Abstract
B-cell receptor (BCR) diversity is achieved centrally by rearrangement of Variable, Diversity, and Joining genes, and peripherally by somatic hypermutation and class-switching of the rearranged genes. Peripheral B-cell populations are subject to both negative and positive selection events in the course of their development that have the potential to shape the BCR repertoire. The origin of IgM+IgD+CD27+ (IgM memory) cells is controversial. It has been suggested that they may be a prediversified, antigen-independent, population of cells or that they are a population of cells that develop in response to T-independent antigens. Most recently, it was suggested that the majority of IgM memory cells are directly related to switched memory cells and are early emigrants from the germinal center reaction. Advances in sequencing technology have enabled us to undertake large scale IGH repertoire analysis of transitional, naive, IgM memory and switched memory B-cell populations. We find that the memory B-cell repertoires differ from the transitional and naive repertoires, and that the IgM memory repertoire is distinct from that of class-switched memory. Thus we conclude that a large proportion of IgM memory cells develop in response to different stimuli than for class-switched memory cell development.
Introduction
B-cell development in the periphery is a crucial process in the humoral immune response, where the immunoglobulin (Ig) gene repertoire is changed by processes of somatic hypermutation (SHM), class-switching (CSR), and selection in response to stimulation. Thus, hypermutated and class-switched Ig genes are characteristic of memory B cells along with loss of IgD expression, and gain of the activation marker CD27. It was originally thought that CSR and SHM were 2 interlinked processes in the germinal center (GC). However, the discovery of hypermutated IgM marginal zone B cells in the spleen and the IgM memory B cells in the peripheral blood suggest that the 2 processes could be separated.1,2 The existence of such IgM memory cells, that in the blood are IgM+, IgD+, CD27+, has been the cause of much debate about the peripheral B-cell development process.3,4 IgM memory cells may represent early emigrants from a classical T-dependent (TD) GC reaction, because SHM has been shown to precede CSR in the GC.5 Alternatively, some GC reactions, such as the GCs formed in response to T-independent (TI) antigen,6,7 may proceed without significant CSR events. It is thought that the splenic marginal zone and IgM memory cells are equivalent populations of cells in humans and are important in responses to TI antigens.8,9 IgM memory cells play a key role in the protection of people against encapsulated bacteria, such as Streptococcus.10
There are also several lines of evidence to suggest that SHM events need not be confined to the GC and that IgM memory cells arise from GC-independent events.4 Mice deficient in Bcl6, lymphotoxin α, or CD28 are unable to properly form GCs but can still produce both memory cells and cells with hypermutated Ig genes, albeit at much lower frequencies than normal.11-13 Similarly, low levels of hypermutated IgM memory cells can be found in human immunodeficiency states where GC formation is thought to be completely absent.8 Activation-induced cytidine deaminase (AID) is necessary for SHM and CSR14 and was thought to be confined to expression in GC B cells. However, there are reports that AID can be found in B cells outside the GC,15,16 which also calls into question the requirement for GC involvement in SHM. In view of the association of IgM memory cells with TI responses, and because classical GC formation is associated with TD responses, it has been suggested that IgM memory cells arise through extrafollicular SHM in TI responses.9 An alternative suggestion is that IgM memory B cells are the result of antigen-independent diversification, such as occurs in chicken, sheep, and rabbits. Compelling evidence for this is the existence of a small population of IgM+IgD+CD27+ B cells with mutated Ig genes in cord blood17 and in fetal tissues.18 AID transcripts have been found in human transitional cord blood B cells,19 and stimulation of these cells with the Toll-like receptor ligand CpG can result in the expression of AID and BLIMP-1, acquisition of an IgM memory phenotype, and the production of antibodies with antipneumococcal specificity.10 However, it was recently found that IgM memory cells can have mutations in BCL6 that are characteristic of B cells from GCs, and that some IgM memory cells are related to IgG+ cells in a manner that suggests the development of the latter from the former,20 thus providing convincing evidence that some IgM memory cells are the product of a GC reaction prior to class-switching. These authors suggest that numbers of IgM memory cells and IgG memory cells are equivalent, and therefore finding related IgM+ and IgG+ cells that are also roughly equivalent in number indicates that the majority of IgM memory cells are formed in GC reactions.
Selective processes are key in shaping the B-cell repertoire, and the influence of selection is of concern in a number of fields of study. An examination of specificities in the repertoire of different B-cell populations indicates that negative selection checkpoints exist in the transitional and IgM memory populations to control the numbers of cells with potential autoimmune specificities.21 Conventional positive selection pressures influence the shape of the repertoire, where antigen-specific cells are expanded in a classical adaptive response to antigen challenge. Clonal expansions are identifiable by the presence of cells with identical V-D-J (Variable, Diversity, and Joining) junctional sequences, because this complementarity-determining region (CDR) 3 is unique to each different Ig gene rearrangement and is a crucial part of the antigen binding region. Expansions of B cells with a particular IGHV gene, independent of the specificity conferred by the complementarity determining regions of the gene, may occur. For example, it has been suggested that N-glycosylation of IGHV regions such as IGHV4-34 might confer a selection advantage via interactions of the glycosylated BCR with mannose binding lectins in the GC and thus help account for the prevalence of IGHV4-34 usage in follicular lymphoma.22,23
Here we use deep sequencing technologies to study human B-cell Ig heavy chain repertoires, and have compared the characteristics of transitional, naive, IgM memory, and switched memory B cells. There are some small differences between transitional and naive cells, but the most significant changes in repertoire occur between naive and memory populations. Crucially, we report evidence for highly significant differences in repertoire between switched and IgM memory, indicating that a large proportion of IgM memory B cells are not derived from the same developmental pathway as switched memory.
Methods
B-cell isolation and cell sorting
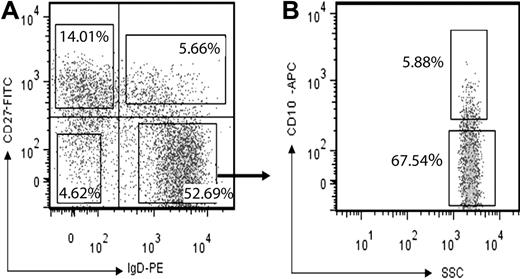
Peripheral blood mononuclear cells were isolated from 3 young, healthy volunteers (21 to 26 years; written consent was obtained in accordance with the Declaration of Helsinki after approval from the Guy's Hospital research ethics committee), using Ficoll-Paque Plus (GE Healthcare) and Leucosep tubes (Greiner Bio-One Ltd). CD19+ B cells were then positively selected from peripheral blood mononuclear cells, using the MACS B cell Isolation Kit (Miltenyi Biotec), stained with CD27-FITC, CD10-APC (Miltenyi Biotec) and IgD-PE (BD Biosciences PharMingen) at 4°C (15 minutes), and analyzed on a FACSAria machine (BD Biosciences PharMingen). Five subsets (Figure 1) were separately collected into 180 μL of Sort-Lysis RT buffer (SLyRT). SLyRT comprises 150 ng/μL pd(N)6 (Invitrogen), 2.5 U/μL RNAse inhibitor (Bioline), 0.13% Triton X-100 (Sigma-Aldrich), 12.5mM DTT and 500μM deoxyribonucleotide triphosphate (dNTP) mix (Promega) in 1× First-Strand RT buffer (Invitrogen) final concentration (ie, in 200 μL). The estimated numbers of cells used to generate the sequences for each sample are given in supplemental Table 2 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
cDNA synthesis and Ig PCR
To synthesise cDNA, 500U SuperScript III reverse transcriptase (RT; Invitrogen) in 20 μL were added to 180 μL of cells in SLyRT buffer. RT was performed at: 42°C (10 minutes), 25°C (10 minutes), 50°C (60 minutes), and 72°C (15 minutes). Ig genes were amplified using a seminested, isotype-specific, polymerase chain reaction (PCR): PCR1 & 2. IgG and IgA reactions were performed for the IgD− populations only. In PCR1, a 25-μL reaction mix contained 6.25 μL of cDNA, 0.625 U Phusion DNA polymerase (NEB), 200μM each dNTPs, 41.75nM each upstream IGHV1-6 primers, 250nM downstream primers (either CHA″, CHG″ or CHM″ for IgA, IgG, and IgM, respectively) in 1× reaction buffer. After a hotstart at 98°C (for 30 seconds, hold at 50°C) Phusion DNA polymerase was added, followed by 15 cycles of 98°C, (10 seconds); 58°C (15 seconds); 72°C (30 seconds), and 1 cycle of 72°C (5 minutes). PCR2 was then used to amplify from PCR1 products using primers having 10-base multiplex-identifier (MID) tails. MID tags enable 12 different samples to be pooled into 1 sequencing sample for more cost-efficient results. The individual experimental samples can later be separated by sequence analysis of the MID tags. Twenty microliters of PCR2 reaction mix contained 2 μL of PCR1 products, 0.5U Phusion DNA polymerase, 200μM each dNTPs, 41.75nM each upstream MID: IGHV 1-6 primers, 250nM downstream primers (MID: CHA, MID: CHG or MID: CHM for IgA, IgG, and IgM, respectively) in 1× reaction buffer. PCR2 were performed at: 98°C for (30 seconds), 20 cycles of 98°C (10 seconds); 58°C (15 seconds); 72°C (30 seconds), and 1 cycle of 72°C (5 minutes). Primer sequences are given in supplemental Table 1. A single control Ig gene of known sequence was also included, using the same PCR conditions but with IGHJ region primers, to evaluate the error rate for the method.
Preparation of MID PCR products for deep sequencing
To produce sufficient DNA for sequencing, while minimising PCR amplification, repeats of PCR1 (×8) and PCR2 (×2) were performed for each individual experimental sample. PCR Primers were removed from the pooled (×16) products by electrophoresis and using QIAquick Gel Purification Kit. Samples to be pooled for sequencing were mixed in equal quantities and concentrated using QIAquick PCR Purification Kit (QIAGEN) before sequencing on the GS FLX Titanium Sequencer (Agowa GmBH). Accuracy of the method as a whole was determined using analysis of results from the control Ig gene and was less than 1 error per 300 bp, or 1 per 1300 bp if indels (a known issue with the sequencing platform) were discounted.
Sequence analysis
Sequences were assigned to the corresponding samples based on the terminal MID sequence. Sequences that contained a second MID sequence that was either different, or located internally, were excluded. Sequences were then assigned an isotype (IgA, IgM, IgG) and V gene by use of the original PCR primer sequences and additional isotype motifs. Using this information, a series of stringent quality control criteria were applied to exclude biologically implausible sequences. Sequences with an isotype primer at both ends, or with a mismatch between an internal and primer-based isotype assignment, were excluded, as were sequences with multiple V gene primer motifs. Sequences that had terminal isotype and V gene primer motifs but were too short to be biologically plausible (IgM:V < 333nt; IgA:V < 389nt; IgG:V < 409nt) were also excluded.
Sequences that passed the quality control criteria but whose reads were too short to span the CDR3 region were excluded as uninformative. For the remaining sequences, the amino acid sequence of each CDR3 junction region was determined using V-QUEST,24 allowing for indels. The physicochemical properties of the peptide between the conserved first and last amino acid positions were determined using ProtParam.25
Data were combined and subsequent analyses performed in Excel (Microsoft). Clonally related sequences were identified by sorting based on their CDR3 amino acid sequences with IGHV and IGHJ gene use as secondary identifiers. Proportions were compared using χ2 tests and continuous variables (CDR3 characteristics) were compared using Mann-Whitney U test.
Results
Ig repertoire clonality in different populations
The CD27+ memory cells, with hypermutated Ig genes, can be divided into 4 populations; the classical IgD- switched memory population, the rare IgM+ IgD− and IgM− IgD+ populations and the IgM+IgD+ population (also known as IgM memory cells). The IgD+ CD27− population is divided into the transitional (CD10+) and naive populations (CD10−). We used IgD, CD27 and CD10 to sort cells into transitional, naive, switched memory, and IgM memory populations (Figure 1). The numbers of cells used to produce the Ig genes for sequencing, the efficiency of sequencing and the number of sequences obtained from each sample are given in supplemental Table 2. The different isotypes in the IgD negative populations were distinguished using constant region-specific primers.
Sorting human B-cell subsets. (A) Human peripheral B cells (CD19+) were stained and sorted into 4 populations, according to their expression of CD27 and IgD: IgD+CD27– (naive and transitional), IgD+CD27+ (IgM memory), IgD–CD27+, and IgD–CD27– B cells. Numbers indicate the percentage of subsets in boxes. (B) From the IgD+CD27– gate, naive and transitional cells were further divided into: CD10+ transitional and CD10– naive cells. Numbers indicate the percentage within the IgD+CD27– gate. Data are representative of 3 separate experiments.
Sorting human B-cell subsets. (A) Human peripheral B cells (CD19+) were stained and sorted into 4 populations, according to their expression of CD27 and IgD: IgD+CD27– (naive and transitional), IgD+CD27+ (IgM memory), IgD–CD27+, and IgD–CD27– B cells. Numbers indicate the percentage of subsets in boxes. (B) From the IgD+CD27– gate, naive and transitional cells were further divided into: CD10+ transitional and CD10– naive cells. Numbers indicate the percentage within the IgD+CD27– gate. Data are representative of 3 separate experiments.
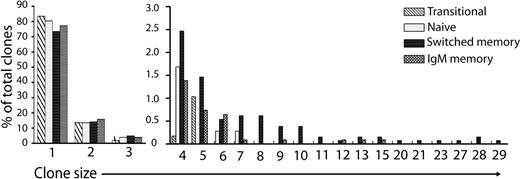
A total of 6706 Ig gene sequences were generated. The amino acid sequence across the CDR3 region was used to identify unique Ig gene rearrangements. Ig genes with identical, or related, CDR3 regions and the same IGHV, IGHD, IGHJ usage were seen, representing clonal families of genes. Because the method of isolation involves PCR from cDNA, it is possible that some “clones” will arise from PCR amplification of a single sequence, or isolation of multiple cDNA copies of the sequence from 1 cell. This is particularly the case for the smaller clonal groups, so clonality comparisons are comparative rather than absolute (supplemental Table 2). Antigen-experienced populations had significantly fewer unique Ig gene rearrangements, and larger clone sizes, than in the transitional and naive populations (Figure 2). In addition, more than 55% of the total (1741) Ig gene sequences from IgM memory cells were unique Ig gene rearrangements, compared with 41% of the total (2067) IgG and IgA sequences (P < .0001).
Frequency of clonal expansion in different B-cell populations. Large clonal expansions were observed in antigen-experienced memory populations. Sequencing of Ig gene rearrangements showed that some occurred more than once, and were considered to belong to a clonal family. Members of the same clone (with the same junctional CDR3 sequence) were identified. The size of each family was noted but only 1 member of each family was used in subsequent analyses. We plotted the frequency distribution of sequences between clones of different size for transitional (IgD+CD27−CD10+, n = 433), naive (IgD+CD27−CD10−, n = 662), switched memory (IgD− and either IgG+ or IgA+, n = 1293) and IgM memory (IgD+CD27+, n = 1209) populations. Clone size of 1 indicates no related sequences were found.
Frequency of clonal expansion in different B-cell populations. Large clonal expansions were observed in antigen-experienced memory populations. Sequencing of Ig gene rearrangements showed that some occurred more than once, and were considered to belong to a clonal family. Members of the same clone (with the same junctional CDR3 sequence) were identified. The size of each family was noted but only 1 member of each family was used in subsequent analyses. We plotted the frequency distribution of sequences between clones of different size for transitional (IgD+CD27−CD10+, n = 433), naive (IgD+CD27−CD10−, n = 662), switched memory (IgD− and either IgG+ or IgA+, n = 1293) and IgM memory (IgD+CD27+, n = 1209) populations. Clone size of 1 indicates no related sequences were found.
Where a clonal expansion was identified, one of the sequences was chosen at random as a representative of the clone to look at the repertoire in the absence of any contribution by clonal expansion. Hence for repertoire analysis we used a total of 3597 unique Ig genes from 4 different B-cell populations (Table 1). IgG and IgA cells were found to have a very similar repertoire and therefore the data were pooled together for future comparisons (Table 1 and D.K.D.-W., unpublished data, 2010). The contribution to the repertoire from different donors is shown in supplemental Figure 1.
The number of unique sequences incorporated into the analyses
| . | IGHV1 . | IGHV2 . | IGHV3 . | IGHV4 . | IGHV5 . | IGHV6 . | IGHV7 . | Total unique . | Overall total* . |
|---|---|---|---|---|---|---|---|---|---|
| Naive | 113 | 13 | 328 | 182 | 14 | 9 | 3 | 662 | 806 |
| Transitional | 40 | 12 | 232 | 135 | 8 | 6 | 0 | 433 | 551 |
| IgM memory | 21 | 80 | 762 | 279 | 27 | 40 | 0 | 1209 | 1741 |
| IgG | 220 | 21 | 249 | 151 | 22 | 12 | 4 | 679 | 1065 |
| IgA | 197 | 28 | 216 | 141 | 15 | 15 | 2 | 614 | 1002 |
| IgG+ or IgA+ | 417 | 49 | 465 | 292 | 37 | 27 | 6 | 1293 | 2067 |
| . | IGHV1 . | IGHV2 . | IGHV3 . | IGHV4 . | IGHV5 . | IGHV6 . | IGHV7 . | Total unique . | Overall total* . |
|---|---|---|---|---|---|---|---|---|---|
| Naive | 113 | 13 | 328 | 182 | 14 | 9 | 3 | 662 | 806 |
| Transitional | 40 | 12 | 232 | 135 | 8 | 6 | 0 | 433 | 551 |
| IgM memory | 21 | 80 | 762 | 279 | 27 | 40 | 0 | 1209 | 1741 |
| IgG | 220 | 21 | 249 | 151 | 22 | 12 | 4 | 679 | 1065 |
| IgA | 197 | 28 | 216 | 141 | 15 | 15 | 2 | 614 | 1002 |
| IgG+ or IgA+ | 417 | 49 | 465 | 292 | 37 | 27 | 6 | 1293 | 2067 |
The numbers in the table refer to the number of unique sequences, where only 1 example of a clonal expansion is counted, the overall total is the total number of sequences obtained before each adjustment was made. There is a high rate of attrition of sequence reads from the 454. The numbers here represent, on average, 20% of the initial data as 60% of sequences either do not pass our quality control checks or do not return results from V-QUEST.24 A further 20% were removed as they lacked some component of the full immunoglobulin VDJC gene rearrangement, for example, some of the sequences in the IgD-CD27- or IgD- CD27+ populations could not be identified as being switched due to lack of C region sequence so were not included.
Comparison of transitional and naive B-cell repertoires
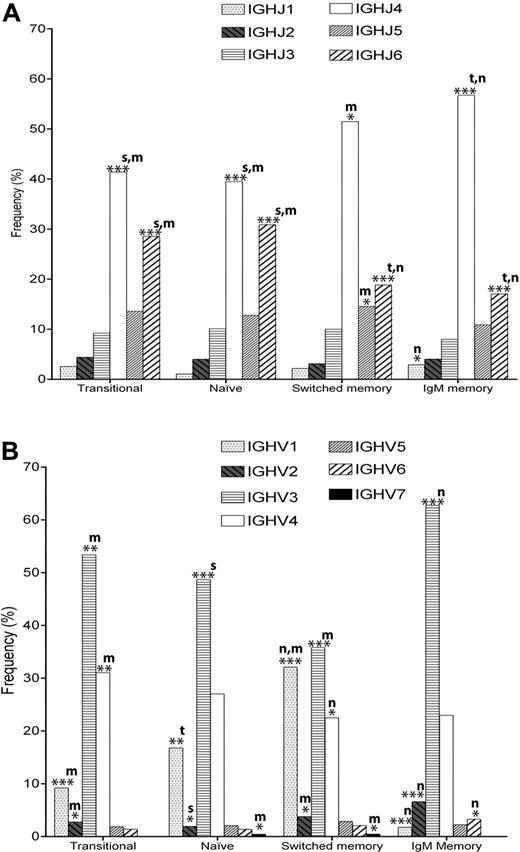
The transitional B-cell repertoire was found to be similar to that of naive B cells. We see no significant difference in overall IGHJ gene usage (Figure 3A); however, the IGHV gene family usage varies slightly (Figure 3B)—with naive cells having significantly more IGHV1 than transitional cells (16.7% vs 9.1%, P < .005). The most commonly used IGHV3 genes are altered, with IGHV3-23 use being increased in naive cells and other IGHV3 genes being decreased (Figure 4). The ratio of IGHV3-23 to IGHV3-30 + IGHV3-33 is 0.55 for transitional cells compared with 1.0 for naive cells. If the IGH genes compared are restricted to those that also use the IGHJ4 gene then this difference becomes even greater, the ratio being 0.4 for transitional cells compared with 1.2 for naive cells. These ratio differences are consistent across the 3 different donors (supplemental Figure 2).
IGHJ gene and IGHV family usage. IGHV family and IGHJ genes were identified from the Ig gene sequences using V-QUEST.24 (A) The relative frequency of 6 IGHJ genes, and (B) 7 IGHV gene families in transitional (IgD+CD27−CD10+, n = 433), naive (IgD+CD27−CD10−, n = 662), switched memory (IgD− and either IgG+ or IgA+, n = 1293) and IgM memory (IgD+CD27+, n = 1209) populations. “m,“ “s,” “n” and “t” indicate significant differences compared with IgM memory, switched memory, naive, and transitional cells, respectively. *P < .05; **P < .005; ***P < .0005.
IGHJ gene and IGHV family usage. IGHV family and IGHJ genes were identified from the Ig gene sequences using V-QUEST.24 (A) The relative frequency of 6 IGHJ genes, and (B) 7 IGHV gene families in transitional (IgD+CD27−CD10+, n = 433), naive (IgD+CD27−CD10−, n = 662), switched memory (IgD− and either IgG+ or IgA+, n = 1293) and IgM memory (IgD+CD27+, n = 1209) populations. “m,“ “s,” “n” and “t” indicate significant differences compared with IgM memory, switched memory, naive, and transitional cells, respectively. *P < .05; **P < .005; ***P < .0005.
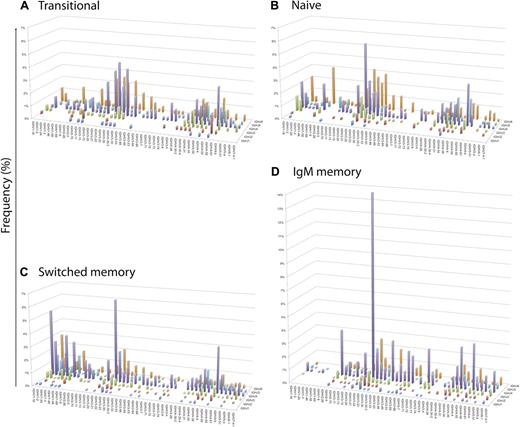
Three-dimensional landscapes of IGHV gene usage in conjunction with IGHJ usage show variation between antigen-inexperienced and memory B-cell populations. Landscapes illustrate the pairing of individual IGHV and IGHJ gene combinations in the Ig gene sequences, as identified by V-Quest. The relative frequency (y-axis) of individual IGHV genes (x-axis) using IGHJ genes (z-axis) in (A) Transitional (IgD+CD27−CD10+, n = 433), (B) Naive (IgD+CD27−CD10−, n = 662), (C) Switched memory (IgD− and either IgG+ or IgA+, n = 1293), and (D) IgM memory (IgD+CD27+, n = 1209) populations is shown. Allelic variations of each IGHV gene are not shown. P values for χ2 comparisons of individual IGHV gene usage between the different groups are indicated in Table 2.
Three-dimensional landscapes of IGHV gene usage in conjunction with IGHJ usage show variation between antigen-inexperienced and memory B-cell populations. Landscapes illustrate the pairing of individual IGHV and IGHJ gene combinations in the Ig gene sequences, as identified by V-Quest. The relative frequency (y-axis) of individual IGHV genes (x-axis) using IGHJ genes (z-axis) in (A) Transitional (IgD+CD27−CD10+, n = 433), (B) Naive (IgD+CD27−CD10−, n = 662), (C) Switched memory (IgD− and either IgG+ or IgA+, n = 1293), and (D) IgM memory (IgD+CD27+, n = 1209) populations is shown. Allelic variations of each IGHV gene are not shown. P values for χ2 comparisons of individual IGHV gene usage between the different groups are indicated in Table 2.
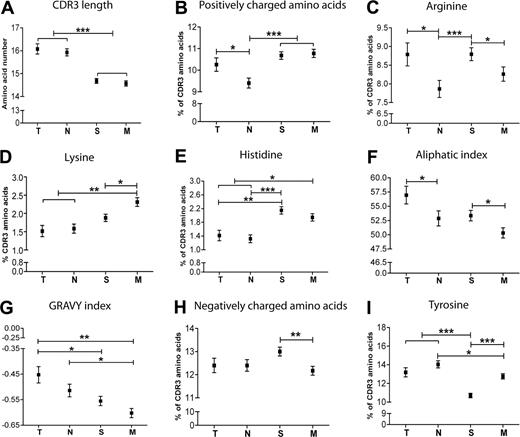
Because the CDR3 region of the Ig gene is thought to have the most influence over antigen specificity, we also studied CDR3 characteristics of the IGH repertoire in transitional and naive cells (Figure 5). Average CDR3 sizes were comparable in both populations (Figure 5A). However, there was a decreased proportion of positively charged amino acids in naive B cells (Figure 5B), which was due to a difference in arginine composition (Figure 5C) because there was no difference in the composition of lysine or histidine (Figure 5D-E). There was also a decrease in aliphatic index in naive B cells, with an accompanying downward trend in hydrophobicity (Figure 5F-G).
Four subsets of B cells are distinguished from one another in their Ig CDR3 properties. The amino acid sequence of the Ig CDR3 region was determined from V-Quest for each sequence and subjected to ProtParam25 analysis to determine the peptide characteristics for transitional (T, n = 433), naive (N, n = 662), switched memory (S, n = 1293), and IgM memory (M, n = 1209) populations. Error bars are SEM. *P < .05; **P < .005; ***P < .0005.
Four subsets of B cells are distinguished from one another in their Ig CDR3 properties. The amino acid sequence of the Ig CDR3 region was determined from V-Quest for each sequence and subjected to ProtParam25 analysis to determine the peptide characteristics for transitional (T, n = 433), naive (N, n = 662), switched memory (S, n = 1293), and IgM memory (M, n = 1209) populations. Error bars are SEM. *P < .05; **P < .005; ***P < .0005.
B-cell memory repertoire selection
IGHJ4 and IGHJ6 usage is changed between naive cells and the switched or IgM memory cells (Figure 3A). IGHJ4 usage is some 15% higher in memory than in naive cells. Conversely, IGHJ6 usage was significantly higher in naive B cells compared with memory cells (P < .0001). As expected, in view of the similarity of the IGHJ repertoire in naive and transitional cells, there is also a similar highly significant increase in IGHJ4 usage and decrease in IGHJ6 usage in memory cells compared with transitional cells.
IGHV usage revealed a modest decrease in IGHV4 family in switched memory cells compared with naive cells (P < .05). Similarly, the differences in other families are relatively modest compared with the dramatic repertoire differences observed in the IGHV1 and IGHV3 families. Compared with naive cells IGHV1 family usage increases by 15% in switched cells but decreases by 15% in IgM memory cells (P < .0001; Figure 3B). The increased IGHV1 usage in the switched cells was particularly associated with the IGHV1/IGHJ4 (P < .0001) gene family combination (Figure 4 and supplemental Figure 5). Conversely, compared with naive cells, the IGHV3 family usage was decreased by 13% in switched cells, attributable to a loss of the IGHV3/IGHJ6 gene combination (Figure 4 and supplemental Figure 5), and increased by 14% in IgM memory cells (P < .0001 all comparisons; Figure 3B). The increased IGHV3 representation in IgM memory was solely due to IGHV3/IGHJ4 genes (P < .0001), because the IGHV3/IGHJ6 gene combination was actually decreased compared with the naive repertoire (P < .005; Figure 4 and supplemental Figure 5). Comparisons between transitional cells and memory cells were very similar to those between naive cells and memory cells. Individual IGHV gene analysis revealed a large number of genes where highly significant differences between 2 or more subsets of cells were seen (Figure 4 and supplemental Figure 3; Table 2). Several genes of the IGHV1 family were decreased, and IGHV3-23 and IGHV3-66 genes were overrepresented, in IgM memory compared with both the naive and switched subsets. Naive B cells had more IGHV3-30-3 and IGHV3-33 than both memory subsets, switched B cells had higher IGHV1-3 and IGHV1-46 than both naive and IgM memory, and IgM memory had more IGHV3-48 and IGHV3-7 than did switched memory. The overall pattern is that of a very different repertoire of IGHV genes in naive, switched memory and IgM memory B-cell subsets, with only 11 of the 48 IGHV genes showing no significant difference in use between the different populations.
χ2 comparisons of individual IGHV genes between four different populations of B cells†
| IGHV names . | T versus N . | T versus S . | T versus M . | N versus S . | N versus M . | S versus M . |
|---|---|---|---|---|---|---|
| IGHV1-18 | ** | *** | * | *** | *** | |
| IGHV1-2 | ** | *** | ** | *** | *** | |
| IGHV1-24 | * | |||||
| IGHV1-3 | * | *** | *** | |||
| IGHV1-46 | *** | *** | * | *** | ||
| IGHV1-58 | * | * | ** | |||
| IGHV1-69 | * | ** | *** | *** | *** | |
| IGHV1-8 | * | ** | * | |||
| IGHV2-26 | ||||||
| IGHV2-5 | * | *** | *** | |||
| IGHV2-70 | * | |||||
| IGHV3-11 | * | ** | * | |||
| IGHV3-13 | *** | * | * | * | ||
| IGHV3-15 | * | |||||
| IGHV3-20 | * | |||||
| IGHV3-21 | ||||||
| IGHV3-23 | * | *** | *** | *** | ||
| IGHV3-30 | * | ** | ||||
| IGHV3-30-3 | ** | *** | ** | |||
| IGHV3-33 | *** | *** | *** | *** | ||
| IGHV3-43 | * | |||||
| IGHV3-48 | * | ** | *** | |||
| IGHV3-49 | * | ** | ||||
| IGHV3-53 | ||||||
| IGHV3-64 | ||||||
| IGHV3-66 | ** | *** | *** | |||
| IGHV3-7 | * | * | *** | |||
| IGHV3-71 | ||||||
| IGHV3-72 | * | |||||
| IGHV3-73 | ||||||
| IGHV3-74 | * | *** | ** | |||
| IGHV3-9 | ** | |||||
| IGHV3-h | ||||||
| IGHV4-28 | ||||||
| IGHV4-30-2 | * | * | * | |||
| IGHV4-30-4 | ** | *** | ** | |||
| IGHV4-31 | ** | *** | ** | |||
| IGHV4-34 | ** | * | ||||
| IGHV4-39 | ||||||
| IGHV4-4 | ||||||
| IGHV4-55 | ||||||
| IGHV4-59 | ||||||
| IGHV4-61 | * | * | ||||
| IGHV4-b | * | * | ||||
| IGHV5-51 | * | |||||
| IGHV5-a | ** | * | ||||
| IGHV6-1 | * | * | ||||
| IGHV7-4-1 | * | * |
| IGHV names . | T versus N . | T versus S . | T versus M . | N versus S . | N versus M . | S versus M . |
|---|---|---|---|---|---|---|
| IGHV1-18 | ** | *** | * | *** | *** | |
| IGHV1-2 | ** | *** | ** | *** | *** | |
| IGHV1-24 | * | |||||
| IGHV1-3 | * | *** | *** | |||
| IGHV1-46 | *** | *** | * | *** | ||
| IGHV1-58 | * | * | ** | |||
| IGHV1-69 | * | ** | *** | *** | *** | |
| IGHV1-8 | * | ** | * | |||
| IGHV2-26 | ||||||
| IGHV2-5 | * | *** | *** | |||
| IGHV2-70 | * | |||||
| IGHV3-11 | * | ** | * | |||
| IGHV3-13 | *** | * | * | * | ||
| IGHV3-15 | * | |||||
| IGHV3-20 | * | |||||
| IGHV3-21 | ||||||
| IGHV3-23 | * | *** | *** | *** | ||
| IGHV3-30 | * | ** | ||||
| IGHV3-30-3 | ** | *** | ** | |||
| IGHV3-33 | *** | *** | *** | *** | ||
| IGHV3-43 | * | |||||
| IGHV3-48 | * | ** | *** | |||
| IGHV3-49 | * | ** | ||||
| IGHV3-53 | ||||||
| IGHV3-64 | ||||||
| IGHV3-66 | ** | *** | *** | |||
| IGHV3-7 | * | * | *** | |||
| IGHV3-71 | ||||||
| IGHV3-72 | * | |||||
| IGHV3-73 | ||||||
| IGHV3-74 | * | *** | ** | |||
| IGHV3-9 | ** | |||||
| IGHV3-h | ||||||
| IGHV4-28 | ||||||
| IGHV4-30-2 | * | * | * | |||
| IGHV4-30-4 | ** | *** | ** | |||
| IGHV4-31 | ** | *** | ** | |||
| IGHV4-34 | ** | * | ||||
| IGHV4-39 | ||||||
| IGHV4-4 | ||||||
| IGHV4-55 | ||||||
| IGHV4-59 | ||||||
| IGHV4-61 | * | * | ||||
| IGHV4-b | * | * | ||||
| IGHV5-51 | * | |||||
| IGHV5-a | ** | * | ||||
| IGHV6-1 | * | * | ||||
| IGHV7-4-1 | * | * |
Transitional (T, n = 433), naive (N, n = 662), switched memory (S, n = 1293) and IgM memory (M, n = 1209) populations were compared with each other.
P < .05; **P < .005; ***P < .0005. Note that if Bonferroni correction is applied the comparisons marked * are no longer considered to be significant. Some of these P values are the result of a trend in the 3 separate donors that individually did not reach significance. Those that are bold indicate that the P values of more than 1 of the donors reached significance individually (supplemental Table 3).
A significant reduction in average CDR3 size was observed in switched memory cells compared with naive (Figure 5A). There were also significant changes in CDR3 composition, where the GRAVY index of hydrophobicity and the number of tyrosine residues decrease (Figure 5G,I) and the number of positively charged amino acids, especially arginine and histidine, increases (Figure 5B-E).
Switched memory versus IgM memory B-cell populations
We found major differences in the repertoires from the switched memory and IgM memory populations, particularly in the IGHV1 and IGHV3 families (Figure 3B). There are 10 individual IGHV genes showing highly significant (P < .0005) differences between the 2, and a further 12 showing differences of less significance (Figure 4; Table 2). Particularly noteworthy (P < 10−11) are decreased IGHV1-18, IGHV1-2, IGHV1-46, IGHV1-69 and increased IGHV3-23 in IgM memory compared with switched memory (Figures 4; supplemental Figure 1). Some differences are also seen in the IGHJ repertoire (Figure 3A), with the IgM memory cells having 5% more IGHJ4 and 8% less IGHJ5 than the switched memory (P < .01).
There were also significant differences in CDR3 characteristics between the 2 different memory populations. The IgM memory population had a lower aliphatic index than switched memory (Figure 5F). Although the total number of positively charged amino acids was comparable between the 2 populations, IgM memory cells had less arginine and more lysine (Figure 5B-D). In addition, IgM memory cells had levels of negatively charged amino acids similar to those in transitional and naive cells, whereas the switched cells had more negatively charged residues (Figure 5H). Switched memory cells are also distinguished by their low levels of tyrosine compared with IgM memory (Figure 5I).
Discussion
It is well accepted that exogenous antigen challenge will result in a postactivation repertoire that is shaped by the types of antigens that have been encountered. This could result in a skew of the repertoire in favor of gene combinations that preferentially recognize the challenging antigen, or it may be that a history of multiple and diverse challenges, such as is expected in humans, would not result in a skewed repertoire. A previous study was unable to detect significant changes in repertoire between naive and memory cells.26 However, their single-cell approach to repertoire analysis meant that the numbers were necessarily limited to less than 100 from each group. We have therefore used recent developments in deep sequencing technology to perform a novel repertoire analysis with sufficient power to critically address the differences between naive and memory cells. As expected, clonal families of Ig genes were more often seen in the antigen-experienced populations than in the transitional and naive populations (Figure 2). Furthermore, there was also a highly significant increase in the prevalence of clonal expansions in class-switched B cells compared with IgM memory cells. Finding clonally related cells in a population is dependent on the frequency of related cells in the total population, that is, the larger the clone sizes the more chance of finding them in any given sample. Thus, the finding of more clonally related sequences in switched memory indicates that there are fewer large clones in the IgM memory population.
Signals from the BCR have an important regulatory influence over B cells in response to exogenous antigen challenge, they can also be important for survival even in the absence of a specific exogenous challenge.27 Thus positive endogenous selective forces could influence the Ig gene repertoire, although precisely what stimulatory molecules are involved in this BCR signaling has not been determined. There is strong evidence that negative selection events occur at the transitional B-cell stage, being 1 of 2 peripheral tolerance checkpoints.28 Clearly signaling thresholds of the BCR are important in deciding the fate of B cells at the transitional-naive developmental stage, and therefore in a healthy person one might expect to see some Ig gene repertoire differences between the 2 B-cell populations. We see that the transitional B-cell repertoire resembles that of naive B cells, although there are some distinguishing features that can be used individually, or in combination, to differentiate the 2. A key characteristic of naive cells compared with transitional cells is an alteration in the ratios of IGHV3-23 to IGHV3-30+IGHV3-33 gene usage. Because these IGHV3 genes make up 24% of the total population, they may be useful in distinguishing between the 2 subsets of cells.
Longer CDR3 lengths, or increased levels of positively charged amino acids such as arginine, have previously been associated with DNA binding antibodies.29-31 We observed that the repertoire moves toward having less positively charged residues in CDR3 in the transition to mature naive cells, along with a decrease in aliphatic index. It is difficult to predict what kinds of antigen might select for this characteristic. Aliphatic index is related to hydrophobicity and is a measure of the relative volume occupied by aliphatic side chains (valine, isoleucine, and leucine) that has been regarded as a positive factor for the increase of thermostability of globular proteins. Lower thermostability and more hydrophilic antigen binding sites are qualities that have previously been linked to increased antibody polyreactivity.32,33
Changes in IGHV4 gene usage between naive and switched memory populations have previously been reported,34 however this study was restricted to the IGHV4 family and so the overall usage of the IGHV4 repertoire compared with other families was not determined. Here we observed a slightly decreased IGHV4 family use in switched memory compared with naive cells, with a 2-fold difference in the IGHV4-34 gene in particular. However, the changes in IGHV4 genes appear subtle compared with the dramatic repertoire differences in the IGHV1 and IGHV3 families, where the former is more frequent and the latter less frequent in switched memory compared with naive and transitional cells. Such differences in the IGHV1 and IGHV3 families are attributable to changes in many individual genes, particularly IGHV1-3, IGHV1-46, IGHV3-30-3 and IGHV3-33. (supplemental Figure 3; Table 2). As has been previously documented,26,35 we see a significant reduction in IGHJ6 and an increase in IGHJ4 genes in the transition to memory cells. Such changes in the IGHJ families are largely responsible for a shorter CDR3 size in memory cells, because IGHD gene contributions are comparable between naive and memory populations (D.K.D.-W., unpublished data, 2010). This is in line with previous findings that higher affinity antigen-experienced B cells harbour a shorter CDR3.35,36 There are also a number of changes in the amino acid composition of the CDR3 region with the transition from naive into switched memory cells, the most significant of these being an increase in the positively charged amino acids arginine and histidine. The reduction in tyrosine residues is probably a result of decreased IGHJ6 usage, because IGHJ6 contains a large proportion of tyrosine residues.
The origins of class-switched memory cells are thought to be well understood, being the B cells that have encountered antigen in a T cell–dependent manner, because T helper cells are required for class-switch events to take place in the GC. However, as outlined in the introduction the origins and functions of IgM memory cells remain controversial. On one side it has been said that IgM memory cells originate from a T-dependent GC reaction before class-switching has occurred,20 while the alternative view is that the development of these cells is GC-independent and they are T-independent antigen responders.4 To address this we made a direct comparison of the Ig gene repertoires of the 2 different populations. If most IgM memory cells were formed as a first step in the process toward class-switched cells, as suggested by Seifert and Kuppers,20 then the repertoire would be expected to have much in common with the switched memory cells. However, we have shown major differences in the repertoires from the 2 different populations (Figure 4), with IgM memory cells having much less IGHV1 and more IGHV3 family genes. Individual IGHV gene analysis revealed striking variations in IGHV1-18, IGHV1-2, IGHV1-46, IGHV1-69 and IGHV3-23 genes (P < 10−11). Although a previous comparison of IgM memory and switched memory reported that there were no significant differences in repertoire,26 their data did show a trend toward increased IGHV3-23 in IgM memory cells and it seems probable that it was their lower numbers (63 vs 85 Ig genes) that accounted for the failure to reach significance.
Although CDR3 sizes are comparable between switched and IgM memory populations, their characteristics vary between the two. IgM memory cells had less negatively charged amino acids than switched memory cells, while the levels of positively charged amino acids did not appear to vary. Interestingly, in the light of comparable IGHJ6 usages, IgM memory cells are higher in tyrosine. High tyrosine and lysine levels have previously been correlated with polysaccharide binding ability.37 Furthermore, there is a tendency of IgM memory cells toward a lower hydrophobic and aliphatic index, perhaps indicating a less thermostable and more polyspecific, activity than in switched memory.
These repertoire differences provide strong evidence against the hypothesis by Seifert and Kuppers that the majority of IgM memory cells are related to IgG memory cells and are formed as part of a GC reaction.20 We also looked for matching clones between switched and IgM memory cells in our data and saw no examples of related IgG and IgM memory Ig genes between the 1829 switched memory Ig genes (isolated from a pool of ∼ 22 000 cells) and 1741 IgM memory Ig genes (isolated from ∼ 37 000 cells), although we did see many examples of expanded Ig gene families that were of a single isotype. In agreement with previous reports of clonal expansions in the memory populations of children,38 we found that clonal expansion was much more evident in the switched memory population than in the IgM memory population. The clonal relationships previously found between IgM memory and IgG genes were sound evidence that some IgM memory cells are formed as part of a GC reaction. However, our results indicate that a larger proportion of IgM memory cells must derive from some other developmental pathway.
If the majority of IgM memory cells are not directly related to switched memory cells then perhaps they are from a separate population that develops by antigen-independent processes, analogous to the mouse marginal zone lineage.4 However, it is difficult to reconcile the idea that B cells might have a BCR-independent mode of development from transitional cells with our observation that IgM memory cells have a repertoire that is distinct from transitional and naive B cells—some selective process involving the BCR must be involved in the formation of the majority of cells. This does not discount the theory that antigen-independent diversification is possible, but rather indicates that only a small proportion of IgM memory cells would originate in this fashion in a healthy population and are not enough to mask the skewing of the repertoire that is seen. The alternative consideration is that development of IgM memory could be an exogenous antigen-independent process but might still rely on positive signals from autoantigens for development, and that these endogenous signals shape the repertoire.
It may be more likely that the skewed repertoire seen in the IgM memory cells reflects developmental antigen selection by a different type of exogenous antigen than the switched memory cells, that is, a T-independent compared with a T-dependent antigen. We cannot exclude the possibility that peripheral autoantigen may still have some selective effect, perhaps complementing signals from exogenous Toll-like receptor ligands or other stimulators. In this context it is interesting that the overexpressed gene, IGHV3-23, has been reported as having a wide range of specificities, including against DNA and rheumatoid factor.39 However, it has been shown that a second peripheral checkpoint against autoreactivity in the transition between naive and IgM memory cells exists,40 which makes this scenario somewhat less likely. It is not easy to explain why there might be such a dramatic decrease in IGHV1 family genes alongside the increased IGHV3-23 use. Perhaps IGHV1 genes do not predispose toward the polyreactivity for which IgM memory cells are renown. CD5+ B cells have long been associated with polyspecific antibody responses. It has been previously shown that, in the same response to rotavirus infection in children, IGHV usage of a CD5+ population is skewed toward IGHV3 usage and away from IGHV1 usage compared with the CD5− population.41
In conclusion, high-powered analyses of Ig gene repertoire can be performed using a deep sequencing platform that enables longer sequence reads, and these analyses can provide information to distinguish different B-cell populations. We have shown distinguishing features between transitional and naive cells, and between naive and memory cells. We can also easily distinguish between different types of memory cells, IgM memory cells having a distinct repertoire that indicates a different set of selective processes than switched memory cells. Therefore, IgM memory cells cannot all be earlier relatives of switched memory clones and the IgM memory population is likely of heterogeneous origin.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank Richard van Gelder for help with sample collection and all our volunteers for their blood donation.
This work was funded by the Human Frontiers Science program (Y.-C.W.), Research into Ageing (A.A.A.), and the Biotechnology and Biological Research Council (V.M.).
Authorship
Contribution: Y.-C.W. designed and carried out experiments, analyzed data, and wrote the manuscript; D.K. designed and wrote data-handling and analysis scripts and wrote the manuscript, H.S.L. wrote data-handling scripts; V.M. collected samples and wrote the manuscript; A.A.A. collected samples and designed experiments; and D.K.D.-W. oversaw the project, designed experiments and analytical tools, carried out data analysis, and wrote the paper.
Conflict-of-interest: The authors declare no competing financial interests.
Correspondence: Deborah Dunn-Walters, Peter Gorer Department of Immunobiology, 2nd Fl Borough Wing, King's College London School of Medicine, Guy's Campus, London SE1 9RT, United Kingdom; e-mail: deborah.dunn-walters@kcl.ac.uk.
References
Author notes
Y.-C.W. and D.K. contributed equally to this paper.