Abstract
BCR-ABL negative myeloproliferative neoplasms (MPNs; polycythemia vera, essential thrombocythemia, primary myelofibrosis) are malignant diseases arising from a multipotent hematopoietic progenitor, frequently altered by JAK2 V617F or other JAK/STAT activating mutations. The thrombopoietin receptor (TpoR, MPL) is one of the major dimeric cytokine receptors that use JAK2 in the myeloid lineage, and was found to be down-modulated in certain MPN patients. We searched for negative regulators of MPL expression. Here we report that miR-28 targets the 3′ untranslated (3′UTR) region of MPL, inhibiting its translation, as well as other proteins potentially involved in megakaryocyte differentiation, such as E2F6. Expression of miR-28 in CD34-derived megakaryocytes inhibited terminal differentiation. miR-28 was found to be overexpressed in platelets of a fraction of MPN patients, while it was expressed at constant low levels in platelets from healthy subjects. Constitutive activation of STAT5 leading to autonomous growth of hematopoietic cell lines was associated with increased miR-28 expression. We discuss how down-modulating MPL and other targets of miR-28, and of related miR-708 and miR-151, could contribute to MPN pathogenicity.
Introduction
The BCR-ABL negative myeloproliferative neoplasms (MPNs), polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF) are malignant diseases arising from a mutant multipotent hematopoietic stem cell (HSC),1,2 that are associated with constitutively active JAK-STAT signaling. The JAK2 V617F mutation is present in 95% of PV and 50% of ET and PMF.3-6 JAK2 exon 12 mutations are found in a minority of PV patients.7 5% of PMF and 1% of ET harbor thrombopoietin receptor (TpoR, MPL) W515 mutations,8,9 which constitutively activate JAK2 signaling.10
Moliterno et al have reported diminished platelet MPL expression in MPNs11 and an inverse correlation between JAK2 V617F allele burden and MPL expression, although down-modulation of MPL was observed also in JAK2 V617F-negative patients.12 This suggests that mechanisms that limit the expression, surface localization, and function of MPL might operate during the establishment of MPNs.
We searched for putative microRNAs13 that target the 3′UTR of the MPL mRNA. We found that miR-28 is an inhibitor of MPL translation, which is also the case for 2 close relatives of miR-28, miR-151 and miR-708. We identified several miR-28 targets, besides MPL, such as E2F6, a transcription factor involved in the control of proliferation and apoptosis, and the MAP-kinase MAPK1/ERK2. We then detected induction of miR-28 in cell lines transformed by JAK2 V617F or activated MPL mutants. We have also investigated levels of expression of miR-28 in platelets from healthy subjects and MPN patients.
Methods
Cell lines, plasmid/luciferase constructs, and reagents
Human erythroleukemia (HEL), JAK2-deficient cell γ2A, UT-7, Mo-7e, and Ba/F3 cells were maintained as described.14-18 The UKE-1 cell line was a kind gift of Dr Walter Fiedler (University Hospital Eppendorf, Hamburg, Germany). Translational inhibition by microRNAs was measured using the psi-CHECK-2 reporter vector (Promega). Briefly, 3′UTR specific primers extended with adaptor sequences corresponding either to a NotI or XhoI site were used in a polymerase chain reaction (PCR). PCR products were cloned in the TOPO vector (Invitrogen) and positive clones were digested by NotI and XhoI and ligated in the digested psi-CHECK-2 vector. After sequencing, the microRNA target sites were mutated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer's instructions. These plasmid constructs (0.8 μg) were transfected with 40nM pre-miR (Ambion) in γ2A cells using Lipofectamine (Invitrogen). Luciferase activities were assayed 48 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega).
Pri-miRs were amplified with pri-miR-28 primers (28s GGCAACATCTAAATATGGCTTG; 28a TGAGGTAGGCAGTATTAGCTCTGA), pri-miR-708 primers (708s GAAACCTAACCCCCATGGTT; 708a TAGAGGGTCCTCAGGTGGTG) and pri-miR-151 primers (151s AGCTGAGCCTGGTGCTAGTC; 151a ATTCAGTGCCTGGGTGACTC), cloned in Topo TA vector (Invitrogen), digested by EcoRI and ligated in pMEGIX-IRES–green fluorescent protein (GFP). All constructs were verified by sequencing. Retroviral transduction were performed as described.19 The dominant negative STAT5 (STAT5AΔ749; originally described by Dr T. Kitamura, University of Tokyo) was cloned in pREX-IRES-CD2 and used for Ba/F3 TpoR JAK2 V617F or HEL cells electroporations. JAK2 inhibitors were from Calbiochem (JAK inhibitor 1), Astra Zeneca (AZD1480), Sigma-Aldrich (AG490) and SYN thesis med chem (TG101209).
Patient samples
Platelets were isolated from MPN patients of Hôpital Henri Mondor, Paris, France, and from St Luc Hospital, Brussels, Belgium. Blood samples were collected with approval of all participating institutions' ethical committees and informed consent was obtained in accordance with the Declaration of Helsinki. The JAK2 V617F and MPL W515 status was determined by TaqMan PCR and sequencing as described.12
Real-time quantitative PCR
Total RNA, including miRNA and other small RNA molecules, was isolated using TRIzol (Invitrogen) and subsequently purified with the miRNeasy kit (QIAGEN) following the manufacturer's instructions. Stem-loop reverse transcription and quantitative real-time PCR have been performed as described,20 using the following RT-Primers: GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACC TCA AT (RT-miR28); GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA AAA ATA TG (RT-U6); Forward primers: CGTCAAGGAGCTCACAGTCTA (FW-miR28); AAATTCGTGAAGCGTTCCAT (FW-U6); the universal reverse primer GTGCAGGGTCCGAGGT (RC); and the miR-28 probe 5′(6-FAM)-ACTGGATACGACCTCAA-3′(BHQ1). Quantities of miR-28 are normalized to U6 levels in each sample and are depicted as relative quantities to the normalized miR-28 levels of a control cell line, that is, UT7, Mo7e, Ba/F3, or HEL, as indicated on the y-axis.
Expression analysis of the LPP gene was performed by real-time PCR with the following primers: LPPs GTGCAATGTGTGTTCCAAGC; LPPa TGGCATAATAGGCTCCTTGC. Data were normalized to β-actin amplified with the following primers: ACTBs CCTGGCACCCAGCACAAT; ACTBa GGGCCGGACTCGTCATACT.
5′-Rapid amplification of cDNA ends
Amplification of 5′ ends of LPP mRNAs was performed using the SMART RACE cDNA Amplification Kit protocol (Clontech) and LPP gene specific primers: LPP-GSP1 TGGCAGCCAAGATGGGTGAGACATT and LPP-GSP2 CCTCTGATGAGCGTCCAGTGGAACA. The PCR products were analyzed on a 1.2% agarose gel. PCR bands were excised and purified with the QIAquick gel extraction kit (QIAGEN). Purified DNAs were cloned with the TOPO TA cloning kit (Invitrogen) and 8 positive clones from each transformation were sequenced.
Chromatin immunoprecipitation
Chromatin cross-linking, cell lysis, and DNA shearing were performed as described.21 Immunoprecipitation was performed with the OneDay chromatin immunoprecipitation (ChIP) kit (Diagenode), as recommended by the manufacturer. Antibodies for immunoprecipitation of STAT5B and RNApolII were purchased from Millipore (no. 06-969) and Diagenode (no. AC-055-100), respectively. Primers for ChIP qPCR analysis were designed with the primer3 software (http://frodo.wi.mit.edu/primer3/) and tested for amplification efficiency and specificity on genomic DNA. Validated primers were used for real-time PCR on purified DNA after ChIP. The primers used for qPCR are ProAs TTGAGCACAGGACAGAGGAA; ProAa TTTTAGCCCTGAGCCTTGAA; Pro1s TTGCATTCTGGATGGTCTCA; Pro1a AGGCTGGAGGATGTCAGAAA; 3's AAAAGCAAACCTTGCCTGAA; 3′a TGAAGAGAGCCAATGAACGA. Primers for actin were identical to those described in “Real-time quantitative PCR,” and the PCR product does not contain any putative STAT-binding sites.
Retroviral transductions
Retroviruses produced after BOSC packaging cells transfections were used to infect Ba/F3 cells as described.19 GFP positive cells were sorted 72 hours after infection. Cell numbers were recorded with a Coulter cell counter.
CD34 cultures and megakaryocyte differentiation in liquid culture
Liquid cultures using human CD34+ cells were retrovirally transduced with bicistronic viruses coding for miR-28, empty vector (pMegix), and GFP. Cells were maintained in serum-free medium containing Tpo and proplatelet counting was performed on GFP/CD41 double positive megakaryocytes, as described.22 Megakaryocyte micrographs (×40 objective) were taken with a Nikon TE300 microscope at 37°C in serum-free medium containing Tpo. Photos were taken with an AxioCamMR camera and processed with AxioVisionRel.4.6 (Carl Zeiss).
Western blotting
MPL protein levels were evaluated in Mo7e cell using the Anti-TpoR/c-Mpl antibody (Millipore; 06-944). Cells were harvested, washed in cold PBS, and resuspended in cold NP40 Lysis Buffer (50mM Tris-Cl pH8, 150mM NaCl, 1% NP40, 0.1mM EDTA, 10mM NaF, 1mM Na3VO4, 1mM PMSF, 1mM DTT). Upon incubation on ice for 30 minutes with occasional vortexing, lysates were centrifuged for 20 minutes at 20 000g and 4°C. Supernatants were collected and protein quantification were performed with the BCA Protein Assay Kit (Pierce). Protein extracts (50 μg) were loaded on NuPage 4-12% Bis-Tris gels (Invitrogen) and Western blot analysis was performed using a 1:1000 MPL antibody and 1:10 000 dilution anti–rabbit-horseradish peroxidase antibodies (GE Healthcare UK). The membranes were stripped and reprobed with anti-actin beta (monoclonal anti-ACTB; Sigma-Aldrich).
Results
The MPL 3′UTR is a target of miR-28
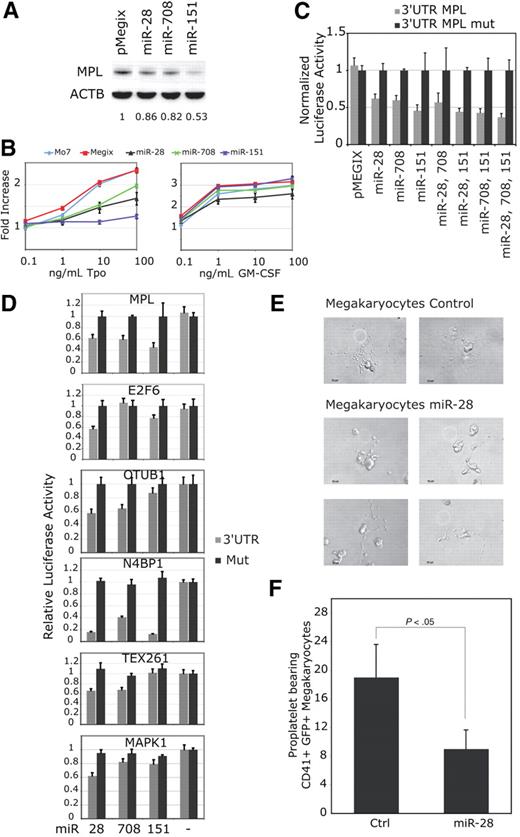
Five potential microRNA targets were identified with the miranda (http://www.microrna.org) and miRBase (http://microrna.sanger.ac.uk) programs in the 3′UTR MPL mRNA, starting at nucleotides 2157; 2531; 2711; 2836 and 3524. The MPL 3′UTR was cloned downstream the luciferase coding region of a reporter vector and used to test the ability of microRNAs to inhibit translation. None of the let-7 family (let-7a, c, d, e, f and miR-98) pre-miRs (precursor of microRNA) inhibited translation (Figure 1A). miR-28 inhibited the luciferase activity by approximately 30%, compared with the activity of a control microRNA, that does not recognize any potential target. Because miR-28 could potentially bind to 2 distinct sites (starting at nucleotides 2157 and 3524, respectively), we performed mutagenesis of each of these sites (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We observed that mutagenesis of nucleotides 3530-3542 relieved the inhibition of luciferase activity by miR-28, while mutagenesis of the region encompassing nucleotides 2168-2174 did not. Thus, miR-28 inhibited luciferase expression by binding to MPL 3′UTR nucleo-tides 3524-3545.
miR-28 recognizes the MPL 3′UTR and inhibits its translation. (A) γ2A cells were cotransfected either with 0.8 μg MPL 3′UTR luciferase reporter (gray histograms) or MPL 3′UTR luciferase reporter mutated for the site of interest (black histograms) and either 40nM Pre-miR-28, Pre-let-7a, 7c, 7d, 7e, 7f, Pre-miR-98 or Pre-miR-control. Values represent the mean luciferase activity ± SD of 3 independent experiments relative to Pre-miR-control transfected cells. The 1680 nt MPL 3′UTR sequence was cloned after the stop codon of the luciferase and is represented with the mRNA coordinates of the putative miR binding sites (gray boxes). (B) Mo7e cells were transduced with a bicistronic retroviral vector expressing miR-28 together with the green fluorescent protein (GFP). After sorting for equivalent GFP levels, cell lines expressing either the empty vector (pMegix) or the miR-28 expressing vector (miR-28) were subjected to Western blotting for MPL protein levels. The histogram reports MPL relative bands intensities measured on Western blot.
miR-28 recognizes the MPL 3′UTR and inhibits its translation. (A) γ2A cells were cotransfected either with 0.8 μg MPL 3′UTR luciferase reporter (gray histograms) or MPL 3′UTR luciferase reporter mutated for the site of interest (black histograms) and either 40nM Pre-miR-28, Pre-let-7a, 7c, 7d, 7e, 7f, Pre-miR-98 or Pre-miR-control. Values represent the mean luciferase activity ± SD of 3 independent experiments relative to Pre-miR-control transfected cells. The 1680 nt MPL 3′UTR sequence was cloned after the stop codon of the luciferase and is represented with the mRNA coordinates of the putative miR binding sites (gray boxes). (B) Mo7e cells were transduced with a bicistronic retroviral vector expressing miR-28 together with the green fluorescent protein (GFP). After sorting for equivalent GFP levels, cell lines expressing either the empty vector (pMegix) or the miR-28 expressing vector (miR-28) were subjected to Western blotting for MPL protein levels. The histogram reports MPL relative bands intensities measured on Western blot.
To assess whether miR-28 inhibits translation of endogenous MPL, we transduced the human Mo7e cell line with a bicistronic retroviral vector expressing miR-28 along with the GFP. Mo7e cells express endogenous MPL and can proliferate in medium supplemented with MPL ligand thrombopoietin (Tpo). Cells infected with the empty retrovirus (pMegix) or miR-28 expressing retrovirus (miR-28) were sorted for equivalent GFP levels. We observed an inhibition of MPL translation by miR-28 (Figure 1B). Importantly, expression of miR-28 did not reduce the level of endogenous mRNA for MPL in Mo7e or in HEL cells (not shown).
miR-28 and sequence related microRNAs target the MPL mRNA and other mRNAs coding for proteins involved in proliferation and apoptosis
We identified 2 microRNAs related to miR-28 (paralogs) with the BLASTN program found on the miRBase Web site (http://microrna.sanger.ac.uk/). miR-151 has 80% sequence identity with miR-28, and miR-708 is 68% identical to miR-28 and 71% with miR-151. Mo7e cells were infected with bicistronic retroviral vectors coding for miR-28, -708 or -151 along with the GFP. After sorting for equivalent GFP levels, we monitored MPL protein levels (Figure 2A) and Tpo induced cell proliferation (Figure 2B). miR-28 and miR-708 were both inhibiting MPL translation by approximately 20%, while miR-151 inhibited it by 50%. miR-151 was also more potent that miR-28 or -708 for inhibition of Tpo mediated cell proliferation. Thus, miR-28 and sequence related miRs inhibit Tpo induced proliferation by MPL down-regulation. Furthermore, as expected from the functional data, all the miRs targeted the MPL 3′UTR for translational inhibition (Figure 2C). The combination of miR-28 with miR-151 and miR-708 did not result in significantly synergic inhibition of luciferase activity, consistent with the notion that they all target the same sequence in the 3′UTR of MPL (Figure 2C).
miR-28 and closely related miR-151 and miR-708 inhibit Tpo-dependent proliferation of Mo7e cells and target mRNAs coding for proteins involved in proliferation and apoptosis. (A) Mo7e cell lines transduced with a bicistronic retrovirus (pMegix) expressing the GFP protein along with indicated microRNAs. Western blot analyses for MPL and ß-actin (ACTB) protein levels were performed. Normalized MPL protein levels are indicated under the blots. (B) Mo7e cell lines expressing miR-28, miR-708 or miR-151 were grown in the presence of indicated amounts of Tpo or GM-CSF (granulocyte macrophage–colony-stimulating factor) for 4 days. Cell proliferation is represented as fold increase compared with cell lines grown without cytokines. Shown are averages of triplicates ± SD of 1 representative experiment of 3. (C-D) 3′UTRs ( ) of indicated mRNAs or 3′UTRs mutated for miR-28 target sites (■) were cloned after the stop codon of the renilia luciferase in the psi-CHECK-2 reporter vector. These luciferase reporter vectors were cotransfected with miR expressing vectors. (E-F) CD34+ hematopoietic progenitors were infected with pMegix bicistronic retrovirused coexpressing miR-28 with the GFP or the control retrovirus expressing only the GFP. After 14 days of culture, proplatelet-bearing CD41/GFP-positive megakaryocytes were counted. Shown in panel E are numbers of proplatelet-bearing CD41/GFP positive megakaryocytes (average of triplicates + SD of 1 representative experiment).
) of indicated mRNAs or 3′UTRs mutated for miR-28 target sites (■) were cloned after the stop codon of the renilia luciferase in the psi-CHECK-2 reporter vector. These luciferase reporter vectors were cotransfected with miR expressing vectors. (E-F) CD34+ hematopoietic progenitors were infected with pMegix bicistronic retrovirused coexpressing miR-28 with the GFP or the control retrovirus expressing only the GFP. After 14 days of culture, proplatelet-bearing CD41/GFP-positive megakaryocytes were counted. Shown in panel E are numbers of proplatelet-bearing CD41/GFP positive megakaryocytes (average of triplicates + SD of 1 representative experiment).
miR-28 and closely related miR-151 and miR-708 inhibit Tpo-dependent proliferation of Mo7e cells and target mRNAs coding for proteins involved in proliferation and apoptosis. (A) Mo7e cell lines transduced with a bicistronic retrovirus (pMegix) expressing the GFP protein along with indicated microRNAs. Western blot analyses for MPL and ß-actin (ACTB) protein levels were performed. Normalized MPL protein levels are indicated under the blots. (B) Mo7e cell lines expressing miR-28, miR-708 or miR-151 were grown in the presence of indicated amounts of Tpo or GM-CSF (granulocyte macrophage–colony-stimulating factor) for 4 days. Cell proliferation is represented as fold increase compared with cell lines grown without cytokines. Shown are averages of triplicates ± SD of 1 representative experiment of 3. (C-D) 3′UTRs ( ) of indicated mRNAs or 3′UTRs mutated for miR-28 target sites (■) were cloned after the stop codon of the renilia luciferase in the psi-CHECK-2 reporter vector. These luciferase reporter vectors were cotransfected with miR expressing vectors. (E-F) CD34+ hematopoietic progenitors were infected with pMegix bicistronic retrovirused coexpressing miR-28 with the GFP or the control retrovirus expressing only the GFP. After 14 days of culture, proplatelet-bearing CD41/GFP-positive megakaryocytes were counted. Shown in panel E are numbers of proplatelet-bearing CD41/GFP positive megakaryocytes (average of triplicates + SD of 1 representative experiment).
) of indicated mRNAs or 3′UTRs mutated for miR-28 target sites (■) were cloned after the stop codon of the renilia luciferase in the psi-CHECK-2 reporter vector. These luciferase reporter vectors were cotransfected with miR expressing vectors. (E-F) CD34+ hematopoietic progenitors were infected with pMegix bicistronic retrovirused coexpressing miR-28 with the GFP or the control retrovirus expressing only the GFP. After 14 days of culture, proplatelet-bearing CD41/GFP-positive megakaryocytes were counted. Shown in panel E are numbers of proplatelet-bearing CD41/GFP positive megakaryocytes (average of triplicates + SD of 1 representative experiment).
To identify targets recognized by miR-28 other than MPL, we used the miRBase target prediction program and the BLASTn program against the Refseq mRNA database. Fourteen targets displaying ΔG ≤ -26 kcal/mol (the miR-28:MPL 3′UTR predicted free energy) were retained for further analysis. We next selected mRNAs expressed in blood cell lineages (http://genome.ucsc.edu). 5 targets were selected: N4BP1, OTUB1, TEX261, MAPK1 and E2F6. We tested using the psi-CHECK-2 luciferase reporter assay the ability of these microRNAs to inhibit translation of the 5 targets (Figure 2D). We observed an up to 80% inhibition of N4BP1 (NEDD4 binding protein 1), a regulator of the E3 Ubiquitin Ligase-Itch,23 by all 3 miRs. OTUB1 (Otubain1), an inhibitor of GRAIL (gene related to anergy in lymphocytes), and TEX261 (testis expressed 261) were both inhibited by miR-28 and -708 by approximately 40%. MAPK1 (mitogen-activated protein kinase 1 Transcript variant 1 also known as Extracellular regulated kinase 2-ERK2) was inhibited by miR-28 by 40%. E2F6 (E2F transcription factor 6) was inhibited by miR-28 and -151 by 40% and 20%, respectively. E2F6 is a negative regulator of E2F1 transcription factor involved in cell-cycle regulation and apoptosis.24 MPL and the last 2 targets are potentially important for megakaryocyte differentiation, since E2F1 was shown to impair megakaryocyte differentiation25 and MAPK1 was shown to be required for megakaryocyte differentiation.26
Effects of miR-28 expression on megakaryocyte proplatelet formation
To assess the functional consequences of miR-28 expression on megakaryocyte proplatelet formation, CD34+ hematopoietic progenitors were infected with bicistronic retroviruses coexpressing miR-28 and GFP, or empty vector and GFP, and then maintained in medium containing Tpo, as described.22 Megakaryocytes were examined after 14 days of culture. The number of proplatelet-bearing CD41/GFP positive megakaryocytes was significantly inhibited (> 50%) by miR-28 expression (Figure 2E-F). Although we did not coexpress JAK2 V617F and miR-28 in these experiments to determine the effect of miR-28 in megakaryocytes expressing JAK2 V617F, our data suggest that miR-28 exerts a negative role on megakaryocyte differentiation. It remains to be determined whether this effect is linked directly to the down-modulation of MPL or to the other targets we identified, such as E2F6 or MAPK1.
Constitutive STAT5 activation in transformed hematopoietic cells is associated with miR-28 expression
We found that the HEL cell line, which has been derived from a patient who relapsed with erythroleukemia after being treated for Hodgkin lymphoma,14 and the UKE-1 cell line derived from a patient with ET, that transformed to acute leukemia27 expressed high levels of miR-28 (Figure 3A); UKE-1 cells are homozygous for the JAK2 V617F mutation,6 while HEL cells have 8 copies of the JAK2 locus and are homozygous for JAK2 V617F28 (Figure 3A). Both HEL and UKE-1 cells express much higher levels of miR-28 than UT-7 cells (Figure 3A), which is a human megakaryocyte leukemia cell line that expresses very high levels of wild type JAK2.
Constitutive signaling by JAK2 V617F induces miR-28 expression. (A) miR-28 relative expression was measured in the HEL and UKE-1 cell lines, which are homozygous for JAK2 V617F and in UT-7 parental cell line, which is negative for JAK2 V617F. The JAK2 V617F and WT expression levels are reported under the histograms. (B) miR-28 relative levels were assessed in UT-7 parental cell line transduced with a bicistronic retrovirus expressing the GFP along with either JAK2 WT or JAK2 V617F and sorted for equivalent GFP levels. *P < .01. (C) Ba/F3 cells were stimulated with 20 ng/mL IL-3 for 3 hours. Ba/F3 EpoR and Ba/F3 TpoR cells coexpressing equivalent JAK2 WT or JAK2 V617F levels (sorted cells) were stimulated for 3 hours with 20 units/mL Epo or 20 ng/mL Tpo, respectively.
Constitutive signaling by JAK2 V617F induces miR-28 expression. (A) miR-28 relative expression was measured in the HEL and UKE-1 cell lines, which are homozygous for JAK2 V617F and in UT-7 parental cell line, which is negative for JAK2 V617F. The JAK2 V617F and WT expression levels are reported under the histograms. (B) miR-28 relative levels were assessed in UT-7 parental cell line transduced with a bicistronic retrovirus expressing the GFP along with either JAK2 WT or JAK2 V617F and sorted for equivalent GFP levels. *P < .01. (C) Ba/F3 cells were stimulated with 20 ng/mL IL-3 for 3 hours. Ba/F3 EpoR and Ba/F3 TpoR cells coexpressing equivalent JAK2 WT or JAK2 V617F levels (sorted cells) were stimulated for 3 hours with 20 units/mL Epo or 20 ng/mL Tpo, respectively.
To test whether miR-28 expression was indeed linked to JAK2 V617F expression, we infected the parental UT-7 cell line, negative for JAK2 V617F and miR-28 expression, with bicistronic retroviral vectors expressing either JAK2 WT or JAK2 V617F together with GFP. After sorting for equivalent GFP levels, JAK2 V617F mRNA represented only approximately 12% (11.88 ± 0.42) of the total JAK2 mRNA cell content. A similar increase (∼ 12%) in the level of JAK2 WT mRNA was observed in UT-7 cells engineered to overexpress JAK2 WT (UT-7 JAK2 WT; supplemental Figure 4). This limited overexpression of JAK2 V617F or JAK2 WT that we could attain is likely because UT-7 cells express very high endogenous JAK2 levels (ie, 5-fold higher than in UKE-1 cells). Nevertheless, miR-28 was expressed at 3-fold higher levels in JAK2 V617F UT-7 cell line, compared with the JAK2 WT cell line (Figure 3B). These data indicate that expression of the constitutive active JAK2 V617F leads to expression of miR-28.
Next, we stably transduced the erythropoietin receptor (EpoR) or MPL (TpoR) in Ba/F3 cells together with JAK2 WT or JAK2 V617F and sorted cells for receptor expression and for equivalent levels of JAK2 WT and JAK2 V617F. Ba/F3 cells are IL3-dependent murine proB cells.17 Neither IL-3, Epo or Tpo stimulations increased miR-28 expression in parental Ba/F3, Ba/F3-EpoR or Ba/F3-TpoR, respectively. Similarly, Ba/F3 EpoR JAK2 WT and Ba/F3 TpoR JAK2WT cell lines did not respond by miR-28 induction after cytokine stimulation. In contrast, the Ba/F3 EpoR JAK2 V617F and Ba/F3 TpoR JAK2 V617F cell lines, expressed 6- and 8-fold more miR-28, and these levels were significantly increased in a dose-dependent manner (not shown) by further cytokine stimulation (Figure 3C). Thus, miR-28 is specifically induced by the constitutive JAK2 V617F signaling.
The Ba/F3 cells coexpressing TpoR and JAK2 V617F were then selected for autonomous growth. As expected, miR-28 level further increased when the JAK2 V617F constitutive activity was enhanced (by 4-fold; not shown). The JAK2 inhibitors: AG490, JAK Inhibitor I, AZD1480 and TG101209 were all potent to reduce miR-28 expression (Figure 4A), suggesting that miR-28 expression is dependent on the catalytic activity of JAK2 V617F.
miR-28 is induced by JAK2 V617F, MPL W515 mutants, Bcr/Abl and STAT5 1*6. Ba/F3 cell lines expressing indicated proteins were tested for miR-28 relative expression. (A) JAK2 V617F human TpoR-Ba/F3 cells selected for autonomous growth were treated with the JAK2 inhibitors: AG490 (10μM); JAK Inhibitor I (0.5μM); AZD1480 (3μM), TG101209 (3μM) or vehicle only (-) for 24 hours (*P < .05; **P < .001). Absence of toxicity was demonstrated by Trypan blue staining. (B) Ba/F3 cells expressing the indicated human TpoR mutants, Bcr/Abl or STAT5 1*6 were selected for autonomous growth in absence of cytokines and tested for miR-28 expression (*P < .0001). (C) The indicated cell lines were electroporated either with an empty expression vector expressing the CD2 surface marker (pREX-CD2), or the same bicistronic expression vector coexpressing the STAT5DN (dominant negative) mutant with the CD2 surface marker. Electroporation levels were verified by FACS and the miR-28 expression was quantified by stem-loop qRT-PCR. The presented results are representative of 3 indpendent experiments.
miR-28 is induced by JAK2 V617F, MPL W515 mutants, Bcr/Abl and STAT5 1*6. Ba/F3 cell lines expressing indicated proteins were tested for miR-28 relative expression. (A) JAK2 V617F human TpoR-Ba/F3 cells selected for autonomous growth were treated with the JAK2 inhibitors: AG490 (10μM); JAK Inhibitor I (0.5μM); AZD1480 (3μM), TG101209 (3μM) or vehicle only (-) for 24 hours (*P < .05; **P < .001). Absence of toxicity was demonstrated by Trypan blue staining. (B) Ba/F3 cells expressing the indicated human TpoR mutants, Bcr/Abl or STAT5 1*6 were selected for autonomous growth in absence of cytokines and tested for miR-28 expression (*P < .0001). (C) The indicated cell lines were electroporated either with an empty expression vector expressing the CD2 surface marker (pREX-CD2), or the same bicistronic expression vector coexpressing the STAT5DN (dominant negative) mutant with the CD2 surface marker. Electroporation levels were verified by FACS and the miR-28 expression was quantified by stem-loop qRT-PCR. The presented results are representative of 3 indpendent experiments.
Similarly, when Ba/F3 were transformed to autonomous growth by expression of TpoR W515A/L/K or TpoR Δ5 (TpoR deleted in the juxtamembrane RWQFP motif that includes W515), which are constitutively active mutants of MPL,10 we observed an increase in miR-28 expression (Figure 4B).
Another means to induce cytokine-independent growth of Ba/F3 cells via constitutive STAT5 activation is to express the Bcr-Abl oncogene; indeed Bcr-Abl also increased miR-28 levels. Because JAK2 V617F, TpoR W515 mutants and Bcr-Abl are all activating STAT5, we then tested a constitutively active mutant of STAT5, STAT5 1*6, which contains 2 point mutations, H299R and S711F, that lead to strong constitutive activation.29 Cells expressing STAT5 1*6 displayed a 30-fold increase in miR-28 levels, indicating that STAT5 constitutive activation is linked to induction of miR-28 expression. Finally, we electroporated Ba/F3 TpoR JAK2 V617F cells, selected for autonomous growth, with a vector expressing a dominant negative mutant of STAT5 (STAT5DN) together with the CD2 surface marker. After monitoring CD2 surface expression by FACS, we observed an inhibition of miR-28 expression in STAT5DN expressing cells compared with control cells electroporated with an empty vector (Figure 4B). A similar decrease in miR-28 expression could be noted when the STAT5DN was electroporated in HEL cells (Figure 4C). These results demonstrate that miR-28 expression is dependent on constitutive STAT5 activity induced by JAK2 V617F.
The constitutive STAT5 activity in transformed cells is inducing miR-28 host gene transcription
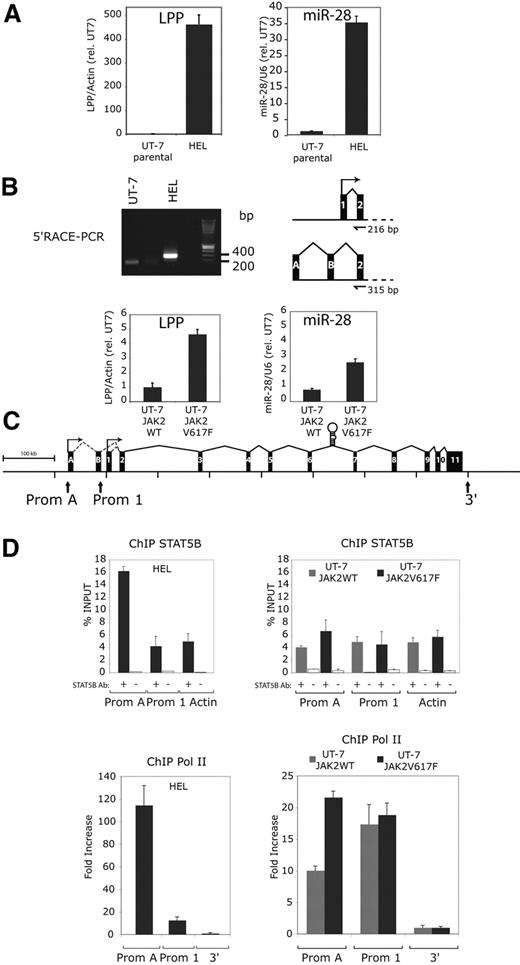
miR-28 is encoded by the sixth intron of the LIM domain lipoma-preferred partner (LPP) gene. This gene was first isolated as part of a fusion protein created by chromosomal translocations in lipomas and certain pulmonary chondroid hamartomas.30,31 Chromosomal rearrangements were also discovered with the mixed lineage leukemia (MLL) gene in secondary acute leukemia (sAML). The breakpoint t(3;11)(q28;q23) occurred in MLL and LPP intron 8.32 The physiologic function of LPP is unknown. Because miR-28 is expressed at 35-fold higher levels in HEL than in UT-7 cells (Figure 5A), we tested whether the LPP gene was expressed at higher levels in HEL versus UT-7 cells, in the scenario that miR-28 expression is induced via expression of its host gene, and not driven by an internal promoter. Indeed, LPP levels are dramatically increased in HEL, compared with UT-7 cells (Figure 5A). To localize the promoter of the LPP gene and therefore of miR-28 in HEL cells, we performed 5′Rapid Amplification of cDNA ends (RACE) PCR on HEL and UT-7 cells (Figure 5B). While we detected the expected 200 bp band in UT-7 cells (according to the UCSC Genome Browser, http://genomeusc.edu), a larger band (around 300-400 bp) was detected in HEL cells. After cloning of PCR products and sequencing of multiple clones, this 300- to 400-bp band was found to contain the LPP exon 2 and 2 alternative exons (A and B) of 94 bp and 119 bp located 59 kb and 4.7 kb upstream the regular transcription start site, respectively, amounting to 315 bp (Figure 5B). Thus, LPP transcription appears to involve, at least in HEL cells, an alternative upstream promoter, compared with the normal LPP transcription in UT-7 cells. Furthermore, expressing JAK2 V617F in UT-7 cells also induced an increase in LPP mRNA levels, in parallel with induction of miR-28 expression (Figure 5B).
LPP, the miR-28 host gene, is overexpressed in HEL cells through the transcriptional activation of an upstream alternative promoter bound by STAT5. (A) Relative expression of the LPP (LIM domain lipoma-preferred partner) transcript and miR-28 in the indicated cell lines. The LPP level in UT-7 parental and UT-7 JAK2 WT cell lines have been arbitrarily set at 1. (B) 5′-Rapid amplification of cDNA Ends (RACE)–PCR: a nested PCR on cDNA integrating an adaptor sequence at the 5′end of the LPP transcript amplified a 200-bp band in UT-7 and a larger 300- to 400-bp band in HEL cell lines. After sequencing, the 216-bp band amplified from UT-7 contains exon 1 and 2 of the LPP gene. The 300- to 400-bp band amplified from HEL contains 2 alternative exons (A and B) and exon 2 of the LPP gene. (C) A schematic view of the human LPP gene (LIM domain lipoma-preferred partner) represented with its 11 exons (black boxes), the 2 alternative exons (A and B) and the pri-miR-28 stem loop sequence in intron 6. The 2 identified promoters are indicated by arrows and labeled as Prom A (alternative) and Prom 1 (normal start site). (D) Quantitative PCR on indicated targets after chromatin immunoprecipitation (ChIP) relative to the starting DNA quantity before immunoprecipitation (% INPUT ± SD). Gray and black histograms represent ChIP performed on JAK2 WT and V617F UT-7 cells or UT-7 and HEL cells, respectively. White histograms are negative controls. The actin gene amplification (ACTB) was used a negative control for STAT5B binding. Quantitative PCR for RNA pol II ChIP were normalized against a non transcribed region located outside (3′) of the LPP gene.
LPP, the miR-28 host gene, is overexpressed in HEL cells through the transcriptional activation of an upstream alternative promoter bound by STAT5. (A) Relative expression of the LPP (LIM domain lipoma-preferred partner) transcript and miR-28 in the indicated cell lines. The LPP level in UT-7 parental and UT-7 JAK2 WT cell lines have been arbitrarily set at 1. (B) 5′-Rapid amplification of cDNA Ends (RACE)–PCR: a nested PCR on cDNA integrating an adaptor sequence at the 5′end of the LPP transcript amplified a 200-bp band in UT-7 and a larger 300- to 400-bp band in HEL cell lines. After sequencing, the 216-bp band amplified from UT-7 contains exon 1 and 2 of the LPP gene. The 300- to 400-bp band amplified from HEL contains 2 alternative exons (A and B) and exon 2 of the LPP gene. (C) A schematic view of the human LPP gene (LIM domain lipoma-preferred partner) represented with its 11 exons (black boxes), the 2 alternative exons (A and B) and the pri-miR-28 stem loop sequence in intron 6. The 2 identified promoters are indicated by arrows and labeled as Prom A (alternative) and Prom 1 (normal start site). (D) Quantitative PCR on indicated targets after chromatin immunoprecipitation (ChIP) relative to the starting DNA quantity before immunoprecipitation (% INPUT ± SD). Gray and black histograms represent ChIP performed on JAK2 WT and V617F UT-7 cells or UT-7 and HEL cells, respectively. White histograms are negative controls. The actin gene amplification (ACTB) was used a negative control for STAT5B binding. Quantitative PCR for RNA pol II ChIP were normalized against a non transcribed region located outside (3′) of the LPP gene.
We next searched for STAT binding sites, defined by the TTC(N)3GAA consensus, and that are conserved between the mouse and human LPP. No STAT binding site was identified near the promoter 1 (Prom 1) transcription start site, in agreement with lack of miR-28 (or LPP) induction in cells transiently stimulated with cytokines. A number of STAT5 potential sites were predicted along LPP (supplemental Figure 2). Detailed experiments are required to test the relevance of each site for transcription of LPP. One of those STAT5-binding sites was close (2 kb) to the alternative transcription site detected in HEL cells. We asked whether the alternative promoter (Prom A; Figure 5C) might be a binding target for the constitutive active STAT5 via this site. Direct binding of STAT5B to LPP promoter sites, Prom 1 or Prom A (Figure 5C), was then assessed in UT-7 JAK2 V617F, in UT-7 JAK2 WT and in HEL cells by STAT5B ChIP and then by PCR amplification of either Prom 1/Prom A, or of actin as a negative control, since no STAT5 binding sites were contained in the amplified actin sequence. We detected enriched STAT5B binding to Prom A site in HEL compared with UT-7 cells (Figure 5D). The difference in STAT5B binding between UT-7 JAK2 V617F and UT-7 JAK2 WT cells was below significance, although the former expressed higher miR-28 levels. This might be due to long miR half-life and the very low levels of JAK2 V617F that could be transduced in UT7 cells. To seek for transcriptional activity, RNA pol II ChIP was performed on both promoter sites. We detected an increased RNA pol II binding to promoter A site in JAK2 V617F expressing cell lines compared with JAK2 WT expressing cell lines (Figure 5D), suggesting that STAT5 activates the transcription of Prom A site.
Expression of miR-28 in platelets from MPN patients
To evaluate whether increased miR-28 expression might be detected in MPN patients, small-size RNA was isolated from platelets of 18 healthy donors and 53 MPN patients (8 PV, 29 ET, and 16 PMF) were extracted and monitored for miR-28 expression levels by Stem-Loop qRT-PCR (Figure 6). miR-28 was expressed in 3 of 6 JAK2 V617F-positive PV patients, 1 of 9 JAK2 V617F-positive ET patients, 9 of 20 JAK2 WT ET patients, 1 of 6 PMF JAK2 WT patients, and 2 of 4 MPL W515 mutant-positive PMF patients (Figure 6A); the MPL W515 mutations also lead to constitutive JAK/STAT activation.8-10 Taken together, miR-28 is expressed in platelets of approximately 30% of MPN patients. When we evaluated miR-151 expression in patients' platelets, only 2 patients, namely ET JAK2 V617F patient 4 and PMF MPL W515L patient 2, displayed significant up-regulated levels. We were not able to evaluate miR-708 levels in patients' platelets due to cross-reactivity of the tested probes.
Detection of miR-28 overexpression in platelets from MPN patients. (A) miR-28 relative levels are depicted in phe positive control that is represented by Mo7e cells transduced by the pMegix vector overexpressing miR-28 (± SD quantification done in triplicate). Ctrl indicates control platelets from healthy volunteers; PV, polycythemia vera; ET, essential thrombocythemia; PMF, primary myelofibrosis. Status for JAK2 or MPL mutations is indicated under the histograms. (B) Relative miR-28 levels in control and MPN platelets. Mean miR-28 levels are represented on graphs (P = .014; Mann-Whitney test). Positive JAK2 V617F patients were grouped accordingly to their allele burden (< 50% or > 50% JAK2 V617F) and examined for miR-28 levels (P = .003; Mann-Whitney test). (C) miR-28 relative levels in platelets and granulocytes of the same patient. Patient numbers and normalization of relative miR-28 quantity are the same as in panel A.
Detection of miR-28 overexpression in platelets from MPN patients. (A) miR-28 relative levels are depicted in phe positive control that is represented by Mo7e cells transduced by the pMegix vector overexpressing miR-28 (± SD quantification done in triplicate). Ctrl indicates control platelets from healthy volunteers; PV, polycythemia vera; ET, essential thrombocythemia; PMF, primary myelofibrosis. Status for JAK2 or MPL mutations is indicated under the histograms. (B) Relative miR-28 levels in control and MPN platelets. Mean miR-28 levels are represented on graphs (P = .014; Mann-Whitney test). Positive JAK2 V617F patients were grouped accordingly to their allele burden (< 50% or > 50% JAK2 V617F) and examined for miR-28 levels (P = .003; Mann-Whitney test). (C) miR-28 relative levels in platelets and granulocytes of the same patient. Patient numbers and normalization of relative miR-28 quantity are the same as in panel A.
The mean miR-28 platelet level in MPNs was 11-fold greater than in controls (P = .014; Figure 6B). To evaluate whether a relationship can be established between expression of JAK2 V617F and of miR-28 in platelets, we set up a genotyping test for the quantitation of JAK2 V617F and JAK2 WT at the mRNA/cDNA level (as platelets do not have genomic DNA), using the same TaqMan probes used for JAK2 V617F detection at the genomic DNA level. We observed a good correlation between JAK2 V617F mRNA in MPNs granulocytes and the allelic ratio measured from granulocytes DNA (R2 = 0.9607; supplemental Figure 4). Importantly, the JAK2 V617F levels (ratio to total V617F+WT JAK2) at the mRNA level were similar in granulocytes and platelets from the same patient (supplemental Figure 4). This test allowed us to measure levels of JAK2 V617F mRNA in platelets from MPN patients. miR-28 levels were compared between 2 platelet groups, one expressing less than 50% and the other more than 50% JAK2 V617F. The more than 50% JAK2 V617F group displayed a 34-fold increase (P = .003) in miR-28 expression level, indicating a possible correlation between JAK2 V617F allele burden and miR-28 expression. Importantly, miR-28 is expressed in platelets, but not in granulocytes (Figure 6C). Because megakaryocytes are major players in the pathogenicity of MPNs,33,34 we suggest that these data might be relevant for further understanding of molecular anomalies of this lineage in MPNs.
Separately from the analysis of JAK2 V617F-positive patients, we could observe that 9 of 20 ET JAK2 WT patients overexpressed miR-28 (Figure 6A). Because the ET JAK2 WT group is known to have higher platelet counts,35 it is possible that miR-28 represents a marker of megakaryocyte proliferation. To establish this correlation, a larger study is necessary.
Next we examined the MPL protein levels for 21 (5 PV, 12 ET, 4 PMF) of the 53 MPN patients and asked whether a correlation could be found between high miR-28 levels and MPL down-modulation (supplemental Figure 3). Of the 5 PV patients examined, all 5 had down-modulation of MPL, but only 4 had high miR-28; PV patient 6 had an allele burden of JAK2 V617F of 93%, down-modulation of MPL and low miR-28 (see “Discussion”). All 4 MPL W515L/K PMF patients exhibited strong down-modulation of MPL; 2 of the patients had high miR-28 levels, and a third (PMF MPL W515L patient 2) had overexpressed miR-151 levels. Hereby we therefore report that MPL W515 mutation also is associated with MPL down-modulation.
For ET patients, 7 of 12 did not present platelet MPL protein down-modulation (supplemental Figure 3). Two ET patients were positive for JAK2 V617F, ET patients 8 and 7, with allele burdens of 26% and 35%, respectively. No miR-28 or MPL down-modulation could be detected. However, in ET patients normal and mutated clones coexist, and the levels of miR-28 and MPL that we can detect represent averages of normal and mutated clones. For ET patients who did not exhibit JAK2 V617F, it is impossible to know the allele burden of the putative “other mutations”; interestingly, 3 such ET patients had high miR-28 and no MPL down-modulation. Whether this simply reflects the inability of miR-28 to down-modulate platelet MPL in ET or the presence of a mix of normal and high miR-28/ weak MPL down-modulation is not known. The nature of the other events responsible for JAK2 V617F/ negative ET might also affect the induction and effects of miR-28.
Discussion
Our major observations are that: (1) miR-28 and 2 related miRs (miR-708 and miR-151) target the 3′UTR of MPL for down-modulation, and (2) a fraction of MPN patients, especially PV, and ET patients wild type for JAK2, overexpress miR-28 in their platelets. Because the JAK2 WT ET group is known to exhibit higher platelet counts,35 miR-28 might represent a marker of megakaryocyte hyperproliferation.
We identified a number of targets of miR-28. Perhaps the most relevant, after MPL, for megakaryocyte differentiation is E2F6, a transcription factor belonging to the E2F family; E2F6 actually represses E2F-responsive genes,24,36 by inhibiting E2F1, which was previously shown to inhibit megakaryocyte terminal differentiation.25 The mitogen-activated protein kinase 1 (MAPK1; ERK2, extracellular signal-regulated kinase 2) is inhibited by miR-28, but not by miR-708 or miR-151, and this might be relevant as the Ras/MAPK pathway is important for megakaryocyte differentiation.26,37,38 CD34+ derived megakaryocte differentiation assays suggest that miR-28 plays a negative role on differentiation of precursors to proplatelet bearing megakaryocytes. We propose that miR-28 induction might be part of negative feedback mechanisms invoked by megakaryocyte proliferation of MPNs.
In cytokine-dependent and leukemia cell lines, cytokine stimulation and activation of STAT5 did not suffice to induce miR-28 expression; this is mirrored by absence of STAT5-binding sites near the described promoter of LPP, the host gene for miR-28. Additional events are therefore required for miR-28 induction, besides transient STAT5 activation. Constitutive STAT5 activation in Ba/F3 cells is associated with increased miR-28 expression; this effect might reflect cooperation between permanently activated STAT5 and yet to be described transcription factors. In HEL cells, induction of the LPP/miR-28 gene occurs via an upstream alternative promoter that contains a STAT5-binding site. Chromatin changes around the LPP alternative promoter might occur in transformed cells, but more experiments are required to elucidate LPP induction in transformed cells.
A fraction of patients with MPNs overexpress miR-28 in their platelets. This was true for PV (with JAK2 V617F), PMF with MPL W515L/K, and, surprisingly, for ET with JAK2 WT. It would be expected that patients with JAK2 V617F harbor high miR-28 levels in platelets. How can we explain that PV patient 6 does not overexpress miR-28, although the JAK2 V617F allele burden was 93%? As stated in the previous paragraph, miR-28 induction might require additional events besides constitutive STAT5 activation, and might depend on the state of megakaryocyte proliferation. Another possibility would be that the CpG islands of the alternative LPP promoter are methylated in these patient. Finally, constitutive active STAT5 might be diverted from the LPP gene by other proteins.
We could perform the analysis of platelet MPN levels for 21 (5 PV, 12 ET, and 4 PMF) of the 53 MPN patients to test whether the MPL down-modulation11 is correlated with high miR-28 levels. PV and PMF patients exhibited MPL protein down-regulation in platelets (supplemental Figure 3), including the 1 PV patient (patient 6) that expressed low levels of miR-28. All 4 MPL W515 mutant PMF patients exhibited down-modulated MPL, but only 2 had high miR-28 levels, while the third (MPL W515L patient 2) overexpressed miR-151 (not shown). No PV or PMF patient with high miR-28 showed high MPL protein. Thus, most patients with PV and PMF had MPL down-modulation, but not all had high miR-28. In the ET group, 3 patients (9, 13, and 16) exhibited high miR-28 and high MPL protein. Because in ET normal and disease/mutated clones coexist,39 high overexpression of miR-28 could remain detectable, while a low (20%) decrease in MPL translation that would be induced by miR-28 might be masked by the normal clones. These results indicate that miR-28 can contribute to but is not the sole mechanisms responsible for MPL down-modulation in platelets. This is expected because MPL has a long half-life,40,41 and MPL down-modulation by enhanced internalization, ubiquitinylation and degradation can be detected in cells expressing JAK2 V617F (C.P. and S.N.C., unpublished observations, January 2009), a process that is mimicked by Tpo-induced TpoR down-modulation.40
The existence of microRNAs in platelets has recently been documented.42 No miR-28 up-regulation was detected in MPN platelets in a microRNA profiling study,43 since the microarray did not contain any miR-28 probe. Another microRNA-profiling study of PMF granulocytes did not detect miR-28 increases.44 In agreement with this result, we show that miR-28 is specifically expressed in platelets, but not in granulocytes from the same patient. Currently, we attempt to identify the precise stage during the megakaryocytic differentiation where miR-28 induction occurs, given that, normally, miR-28 should not be expressed past the pre-megakaryocyte stage.45
In conclusion, we found that miR-28 is a potent inhibitor of MPL translation and of several other proteins potentially important for megakaryocyte differentiation, such as E2F6 and ERK2. We suggest that expression of miR-28 might play important roles in the pathogenicity of MPNs, either as part of negative feedbacks of myeloproliferation or as regulator of disease phenotype.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr W. Fiedler for UKE-1 cells, AstraZeneca R&D Boston (Drs D. Huszar and M. Zinda) for the gift of compound AZD1480, and Dr G. Bommer for advice. S.N.C. is a Research Associate of the Fonds National de la Recherche Scientifique Belgium.
We are grateful to Fondation Salus Sanguinis, Action de Recherche Concertée (ARC) MEXP31C1 of the Université Catholique de Louvain, Fondation Contre le Cancer, the Atlantic Philanthropies, New York, the PAI Program BCHM61B5, Belgium, and the de Duve Institute for generous support.
Authorship
Contribution: M.G. designed the research, performed experiments, and wrote paper; C.P. produced key cell lines; S.B. performed experiments; L.K., A.F., W.V., and S.G. selected and provided patient samples and wrote paper; and S.N.C. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefan N. Constantinescu, Signal Transduction Unit, Ludwig Institute for Cancer Research Ltd, de Duve Institute, UCL, Ave Hippocrate 74, UCL 75-4, Brussels B-1200, Belgium; e-mail: stefan.constantinescu@bru.licr.org.