In this issue of Blood, Rickles et al conducted a high-throughput screen to identify combinations of known drugs looking for synergy to enhance toxicity in myeloma cells.1 This screen identified the novel combination of phosphodiesterase inhibitors and adenosine receptor agonists that induced cell death in myeloma cells preferentially over normal cells.
Known drugs with previously unrecognized anticancer activity can be rapidly advanced into clinical trial for this new indication by leveraging their prior pharmacology and toxicology data. A classic example of this approach is the repositioning of thalidomide for the treatment of myeloma2 and myelodysplasia,3 but there have been multiple other successes. For example, the antifungal ketoconazole has clinical use in the treatment of metastatic prostate cancer.4 Recently, the broad spectrum antiviral ribavirin was found to suppress oncogenic transformation by disrupting the function and subcellular localization of the eukaryotic translation initiation factor eIF4E.5 In a phase 1 dose escalation study in patients with relapsed or refractory M4/M5 acute myeloid leukemia (AML), there were 1 complete remission and 2 partial remissions, suggesting that ribavirin may be efficacious for the treatment of this disease.6
To date, drug repositioning has been largely serendipitous, but some recent studies, including the report by Rickles et al,1 have taken a more systematic approach by compiling and screening large libraries of compounds. In this issue, Rickles et al have advanced the drug-repositioning screening approach 1 step further. They have used high-throughput screening to identify novel combinations of available drugs that when used together act synergistically to produce an antimyeloma effect. By identifying combinations of known drugs, the timeline to complete a phase 1 combination study is much quicker than what is required for a new, unapproved agent (see figure).
The size of the combination screening effort described in this article was impressive. Although only 2841 unique combinations were tested, a full 6 × 6 matrix of drug concentrations was tested resulting in a screen with over 100 000 assays per cell line tested. Moreover, almost 650 combinations were tested with this matrix in 4 cell lines. From this large-scale screening effort synergistic combinations were identified by statistical analyses. The primary goal of the screen was to detect known drugs that acted synergistically with dexamethasone to enhance cell death in myeloma. From their screens, Rickles et al identified the phosphodiesterase (PDE) inhibitor papaverine and the adenosine receptor agonist chloro-IB-MECA. Both acted synergistically with dexamethasone to induce cell death in myeloma cells. The authors have indicated that follow-up studies are planned to further evaluate various combinations of these agents with dexamethasone. However, this manuscript focused on the synergistic combination of phosphodiesterase inhibitors and adenosine receptor agonists in myeloma. Subsequent studies demonstrated that combinations of inhibitors of PDE2, PDE3, and PDE7—along with A2A adenosine agonists—acted synergistically in myeloma. Interestingly, while synergistic in myeloma and lymphoma cell lines, this combination was minimally cytotoxic to leukemia or solid tumor cell lines. Furthermore, the combination was preferentially cytotoxic to primary myeloma cells versus normal hematopoietic cells, suggesting a therapeutic window.
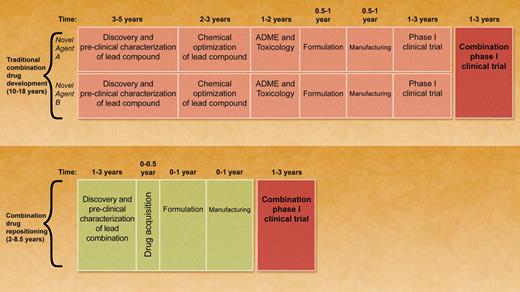
A comparison of traditional combination drug development versus combination drug repositioning. In traditional combination drug development, novel agent 1 and novel agent 2 must be developed in parallel as single agents. Each compound requires independent discovery, preclinical characterization, absorption, distribution, metabolism, and excretion profiling, and toxicology testing. Suitable compounds require formulation and manufacturing. Phase 1 clinical testing is performed in parallel for each novel agent. Combination phase 1 clinical trials can only be conducted after each agent is independently tested in its own phase 1 trial. The timeline to complete the combination phase 1 trial is approximately 10 to 18 years. In combination drug repositioning, known drugs that act synergistically are identified and evaluated in preclinical studies. Drugs are then acquired for clinical testing. New formulations and manufacturing may be required. By levering the prior toxicology and pharmacology of the compounds, a phase 1 combination trial can be rapidly initiated. The timeline to complete the combination phase 1 study using this approach is approximately 2 to 8.5 years. Professional illustration by A. Y. Chen.
A comparison of traditional combination drug development versus combination drug repositioning. In traditional combination drug development, novel agent 1 and novel agent 2 must be developed in parallel as single agents. Each compound requires independent discovery, preclinical characterization, absorption, distribution, metabolism, and excretion profiling, and toxicology testing. Suitable compounds require formulation and manufacturing. Phase 1 clinical testing is performed in parallel for each novel agent. Combination phase 1 clinical trials can only be conducted after each agent is independently tested in its own phase 1 trial. The timeline to complete the combination phase 1 trial is approximately 10 to 18 years. In combination drug repositioning, known drugs that act synergistically are identified and evaluated in preclinical studies. Drugs are then acquired for clinical testing. New formulations and manufacturing may be required. By levering the prior toxicology and pharmacology of the compounds, a phase 1 combination trial can be rapidly initiated. The timeline to complete the combination phase 1 study using this approach is approximately 2 to 8.5 years. Professional illustration by A. Y. Chen.
The high-throughput nature of this study and the large number of combinations analyzed with statistical rigor has advanced the field of drug repositioning. Using their unbiased high-throughput approach, the authors have identified combinations that are active in myeloma. Using traditional drug-discovery approaches, it is highly unlikely that the combination of PDE inhibitors and adenosine analogues would have been tested in combination with each other or with dexamethasone. As the authors indicate in their article, their screening platform also identified additional synergistic combinations active in myeloma that are also being pursued by this group.
The results from this study also open several opportunities for additional investigation. The authors have demonstrated that the combination of PDE inhibitors and adenosine agonists increases cyclic AMP (cAMP) above either agent alone, but additional studies will help clarify whether this increase in cAMP is functionally important for the cytotoxicity of the combination. Moreover, it is unclear why increasing cAMP is preferentially cytotoxic to myeloma and lymphoma cell lines and primary samples. Additional studies that answer these questions would be very interesting and provide further rationale to advance this combination into clinical trial. As part of these additional mechanistic studies, subpopulations of myeloma patients most likely to benefit from this combination could be identified.
The prior toxicology and pharmacology of PDE inhibitors and adenosine analogues suggest that this combination could be rapidly advanced into clinical trial for patients with myeloma. The rationale for this type of clinical study would be enhanced by demonstrating activity in myeloma xenografts. Such animal studies would help address the potential toxicity of this combination.
Finally, this article highlights the general applicability of a high-throughput approach to identify novel combinations. As an extension of this study, one could test the library compiled by Rickles et al, in a similar matrix screen, to identify unexpected synergistic combinations that are active in malignancies other than myeloma. Alternatively, as part of the development of a novel agent for the treatment of myeloma or other hematologic malignancies, one could conduct a focused screening effort to identify agents that act synergistically with the new agent. Rather than testing combinations manually, this rapid high-throughput approach can be used to increase the number of combinations tested.
Thus, in summary, Rickles et al have described a novel and unexpected combination of drugs that act synergistically in myeloma. Furthermore, they offer a path into the future for high-throughput combination drug discovery.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal