ or CD62L− KIR− cells also exist, and differences in their telomere length may not be substantial. On the other hand, CD62L+ KIR+ cells may also represent a heterogeneous cell population, including a contamination of “revertants,” which have reacquired CD62L expression. Thus, CD62L could be up-regulated at least in vitro under selected stimuli14 and in a minor fraction of CD62L− cells in vivo. The idea that CD56dim CD62L− cells might be enriched in terminally differentiated cells fits also to the observation that those cells accumulated with age, while frequencies of CD56bright and CD56dim CD62L+ cells decreased. Our data are also in line with results obtained in a recent study describing NK-cell reconstitution in patients undergoing hematopoietic stem cell transplantation (HSCT).38 Indeed, PB-NK cells reconstituting in patients 30 days after HSCT were enriched in CD56bright cells, as previously shown,39 and almost all CD56dim NK cells expressed CD62L. Percentages of CD56dim CD62L+ NK cells progressively decreased with time and became comparable with those of healthy blood donors only around 100 days after HSCT.38 Our study provides further insights into differentiation history of human NK cells and identifies a unique subset of polyfunctional NK cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Toralf Kaiser, Katharina Raba, and Jenny Kirsch for cell sorting; Heidi Schliemann and Sabine Brösel for excellent technical assistance; Jens Geginat, Christian Münz, and Paola Romagnani for critical discussion; and Dörte Huscher and Anja Weiss for statistical advice.
This work was supported by the Deutsche Forschungsgemeinschaft Grant SFB 650 and TR36.
Authorship
Contribution: K.J. designed and performed experiments, analyzed results, and wrote the manuscript; M.K., M.L.-E., E.P., B.M., G.F., and I.S.-K. performed experiments and analyzed results; J.G. analyzed the microarray data; A.T. interpreted data; and C.R. designed the research, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Chiara Romagnani, Deutsches Rheuma Forschungszentrum, Charitéplatz 1, 10117 Berlin, Germany; e-mail: romagnani@drfz.de.
References
Author notes
M.K. and M.L.-E. contributed equally to this work.