Abstract
Children with Down syndrome (DS) have an increased risk of acute lymphoblastic leukemia (ALL) and an inferior outcome. We reviewed data from 2811 children with ALL enrolled in Children's Oncology Group P9900, which included prospective testing for the major cytogenetic lesions in childhood ALL: ETV6-RUNX1, TCF3-PBX1, BCR-ABL1, and MLL translocations and trisomies of chromosomes 4 and 10. Eighty (3%) B-precursor ALL patients had DS. Age, sex, white blood cell count, and risk group were similar between DS-ALL and non–DS-ALL but significantly more patients with DS-ALL were white (91.2% vs 76.4%, P = .001). Children with DS-ALL had lower rates of the favorable cytogenetic lesions ETV6-RUNX1 (2.5% vs 24%, P < .001) and trisomies 4 and 10 (7.7% vs 24%, P < .001). Five-year event-free (EFS) and overall survival (OS) were inferior in children with DS-ALL: 69.9% ± 8.6% versus 78.1% ± 1.2% (P = .078), and 85.8% ± 6.5% versus 90.0% ± 0.9% (P = .033). However, when children with MLL translocations, BCR-ABL1, ETV6-RUNX1, and trisomies 4 and 10 were excluded, the EFS and OS were similar for children with and without DS (EFS 68.0 %± 9.3% vs 70.5% ± 1.9%, P = .817; and OS 86.7% ± 6.7% vs 85.4% ± 1.5%; P = .852), both overall and adjusted for race. DS-ALL displays a unique spectrum of biologic subtypes with different frequencies of sentinel cytogenetic lesions having a large influence on outcome.
Introduction
Children with Down syndrome (DS) have a substantially increased risk of developing both acute lymphoblastic leukemia (ALL) and acute myeloid leukemia. ALL occurs in 1 in 300 children with DS versus 1 in 3500 children without DS.1 Many groups have reported that children with ALL and DS (DS-ALL) have inferior event-free survival (EFS) and overall survival (OS) compared with children with ALL and without DS (non–DS-ALL).2-6 Factors that have contributed to the inferior outcome for DS-ALL include an increased risk of relapse and increased morbidity and mortality resulting from toxic side effects of therapy.7-9
The inferior survival of children with DS-ALL may also be related to different frequencies of favorable and unfavorable biologic subtype(s). For example, the Children's Cancer Group (CCG) 1952 standard risk (SR) ALL trial found that the absence of favorable lymphoblast cytogenetic features among the DS-ALL cohort contributed to the difference in EFS.10 A recent review of the International Berlin-Frankfurt-Münster (iBFM) cytogenetic register showed that the leukemic blasts of DS patients with ALL had favorable cytogenetic features less often than non-DS patients, with rates of 11% of high hyperdiploidy (51-57 chromosomes) and 10% for ETV6-RUNX1.11 However, neither of these reports included universal screening for specific cytogenetic abnormalities.
To determine more precisely the clinical and biologic variables that account for the inferior outcome of children with DS-ALL, we evaluated data from 2811 children with B-precursor ALL enrolled in Children's Oncology Group (COG) P9900 who underwent uniform prospective testing for the common sentinel cytogenetic lesions that occur in childhood ALL, including ETV6-RUNX1 (formerly termed TEL-AML1), TCF3-PBX1 (formerly E2A-PBX1), BCR-ABL1, MLL translocations, and trisomies of chromosomes 4 and 10. We found markedly different distributions of favorable cytogenetic subtypes in children with DS that contribute significantly to the inferior outcome of children with DS-ALL.
Methods
COG P9900 was a classification and treatment study designed to provide induction therapy for patients with B-precursor ALL and to collect the clinical and laboratory data necessary for risk-stratification and postinduction treatment assignment of children with newly diagnosed ALL. The study was open for accrual between December 13, 1999, and February 28, 2005. Patient eligibility for P9900 included previously untreated, newly diagnosed ALL confirmed by central review of immunophenotype, age less than or equal to 21 years, and submission of the appropriate samples for genetic analysis. DS patients were identified by searching the cytogenetics database and also including others that had answered “Down syndrome” to the questions on the “presence of congenital abnormalities” in the remote data entry system.
The study provided a comprehensive characterization of the biologic features of leukemic blasts by specifying tests on all patients enrolled: immunophenotype and cytochemical stains were performed at the local institution; karyotype was performed at a COG-approved cytogenetics laboratory. Additional centralized testing at the COG reference laboratories included immunophenotype screening for MLL, TCF3-PBX1, and ETV6-RUNX1, immunophenotype detection of minimal residual disease (MRD) by flow cytometry at induction day 8 in peripheral blood (PB) and day 29 in bone marrow (BM), DNA index, fluorescence in situ hybridization (FISH) for MLL rearrangement and trisomies 4 and 10, and reverse-transcribed polymerase chain reaction testing for BCR-ABL1, ETV6-RUNX1, and TCF3-PBX1. Details of these methods were reported previously.12
Patient eligibility for the induction therapeutic study included newly diagnosed precursor B ALL, age more than 1 year and less than 21 years. Patients received either a 3- or 4-drug induction based on their National Cancer Institute (NCI) risk classification by age and initial white blood cell count (WBC) and the presence of central nervous system (CNS) disease.13 The institutional review board at each participating institution approved the protocol before patient enrollment. Written informed consent was obtained from parents or guardians before initiating therapy in accordance with the Declaration of Helsinki.
Induction therapy for NCI SR (age 1-9.99 years and WBC < 50 000/μL) patients consisted of 3-drug therapy with dexamethasone, PEG asparaginase, vincristine, and intrathecal methotrexate (MTX), whereas NCI high-risk (age > 10 years or WBC ≥ 50 000/μL) patients received 4-drug therapy with prednisone, L-asparaginase, vincristine, daunomycin, and intrathecal MTX. At the end of induction, the patients were classified as having low-, standard-, or high-risk ALL and were eligible to enroll in the postinduction therapeutic protocols P9904, P9905, and P9906, respectively. Classification and treatment assignment were based on NCI risk classification, the presence of CNS 3 or overt testicular disease at diagnosis, lymphoblast cytogenetics (simultaneous trisomies of chromosomes 4 and 10 or ETV6-RUNX1 fusion were defined as low-risk features), and morphologic assessment of remission status on day 29. Patients with BCR/ABL1 ALL, hypodiploidy (DNA index < 0.81 or modal chromosome number < 44), M3 (> 25% blasts) BM status at day 29, or M2 (5-25 blasts) at day 43 were excluded from these trials but eligible to participate in a very-high-risk trial (COG AALL0031). The P9904/5 protocols included a randomized comparison between 6 intravenous infusions of MTX 2 g/m2 over 4 hours versus 1 g/m2 over 24 hours, followed by identical schedules of leucovorin rescue. Children with DS received the first course of MTX at 50% dose. If there was no mucositis, then subsequent infusions were administered at full dose. If grade 3 or 4 mucosal toxicity was encountered and/or there was delayed MTX excretion (> 0.2μM level 48 hours after the start of the MTX infusion), subsequent doses were reduced by 25% until tolerated. Patients with an ETV6-RUNX1 translocation were also eligible for randomization to receive or not receive a delayed intensification phase. Patients enrolled on P9905 were eligible for both the MTX and delayed intensification randomization. P9906 was a single-arm trial that used modified augmented BFM therapy.14
Statistical analyses
EFS and OS times were computed for all eligible B-precursor ALL patients identified to be part of this report. EFS time was defined as days from the date of diagnosis until induction failure, first relapse, second malignancy, or death from any cause. Patients not experiencing an event were censored as of the date of last contact. Similarly, OS time was defined as time from diagnosis to death or last contact. The EFS and OS estimates were computed using the Kaplan-Meier method,15 and SEs of the estimates were determined according to Peto and Peto.16 Fisher exact test was used to compare proportions (response, presence of chromosomal abnormalities, MRD), and the log-rank test was used to compare survivor functions. The Cox proportional hazards regression model was used to evaluate the significance of differences in EFS between groups, adjusting for other factors.
Results
Between December 1999 and February 2005, 2811 eligible B-precursor ALL patients were enrolled on COG P9900 for classification and remission induction treatment. Eighty (3%) of the 2811 children had DS. There were no DS patients with T-cell ALL.
Clinical characteristics of the DS and non-DS patients are presented in Table 1. There were no statistically significant differences in sex, age, presenting WBC count, and NCI risk group, between the 2 groups. Distribution of race was significantly different between the 2 groups (P = .001), with children with DS-ALL being largely white (91.2%) with no black and 8.8% “other.” In contrast, children with non–DS-ALL were 76.4% white, 7.7% black, and 15.9% other. The complete remission rates were similar for the children with and without DS (95% vs 97.4%, P = .169). The distribution of induction day 8 PB MRD was significantly different between the 2 groups (P < .017), with DS-ALL patients more likely to have high levels of day 8 PB MRD, whereas the distribution of day 29 BM MRD results was similar (P = .713).
Patient characteristics
| Characteristic . | DS, no. (%) (n = 80) . | Non-DS, no. (%) (n = 2731) . | P . |
|---|---|---|---|
| Sex | .306 | ||
| Male | 38 (47.5) | 1469 (53.8) | |
| Female | 42 (52.5) | 1262 (46.2) | |
| Age, y | .789 | ||
| Less than 10 | 63 (78.8) | 2090 (76.5) | |
| More than or equal to 10 | 17 (21.2) | 641 (23.5) | |
| WBC | .295 | ||
| Less than 50 000 | 70 (87.5) | 2253 (82.5) | |
| More than or equal to 50 000 | 10 (12.5) | 478 (17.5) | |
| Race | .001 | ||
| White | 73 (91.2) | 2087 (76.4) | |
| Black | 0 (0) | 211 (7.7) | |
| Other | 7 (8.8) | 433 (15.9) | |
| NCI risk | .194 | ||
| Standard | 57 (71.2) | 1734 (63.5) | |
| High | 23 (28.8) | 997 (36.5) | |
| DNA index | < .001 | ||
| Less than or equal to 1.16 | 76 (95.0) | 2060 (75.4) | |
| More than 1.16 | 4 (5.0) | 671 (24.6) | |
| Response | .169 | ||
| CR | 76 (95) | 2659 (97.4) | |
| No CR | 4 (5) | 72 (2.6) | |
| MRD day 8 | < .017 | ||
| Less than 0.01% | 17 (24.6) | 713 (29.5) | |
| 0.01%-0.1% | 5 (7.3) | 420 (17.4) | |
| 0.1%-1.0% | 28 (40.6) | 619 (25.6) | |
| More than or equal to 1.0% | 19 (27.5) | 666 (27.5) | |
| MRD day 29 | .713 | ||
| Less than 0.01% | 48 (73.9) | 1885 (78.0) | |
| 0.01%- 0.1% | 8 (12.3) | 220 (9.1) | |
| 0.1%-1.0% | 5 (7.7) | 178 (7.4) | |
| More than or equal to 1.0% | 4 (6.1) | 132 (5.5) |
| Characteristic . | DS, no. (%) (n = 80) . | Non-DS, no. (%) (n = 2731) . | P . |
|---|---|---|---|
| Sex | .306 | ||
| Male | 38 (47.5) | 1469 (53.8) | |
| Female | 42 (52.5) | 1262 (46.2) | |
| Age, y | .789 | ||
| Less than 10 | 63 (78.8) | 2090 (76.5) | |
| More than or equal to 10 | 17 (21.2) | 641 (23.5) | |
| WBC | .295 | ||
| Less than 50 000 | 70 (87.5) | 2253 (82.5) | |
| More than or equal to 50 000 | 10 (12.5) | 478 (17.5) | |
| Race | .001 | ||
| White | 73 (91.2) | 2087 (76.4) | |
| Black | 0 (0) | 211 (7.7) | |
| Other | 7 (8.8) | 433 (15.9) | |
| NCI risk | .194 | ||
| Standard | 57 (71.2) | 1734 (63.5) | |
| High | 23 (28.8) | 997 (36.5) | |
| DNA index | < .001 | ||
| Less than or equal to 1.16 | 76 (95.0) | 2060 (75.4) | |
| More than 1.16 | 4 (5.0) | 671 (24.6) | |
| Response | .169 | ||
| CR | 76 (95) | 2659 (97.4) | |
| No CR | 4 (5) | 72 (2.6) | |
| MRD day 8 | < .017 | ||
| Less than 0.01% | 17 (24.6) | 713 (29.5) | |
| 0.01%-0.1% | 5 (7.3) | 420 (17.4) | |
| 0.1%-1.0% | 28 (40.6) | 619 (25.6) | |
| More than or equal to 1.0% | 19 (27.5) | 666 (27.5) | |
| MRD day 29 | .713 | ||
| Less than 0.01% | 48 (73.9) | 1885 (78.0) | |
| 0.01%- 0.1% | 8 (12.3) | 220 (9.1) | |
| 0.1%-1.0% | 5 (7.7) | 178 (7.4) | |
| More than or equal to 1.0% | 4 (6.1) | 132 (5.5) |
NCI indicates National Cancer Institute.
Table 2 shows the incidence of the sentinel cytogenetic lesions determined prospectively in this study, both overall and by race. ETV6-RUNX1 fusion transcripts were found in 2.5% of DS-ALL versus 24% of non–DS-ALL (P < .001). Similarly, trisomy of both chromosomes 4 and 10 occurred in 7.7% of DS-ALL versus 23.9% of non–DS-ALL (P < .001). The proportion of patients with high hyperdiploidy (DNA index > 1.16) was significantly different in the 2 groups with 5% in DS versus 24.6% in non–DS-ALL patients (P < .001). In contrast, the incidence of TCF3-PBX1 translocations was similar in DS-ALL and non–DS-ALL (3.6% vs 4.3%, respectively, P = .99). No BCR-ABL1 or MLL translocations were found in DS-ALL. When the comparisons were restricted to white patients who made up most of the DS patients (73 of 80), the incidence of ETV6-RUNX1 fusion transcripts was found to be significantly different in DS (2.7%) versus non-DS (24.7%) patients (P < .001). Similarly, the incidence of trisomy of both chromosomes 4 and 10 was 8.4% in DS ALL versus 24.8% non–DS-ALL (P = .001). As with overall rates, among white patients, the incidence of TCF3-PBX1 translocations was similar in DS-ALL and non–DS-ALL (4.1% vs 3.2%, respectively; P = .507).
Chromosomal abnormalities, overall and by race
| Chromosomal abnormalities . | DS, no. (%) . | Non-DS, no. (%) . | P . |
|---|---|---|---|
| ETV6-RUNX1 | |||
| Overall | 2/80 (2.5) | 651/2710 (24.0) | < .001 |
| White | 2/73 (2.7) | 510/2068 (24.7) | < .001 |
| Other | 0/7 (0) | 141/642 (22.0) | .356 |
| Trisomy 4 and 10 | |||
| Overall | 6/78 (7.7) | 643/2689 (23.9) | < .001 |
| White | 6/71 (8.4) | 509/2051 (24.8) | .001 |
| Other | 0/7 (0) | 134/638 (21.0) | .355 |
| TCF3-PBX1 | |||
| Overall | 3/80 (3.6) | 123/2720 (4.3) | .99 |
| White | 3/73 (4.1) | 66/2076 (3.2) | .507 |
| Other | 0/7 (0) | 57/644 (8.8) | > .999 |
| MLL | |||
| Overall | 0/80 (0) | 39/2729 (1.4) | .627 |
| White | 0/73 (0) | 29/2085 (1.4) | .623 |
| Other | 0/7 (0) | 10/644 (1.6) | > .999 |
| BCR-ABL1 | |||
| Overall | 0/79 (0) | 57/2668 (2.1) | .408 |
| White | 0/72 (0) | 42/2032 (2.1) | .399 |
| Other | 0/7 (0) | 15/636 (2.4) | > .999 |
| Chromosomal abnormalities . | DS, no. (%) . | Non-DS, no. (%) . | P . |
|---|---|---|---|
| ETV6-RUNX1 | |||
| Overall | 2/80 (2.5) | 651/2710 (24.0) | < .001 |
| White | 2/73 (2.7) | 510/2068 (24.7) | < .001 |
| Other | 0/7 (0) | 141/642 (22.0) | .356 |
| Trisomy 4 and 10 | |||
| Overall | 6/78 (7.7) | 643/2689 (23.9) | < .001 |
| White | 6/71 (8.4) | 509/2051 (24.8) | .001 |
| Other | 0/7 (0) | 134/638 (21.0) | .355 |
| TCF3-PBX1 | |||
| Overall | 3/80 (3.6) | 123/2720 (4.3) | .99 |
| White | 3/73 (4.1) | 66/2076 (3.2) | .507 |
| Other | 0/7 (0) | 57/644 (8.8) | > .999 |
| MLL | |||
| Overall | 0/80 (0) | 39/2729 (1.4) | .627 |
| White | 0/73 (0) | 29/2085 (1.4) | .623 |
| Other | 0/7 (0) | 10/644 (1.6) | > .999 |
| BCR-ABL1 | |||
| Overall | 0/79 (0) | 57/2668 (2.1) | .408 |
| White | 0/72 (0) | 42/2032 (2.1) | .399 |
| Other | 0/7 (0) | 15/636 (2.4) | > .999 |
Total number varies for each abnormality depending on the number of cases with missing data.
In this series, 4 DS-ALL patients had a DNA index of more than 1.16, 3 (75%) of whom had trisomy of both chromosomes 4 and 10. Similarly, 82% (543 of 660; P < .544) of non–DS-ALL patients with a DNA index of more than 1.16 had simultaneous trisomies of both chromosomes 4 and 10.
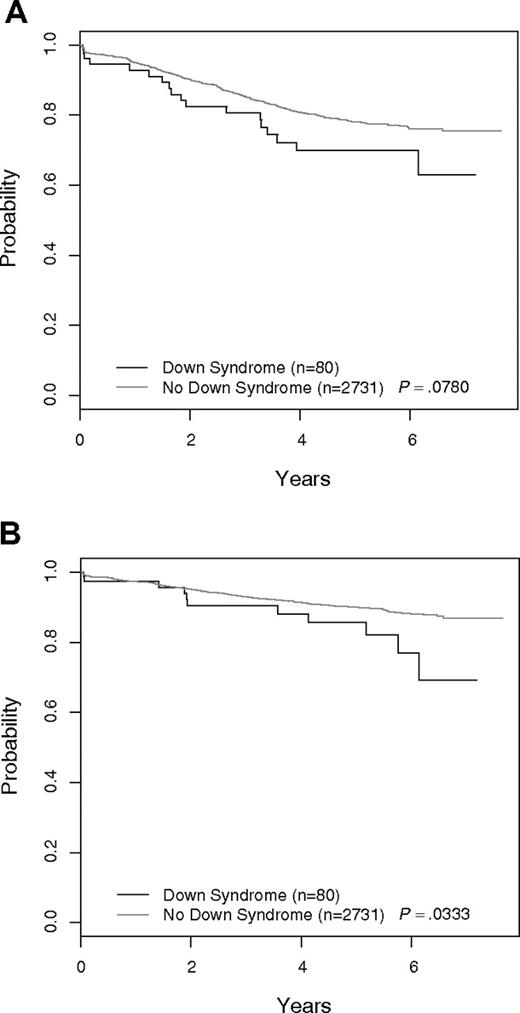
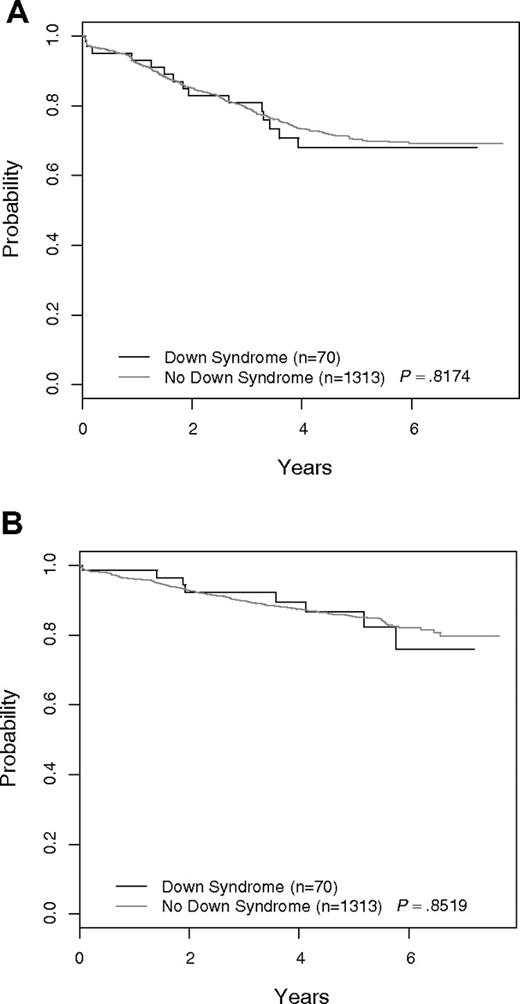
Figure 1A shows the EFS of DS-ALL versus non–DS-ALL for all 2811 COG P9900 patients. The 5-year EFS was 69.9% plus or minus 8.6% for DS-ALL versus 78.1% plus or minus 1.2% for non–DS-ALL (P = .078). Overall survival curves for the 2 groups are given in Figure 1B. The 5-year OS was 85.8% plus or minus 6.5% for DS-ALL versus 90.0% plus or minus 0.9% for non–DS-ALL (P = .033). When patients with either favorable (ETV6-RUNX1 or trisomies 4 and 10) or unfavorable biologic features (BCR-ABL1, MLL translocations) used for risk stratification and treatment assignment in these trials were excluded, the EFS and OS for the remaining DS-ALL (N = 70) and non–DS-ALL (N = 1313) patients were similar (Figure 2). The 5-year EFS excluding the aforementioned biologic features for DS-ALL was 68.0% plus or minus 9.3% versus 70.5% plus or minus 1.9% for non–DS-ALL (P = .817). The 5-year OS for DS-ALL was 86.7% plus or minus 6.7% versus 85.4% plus or minus 1.5% for non–DS-ALL (P = .852). The complete response rates were identical in the 2 groups (97.1% vs 97.3%). There was no significant difference (P = .337) in the distribution of postremission EFS events in the 2 groups (DS: 12 relapses, 0 remission deaths, 1 second malignant neoplasm; non-DS: 235 relapses, 11 remission deaths, 5 second malignant neoplasms). The distribution of day 8 MRD levels was barely significantly different for DS-ALL versus non–DS-ALL (P = .048), whereas that for day 29 MRD was not significantly different between the 2 groups (P = .768). Excluding patients with favorable or unfavorable biologic features, there was no significant difference in EFS between DS-ALL patients with day 8 PB MRD less than 0.01% compared with those with day 8 MRD more than or equal to 0.01% (5-year EFS 92.9% ± 10.1% vs 64.8% ± 12.8%; P = .173), but the number of patients in the 2 groups was very small (14 and 46), and the power to detect a difference is limited. Five-year EFS for DS patients with day 29 MRD less than 0.01% was 81.9% plus or minus 10.1% versus 49.5% plus or minus 24.9% for those with MRD more than or equal to 0.01% (P = .026).
Outcome for all patients: DS-ALL versus non–DS-ALL. (A) Event-free survival. (B) Overall survival.
Outcome for all patients: DS-ALL versus non–DS-ALL. (A) Event-free survival. (B) Overall survival.
Outcome for all patients excluding those with favorable and unfavorable sentinel cytogenetic lesions: DS-ALL versus non–DS-ALL. (A) EFS. (B) OS. Patients with ETV6-RUNX1, BCR-ABL1, MLL translocations, or trisomy of both chromosomes 4 and 10 are excluded.
Outcome for all patients excluding those with favorable and unfavorable sentinel cytogenetic lesions: DS-ALL versus non–DS-ALL. (A) EFS. (B) OS. Patients with ETV6-RUNX1, BCR-ABL1, MLL translocations, or trisomy of both chromosomes 4 and 10 are excluded.
Because there was a significant difference in the racial distribution for DS and non–DS-ALL patients, Cox regression analysis was used to compare EFS between the DS and non-DS patients adjusting for race (white vs others), after excluding patients with the specific favorable and unfavorable biologic features detailed in the previous paragraph. There was no significant difference in EFS between the 2 groups after adjusting for race (hazard ratio = 1.053, P = .846). Restricting the comparison to white patients also showed no significant difference (hazard ratio = 0.974, P = .925).
Discussion
Childhood ALL is a heterogeneous disease composed of distinct molecularly defined subgroups, many of which are associated with treatment outcome. Simultaneous trisomy of chromosomes 4 and 10 (and also 17) or the presence of the cryptic t(12;21) that produces ETV6/RUNX1 fusion was associated with an excellent prognosis in COG trials, whereas severe hypodiploidy (< 44 chromosomes) or the presence of the t(9;22) and BCR/ABL1 fusion conferred an adverse prognosis.17 Many prior studies have shown that DS-ALL is characterized by lower percentages of the favorable genetic features, such as ETV6-RUNX1 and hyperdiploidy, that occur frequently in childhood ALL.6,10,18 A recent review of the iBFM cytogenetic register found that these favorable cytogenetic features were less common in children with DS-ALL compared with non–DS-ALL, although the differences did not reach conventional levels of statistical significance.11
To further investigate how the genetics of DS-ALL compare with that of non–DS-ALL and determine whether any differences in lymphoblast cytogenetics might influence treatment outcome, we compared the distribution of clinical and genetic features in a large cohort of 2811 B-precursor ALL patients enrolled on COG P9900 for classification and remission induction treatment, which included 80 (2.8%) children with DS-ALL. Samples for these patients were collected and analyzed prospectively. All patient samples underwent standardized, validated testing to detect the presence of BCR-ABL1, TCF3-PBX1, MLL, and ETV6-RUNX1 translocations, tumor cell ploidy or DNA index, FISH testing for trisomy of chromosomes 4 and 10, and detection of MRD via flow cytometry.
There were no differences in sex, age, presenting WBC count, or NCI risk group between children with DS-ALL and non–DS-ALL, but those with DS-ALL were more likely to be white. We identified no DS patients with T-ALL, similar to previous studies, which reported a very low incidence of T-ALL in DS-ALL.3,4,6,11,18 Among the B-precursor ALL patients, there were substantial differences in the frequency of sentinel cytogenetic lesions. Taken together, ETV6-RUNX1 translocations or simultaneous trisomy of chromosomes 4 and 10, both of which are associated with a favorable prognosis, occurred in almost half (48%) of the children with non–DS-ALL compared with only approximately 10% of children with DS-ALL. Similar to the significantly lower rates of trisomy 4 and 10, children with DS-ALL had a significantly lower rate of high hyperdiploidy (defined by a DNA index of > 1.16) compared with those with non–DS-ALL (5% vs 24%; P < .001). Among children with a DNA index greater than 1.16, similar percentages of DS-ALL and non–DS-ALL had double trisomy 4 and 10 in COG P9900. To expand this comparison, we reviewed data from children enrolled in the current COG SR-ALL trial AALL0331, which includes determination of DNA index and FISH screening of all patients for trisomies 4, 10, and 17. Among 4365 patients enrolled in COG AALL0331 since 2005, 70% (7 of 10) of DS and 78.3% (1016 of 1297) of non–DS-ALL patients with a DNA index greater than 1.16 had trisomy 4 and 10 (P > = .460) and 60% (6 of 10) of DS and 66% (856 of 1297) of non–DS-ALL patients had trisomy 4, 10, and 17 (P > .742). Thus, we conclude that similar percentages of DS and non–DS-ALL patients with hyperdiploidy have favorable chromosome trisomies. Our study also confirmed previous reports showing a notable absence of high-risk cytogenetic features among children with DS-ALL, including MLL translocations and BCR-ABL1.4,6,7,10,11 However, in contrast to prior studies, we found an equivalent number (3.6% vs 4.3%) of TCF3-PBX1 translocations in DS-ALL and non–DS-ALL.4,6
The low rate of favorable sentinel cytogenetic lesions in DS-ALL is similar to results from the CCG 1952 SR-ALL trial that did not find any ETV6-RUNX1 translocations or high hyperdiploidy (> 51 chromosomes) among 57 cases of DS-ALL.10 However, that study was limited in that only 44% and 37% of DS-ALL cases had samples analyzed for ETV6-RUNX1 fusion or had successful karyotype analysis, respectively, which may have altered the true incidence of these 2 features in their DS cohort. Our findings are also consistent with a recent report of 6 consecutive Italian Association of Pediatric Hematology and Oncology-ALL trials that found the ETV6-RUNX1 fusion transcript in only 1 of 44 (2.2%) children with DS-ALL.18
However, a large study of 215 DS-ALL patients from the iBFM cytogenetic register, 109 of whom had clonal cytogenetic aberrations detected, showed that 10% of children with DS-ALL had ETV6-RUNX1 translocations and 11% had high hyperdiploidy, defined in that study as 51 to 57 chromosomes.11 The comparison group, a control group from the Mitelman Database of Chromosome Aberrations in Cancer, had incidences for ETV6-RUNX1 and high hyperdiploidy of 25% and 23% in non–DS-ALL, respectively. Although there was a clear trend for decreased incidence of favorable genetic subtypes among DS-ALL patients, the differences in the incidence of these 2 favorable cytogenetic features between the DS-ALL and non–DS-ALL groups were not statistically significant. The authors postulated that the true frequency of t(12;21) in DS-ALL might be higher than the 10% they observed, as not all the DS-ALL patients in their register were screened for this abnormality.
Because our study and the iBFM register study did not analyze the same genetic features of high hyperdiploidy, it is challenging to know whether or not there are real differences in incidence between the 2 studies. We found that 5% of DS-ALL patients had a DNA index greater than 1.16 and 7.7% had simultaneous trisomies of chromosomes 4 and 10, whereas Forestier reported that 11% of DS-ALL patients had 51 to 57 chromosomes.11 Given the relatively small numbers of patients analyzed in each group, it appears that both studies find approximately similar percentages of DS-ALL patients to have hyperdiploidy. Overall, simultaneous trisomies of 4 and 10 are found in approximately 80% of hyperdiploidy ALL in COG ALL protocols.19 The differences in the incidence of ETV6-RUNX1 fusion in DS-ALL in the 2 reports seemed to be more striking, with 2.5% in our study and 10% in the iBFM registry. To further explore this question, we reviewed data from children with SR-ALL enrolled in the current COG SR-ALL trial AALL0331, which includes FISH screening of all patients for ETV6-RUNX1. Among 4365 patients enrolled in COG AALL0331 since 2005, the incidence of ETV6-RUNX1 fusion was 30.5% (1296 of 4247) in non–DS-ALL patients compared with 11.9% (14 of 118) in DS-ALL patients (P < .001). The difference in the rate of ETV6-RUNX1 fusion in DS SR-ALL patients between P9900 and AALL0331 (3.5% vs 11.9%) was not the result of a change in ascertainment technique (reverse-transcribed polymerase chain reaction in P9900 and FISH in AALL0331) because the incidence of this lesion in the non-DS SR-ALL patients was almost identical in the 2 studies (P9900, 30.1%; AALL0331, 30.5%). Thus, with screening of thousands of consecutively enrolled patients, we continue to observe a significantly lower incidence of ETV6-RUNX1 fusion in patients with DS-ALL compared with ALL patients without DS, but the incidence in the current trial is similar to that reported by Forestier et al.11
Outcomes in DS-ALL have generally been inferior to those of non–DS-ALL in the majority of published series.2,5-7,20 However, as treatments have improved, EFS and OS for the children with DS-ALL have approached those for non–DS-ALL.7,10 We also examined early response and treatment outcome and analyzed how these differed between patients with and without DS. We found that the complete remission rates for DS-ALL and non–DS-ALL were similar, consistent with prior reports from the Pediatric Oncology Group, iBFM, United Kingdom Acute Lymphoblastic Leukemia, and CCG but different from that reported by Whitlock et al.6,7,10,18 We found that there was a modest increase in the rate of DS-ALL patients with higher levels of MRD in PB at day 8, but that day 29 BM MRD distributions were similar between children with and without DS. Without correction for good- and poor-risk sentinel cytogenetic lesions, the COG P9900 DS-ALL patients had a statistically significantly lower OS and an inferior EFS, approaching statistical significance. However, when patients with the prognostically significant BCR-ABL1, MLL, and ETV6-RUNX1 translocations and trisomies 4 and 10 were excluded, the 5-year EFS and OS for DS-ALL and non–DS-ALL were comparable. Thus, the inferior outcome of DS-ALL is attributable in large part to a much lower incidence of the common favorable genetics subsets ETV6-RUNX1 and trisomies 4 and 10 (48% vs 10% in the COG P9900 trials).
Recently, important new insights have been made into the pathogenesis of DS-ALL that complement findings from the current study. Several groups have reported that lymphoblasts of 50% to 60% of children with DS-ALL overexpress the cytokine receptor CRLF2 caused by either a targeted interstitial deletion of the pseudoautosomal region of the X/Y chromosome that juxtaposes the coding region of CRLF2 with the noncoding first exon of the P2RY8 gene, or a cryptic translocation that fuses CRFL2 with the immunoglobulin heavy chain gene (IgH).21,22 In contrast, CRLF2 activating genetic lesions are only detected in 7% to 14% of non–DS-ALL patients, and the relative frequencies of the activating genetic lesions seem reversed, with IgH-CRLF2 translocations much more common in non–DS-ALL and P2RY8-CRLF2 fusion much more common in DS-ALL.21-23 Previously, JAK2 mutations were reported to occur in 20% to 30% of DS-ALL cases.24-27 The recent reports from Mullighan et al21 and Hertzberg et al22 establish that the JAK mutations occur exclusively in DS-ALL cases with CRLF2 genetic lesions and that these 2 lesions cooperative to produce growth factor independence in vitro. Ultimately, understanding the different spectrum of underlying genetic lesions that occurs in DS-ALL may permit more tailored therapy for children with DS-ALL and the minority of children with non–DS-ALL that have these similar genetic lesions. It is well established that children with DS have a 10- to 15-fold increased incidence of ALL compared with the general population, but the current data as well as previous analyses indicate that this risk varies for certain biologic subtypes and is distinctly different compared with ALL in non-DS-ALL children. Thus, the presence of constitutional trisomy 21 appears to facilitate preferentially the subsequent genetic events operative in nonfavorable leukemia subtypes, such as those containing CRLF2 genomic lesions and/or JAK2 mutations but lacking trisomy 4 and 10 and ETV6-RUNX1.
In conclusion, we report the largest prospective study of DS-ALL patients whose leukemic biologic features were evaluated in a comprehensive manner. We found significantly fewer ETV6-RUNX1 translocations, trisomies of 4 and 10, and high hyperdiploidy as defined by DNA index more than 1.16 in our DS-ALL cohort than in the concurrent non–DS-ALL cohort. After correction for the differing incidences of these sentinel chromosomal features, the EFS and OS are equivalent for DS-ALL and non–DS-ALL. These results support previous observations that DS-ALL has a distinct biology and pathogenesis from that of non–DS-ALL.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (grants CA98543, CA114766, and CA086011). S.P.H. is the Ergen Family Chair in Pediatric Cancer.
National Institutes of Health
Authorship
Contribution: K.W.M. analyzed and interpreted data and wrote the manuscript; W.L.C. analyzed data and wrote the manuscript; A.J.C. and J.P. performed research, analyzed data, and wrote the manuscript; M.D. designed and analyzed data and provided statistical support; M.J.B. and C.L.W. performed research and reviewed the manuscript; P.L.M. conducted clinical trials and reviewed the manuscript; J.A.W. and S.P.H. analyzed data and wrote the manuscript; N.J.W. conducted clinical trials, analyzed data, and wrote the manuscript; and B.M.C. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: J.A.W. is a consultant to the Electric Power Research Institute. The remaining authors declare no competing financial interests.
Correspondence: Kelly W. Maloney, Center for Cancer and Blood Disorders, The Children's Hospital, 13123 E 16th Ave, B115, Aurora, CO 80045; e-mail: Maloney.kelly@tchden.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal