Abstract
Mutations in the IDH1 gene at position R132 coding for the enzyme cytosolic isocitrate dehydrogenase are known in glioma and have recently been detected also in acute myeloid leukemia (AML). These mutations result in an accumulation of α-ketoglutarate to R (2)-2-hydroxyglutarate (2HG). To further clarify the role of this mutation in AML, we have analyzed IDH1R132 in 1414 AML patients. We detected IDH1R132 mutations in 93 of 1414 patients (6.6%) with a clear prevalence in intermediate risk karyotype group (10.4%, P < .001). Although IDH1R132 mutations can incidentally occur together with all other molecular markers, there were strong associations with NPM1 mutations (14.2% vs 5.4% in NPM1wt, P < .001) and MLL-PTD (18.2% vs 7.0% in MLLwt, P = .020). IDH1-mutated cases more often had AML without maturation/French-American-British M1 (P < .001), an immature immunophenotype, and female sex (8.7% vs 4.7% in male, P = .003) compared with IDH1wt cases. Prognosis was adversely affected by IDH1 mutations with trend for shorter overall survival (P = .110), a shorter event-free survival (P < .003) and a higher cumulative risk for relapse (P = .001). IDH1 mutations were of independent prognostic relevance for event-free survival (P = .039) especially in the age group < 60 years (P = .028). In conclusion, these data show that IDH1R132 may significantly add information regarding characterization and prognostication in AML.
Introduction
Cytogenetic aberrations are the most important prognostic parameter in acute myeloid leukemia (AML).1,2 During recent years, it has become clear that gene mutations add important information to the cytogenetic subtypes. AML with normal karyotype can be genetically further characterized by mutations in FLT3 in terms of internal tandem duplications (FLT3-ITD)3 or tyrosine kinase mutations (FLT3-TKD)4,5 and by partial tandem duplication in the MLL gene (MLL-PTD),6-8 NPM1 mutations,9-12 and CEBPA mutations,13-15 the latter being considered disease-defining and resulting in provisional entities in the World Health Organization (WHO) classification. Several studies showed a heterogeneous pattern of certain gene mutations that are associated with different prognostic information. NPM1 mutations are regarded as favorable as long as not accompanied by FLT3-ITD.10-12 In addition, gene mutations are associated with certain cytogenetic aberrations and impact on their primary prognostic significance. Thus, core binding factor leukemias (RUNX1-RUNX1T1- and CBFB-MYH11-positive AML) are associated with a favorable prognosis unless coinciding with KIT mutations, which results in a very unfavorable prognosis.16,17
So far, molecular mutations have been subdivided into 2 different classes: (1) those that accelerate proliferation like mutations of tyrosine kinases (eg, FLT3, KIT) or RAS genes and (2) those that affect differentiation mainly in transcription factors (CEBPA, RUNX1). Very recently, a new kind of mutation has been described affecting an enzyme from the citric acid cycle. These mutations in IDH1 (encoding NADPH-dependent isocitrate dehydrogenase 1), affecting the arginine residue at position 132, were first described in 70% of malignant gliomas18 and have rarely been detected in tumors other than gliomas and AML, such as B cell acute lymphoblastic leukemia and prostate cancer.19 Moreover, novel IDH1 mutations have recently been identified in 5%-11% of thyroid cancers.20 Gliomas with IDH1 mutation were shown to have distinctive genetic and clinical characteristics, and patients with such tumors had a better outcome than those with wild-type IDH1 gene.18 Each of 4 tested IDH1 mutations reduced the enzymatic activity of the encoded protein, resulting in loss of the enzyme's ability to catalyze conversion of isocitrate to α-ketoglutarate18 by dominant-negative inhibition of isocitrate dehydrogenase catalytic activity.21 Instead, mutated IDH1 gains the ability to reduce α-ketoglutarate to R(-)-2-hydroxyglutarate (2HG) resulting in an accumulation of the metabolite 2HG.22
In an even more recent paper describing the analysis of the whole genome of a sole AML patient by next generation sequencing, an IDH1 mutation was detected for the first time in an AML patient.23 Subsequently, these authors analyzed 187 further AML patients and found the mutation recurrently in 15 more cases (8%). Based on their results, the authors suggested an association with normal karyotype and a probable unfavorable impact in AML without NPM1 mutation. Some of these findings were recently confirmed in a cohort of 493 AML patients;24 however, in this cohort, no effect of the IDH1 mutations on survival could be shown.
To further prove these findings in an independent cohort, we analyzed 1414 AML patients. A detailed multivariate analysis for other parameters like morphologic characterization, immunophenotype, karyotype, and molecular genetic markers and prognosis was performed. In our cohort, comprising all AML subtypes, IDH1 mutations were detected with a frequency of 6.6%. There was a very strong association with the intermediate karyotype risk group, a high female prevalence, and an unfavorable impact on survival compared with the IDH1 wild-type group.
Methods
Patients
All 1414 patient samples were referred to our laboratory for first diagnosis of AML between August 2005 and July 2009. AML was diagnosed according to the French-American-British (FAB) and WHO classifications.25,26 In part, the patients were selected according to the availability of complete karyotype information and follow up data. There were 668 female patients and 746 male patients. Median age was 65.8 years (range, 17.1-93.3 years). The patients received different treatment schedules and were in part included into controlled trials of German study groups.27 Therapy consisted of 1 or 2 induction therapy courses and at least one course of consolidation therapy, each with AraC and an anthracycline. A total of 105 patients underwent allogeneic stem cell transplantation, 7 of whom carried an IDH1 mutation. Thus, the incidence of IDH1 mutations in patients undergoing transplantation (6.7%) is within the same range as for the total cohort (6.6%). A total of 63 patients received transplantation in first remission. It was found that the survival was not influenced by transplantation (1-year survival: 72.0% for total group with censoring at time of transplantation vs 72.0% for group not censoring at time of transplantation; P = .866). Therefore, in all analyses, the patients were not censored at time of transplantation. Clinical data for survival analyses were available in 769 patients; of these, 287 (37.3%) were included into controlled trials of German study groups. However, all other patients also received standard AML therapies. Patients with supportive care were excluded only for statistic analysis. Before receiving therapy, all patients gave their informed consent for scientific evaluations (eg, molecular studies) after having been advised about the purpose and investigational nature of the study and of potential risks. The study design was in accordance with the Declaration of Helsinki and was approved by the institutional review board of MLL Munich Leukemia Laboratory before its initiation. The patient characteristics at diagnosis of AML are depicted in Table 1.
Patient characteristics
| . | Total cohort . | IDH1R132 wild-type . | IDH1R132 mutated . | Significant difference . |
|---|---|---|---|---|
| All patients | 1414 | 1321/93.4% | 93/6.6% | |
| Female | 668 (47.2%) | 610/91.3% | 58/8.7% | ↑ P = .003 |
| Male | 746 (52.8%) | 711/95.3% | 35/4.7% | ↓ P = .003 |
| Median age, y (range) | 65.8 (17.1-93.3) | 65.7 (17.1-93.3) | 67.2 (21.8-85.8) | n.s. |
| Median WBC, × 109/L (range) | 8400 (300-600 000) | 8600 (400-600 000) | 5000 (300-255 000) | n.s. |
| Median platelets, × 109/L (range) | 55 000 (1000-1 160 000) | 55 000 (1000-1 160 000) | 74,000 (13 000- 571 000) | n.s. |
| FAB subtype, n/% | ||||
| Total available | 1180 | 1100 | 80 | |
| AML M0 | 66/5.6% | 63/95.5% | 3/4.5% | n.s. |
| AML M1 | 274/23.2% | 232/84.7% | 42/15.3% | ↑ P < .001 |
| AML M2 | 432/36.6% | 405/93.8% | 27/6.2% | n.s. |
| AML M3 | 88/7.5% | 86/97.7% | 2/2.3% | ↓ P = .050 |
| AML M4 | 171/14.5% | 167/97.7% | 4/2.3 % | ↓ P = .013 |
| AML M4eo | 51/4.3% | 51/100% | 0 | ↓ P = .045 |
| AML M5a | 22/1.9% | 22/100% | 0 | n.s. |
| AML M5b | 22/1.9% | 22/100% | 0 | n.s. |
| AML M6 | 51/4.3% | 50/98.0% | 1/2.0% | n.s. |
| AML M7 | 3/0.3% | 2 | 1 | n.s. |
| History | ||||
| De novo | 1170/82.7% | 1090/93.2% | 80/6.8% | n.s. |
| s-AML after MDS | 120/8.5% | 111/92.5% | 9/7.5% | n.s. |
| s-AML after MPN | 37/2.6% | 35/94.6% | 2/5.4% | n.s. |
| t-AML after previous malignancy | 87/6.2% | 85/97.7% | 2/2.3% | n.s. |
| Cytogenetics | 1414 | |||
| Normal karyotype | 673/47.6% | 606/90.0% | 67/10.0% | ↑ P < .001 |
| t(15;17)/PML-RARA | 88/6.2% | 86/97.7% | 2/2.3% | ↓ P = .005 |
| t(8;21)/RUNX1-RUNX1T1 | 84/5.9% | 84/100% | 0 | ↓ P = .005 |
| inv(16)/t(16;16)/CBFB-MYH11 | 57/4.0% | 57/100% | 0 | ↓ P = .029 |
| t(11q23)/MLL | 31/2.2% | 31/100% | 0 | n.s. |
| t(6;9)/DEK-CAN | 5/0.4% | 5/100% | 0 | n.s. |
| t(8;16)/MYST3-CBP | 3/0.2% | 3/100% | 0 | n.s. |
| inv(3)/t(3;3) | 15/1.1% | 15/100% | 0 | n.s. |
| Trisomy 8 | 30/2.1% | 25/83.3% | 5/16.7% | ↑ P = .042 |
| Trisomy 13 | 12/0.8% | 11/91.7% | 1/8.3 % | n.s. |
| Other trisomies | 23/1.6% | 18/78.3 | 5/21.7% | n.s. |
| -7/7q- | 32/2.3% | 28/87.5% | 4/12.5% | n.s. |
| del(5q) | 6/0.4% | 6/100% | 0 | n.s. |
| del(9q) | 4/0.3% | 4/100% | 0 | n.s. |
| del(20q) | 2/0.1% | 2 | 0 | n.s. |
| All others including trisomies, monosomies, deletions or combinations of these | 61/4.3% | 55/90.2% | 6/9.8% | ↑ P = .010 |
| Complex aberrant | 288/20.4% | 285/99% | 3/1.0% | ↓ P < .001 |
| . | Total cohort . | IDH1R132 wild-type . | IDH1R132 mutated . | Significant difference . |
|---|---|---|---|---|
| All patients | 1414 | 1321/93.4% | 93/6.6% | |
| Female | 668 (47.2%) | 610/91.3% | 58/8.7% | ↑ P = .003 |
| Male | 746 (52.8%) | 711/95.3% | 35/4.7% | ↓ P = .003 |
| Median age, y (range) | 65.8 (17.1-93.3) | 65.7 (17.1-93.3) | 67.2 (21.8-85.8) | n.s. |
| Median WBC, × 109/L (range) | 8400 (300-600 000) | 8600 (400-600 000) | 5000 (300-255 000) | n.s. |
| Median platelets, × 109/L (range) | 55 000 (1000-1 160 000) | 55 000 (1000-1 160 000) | 74,000 (13 000- 571 000) | n.s. |
| FAB subtype, n/% | ||||
| Total available | 1180 | 1100 | 80 | |
| AML M0 | 66/5.6% | 63/95.5% | 3/4.5% | n.s. |
| AML M1 | 274/23.2% | 232/84.7% | 42/15.3% | ↑ P < .001 |
| AML M2 | 432/36.6% | 405/93.8% | 27/6.2% | n.s. |
| AML M3 | 88/7.5% | 86/97.7% | 2/2.3% | ↓ P = .050 |
| AML M4 | 171/14.5% | 167/97.7% | 4/2.3 % | ↓ P = .013 |
| AML M4eo | 51/4.3% | 51/100% | 0 | ↓ P = .045 |
| AML M5a | 22/1.9% | 22/100% | 0 | n.s. |
| AML M5b | 22/1.9% | 22/100% | 0 | n.s. |
| AML M6 | 51/4.3% | 50/98.0% | 1/2.0% | n.s. |
| AML M7 | 3/0.3% | 2 | 1 | n.s. |
| History | ||||
| De novo | 1170/82.7% | 1090/93.2% | 80/6.8% | n.s. |
| s-AML after MDS | 120/8.5% | 111/92.5% | 9/7.5% | n.s. |
| s-AML after MPN | 37/2.6% | 35/94.6% | 2/5.4% | n.s. |
| t-AML after previous malignancy | 87/6.2% | 85/97.7% | 2/2.3% | n.s. |
| Cytogenetics | 1414 | |||
| Normal karyotype | 673/47.6% | 606/90.0% | 67/10.0% | ↑ P < .001 |
| t(15;17)/PML-RARA | 88/6.2% | 86/97.7% | 2/2.3% | ↓ P = .005 |
| t(8;21)/RUNX1-RUNX1T1 | 84/5.9% | 84/100% | 0 | ↓ P = .005 |
| inv(16)/t(16;16)/CBFB-MYH11 | 57/4.0% | 57/100% | 0 | ↓ P = .029 |
| t(11q23)/MLL | 31/2.2% | 31/100% | 0 | n.s. |
| t(6;9)/DEK-CAN | 5/0.4% | 5/100% | 0 | n.s. |
| t(8;16)/MYST3-CBP | 3/0.2% | 3/100% | 0 | n.s. |
| inv(3)/t(3;3) | 15/1.1% | 15/100% | 0 | n.s. |
| Trisomy 8 | 30/2.1% | 25/83.3% | 5/16.7% | ↑ P = .042 |
| Trisomy 13 | 12/0.8% | 11/91.7% | 1/8.3 % | n.s. |
| Other trisomies | 23/1.6% | 18/78.3 | 5/21.7% | n.s. |
| -7/7q- | 32/2.3% | 28/87.5% | 4/12.5% | n.s. |
| del(5q) | 6/0.4% | 6/100% | 0 | n.s. |
| del(9q) | 4/0.3% | 4/100% | 0 | n.s. |
| del(20q) | 2/0.1% | 2 | 0 | n.s. |
| All others including trisomies, monosomies, deletions or combinations of these | 61/4.3% | 55/90.2% | 6/9.8% | ↑ P = .010 |
| Complex aberrant | 288/20.4% | 285/99% | 3/1.0% | ↓ P < .001 |
MDS indicates myelodysplastic syndrome; and MPN, myeloproliferative neoplasm; ↑ indicates elevated; and ↓, decreased.
Molecular analysis
Isolation of mononucleated cells, mRNA extraction, and random primed cDNA synthesis was performed as described previously.3 In 1214 cases, bone marrow and in 200 cases peripheral blood was used for the molecular analysis. In cases in which only peripheral blood was available, the median peripheral blast percentage was 56% (range 10%-98%) and thus was within the detection limit for all mutation analysis.
Screening for IDH1 mutations was performed using a melting curve based LightCycler assay with forward primer IDH1-F: GCTTGTGAGTGGATGGGTAA; reverse primer IDH1-R: TATGTACCAGGTATGTCACCTT; hybridization probes IDH1-640 sensor probe TCTGTATTGATCCCCATAAGCATGACGAC-P; and IDH1-F anchor probe TTTTCCAGGCCCAGGAACAACAAAATCAGTT-F. The polymerase chain reaction was carried out in a 20-μL reaction volume with 0.5μM forward and reverse primers, 0.75μM Hyb-Probes, 4mM MgCl2 and 2-μL LightCycler-FastStart DNA Master Hybridization Probes (Roche Diagnostics). LightCycler data were analyzed using the LightCycler 3.0 software (Roche Diagnostics) and the second derivative maximum method. Each 20-μL reaction contained 2 μL of cDNA, an equivalent of approximately 3000 cells. Amplification was performed with 45 cycles using an annealing temperature of 60°C. Final melting curve analysis was started at 40°C up to 95°C with slope of 0.2°C/s and continuous detection with channel F2/F1. All samples with an aberrant melting behavior were subsequently further characterized by direct Sanger sequencing. IDH1 mutations are numbered relative to the ATG (A = 1) in the Ensembl cDNA sequence ENST00000415913
Cytomorphology, cytogenetics, and immunophenotyping
Cytomorphologic assessment was based on May-Grünwald-Giemsa stains, myeloperoxidase reaction, and nonspecific esterase using alpha-naphtyl-acetate as described previously and was performed according to the criteria defined in the FAB and the WHO classifications.25,26,31 Cytogenetic studies were performed after short-term culture. Karyotypes, analyzed after G-banding, were described according to the International System for Human Cytogenetic nomenclature.32 Prognostic classification into favorable, intermediate, and adverse groups was performed according to the refined MRC classification.33 Cytogenetic results were available for all patients in the study. Immunophenotyping was performed as described previously34
Statistical analysis
Survival curves were calculated for overall survival (OS) and event free survival (EFS) according to Kaplan-Meier and compared using the 2-sided log rank test. OS was the time from diagnosis to death or last follow-up. EFS was the time from diagnosis to treatment failure, relapse, death, or last follow-up in complete remission. Relapse was defined according to Cheson et al.35 Cox regression analysis was performed for OS and EFS with different parameters as covariates. Median follow-up was calculated by taking into account the respective final observations in surviving cases and censoring nonsurviving cases at the time of death. Parameters that were significant in univariate analyses were included into multivariate analyses. Dichotomous variables were compared between different groups using the χ2 test and continuous variables by the Student t test. Results were considered significant at P < .05. All reported P values are 2-sided. No adjustments for multiple comparisons were performed. SPSS (version 14.0.1) software was used for statistical analysis.
Results
Characterization of IDH1 mutations
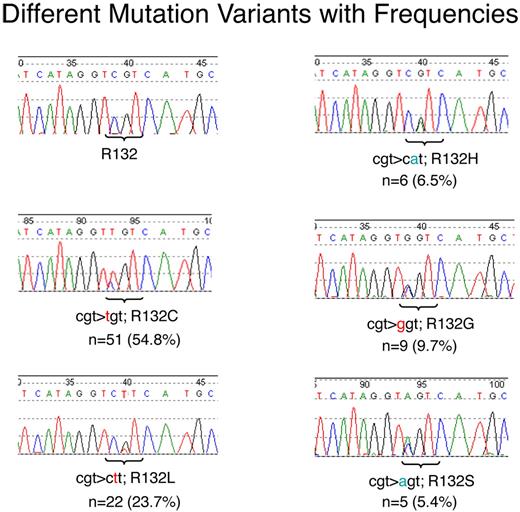
In the total cohort of 1414 patients 93 patients with IDH1 mutations (6.6%) were detected. Five different mutations leading to 5 different amino acid (AA) exchanges were observed with different frequencies: c.C776T; p.R132C in 51 cases (54.8%), c.G777T; p.R132L in 22 cases (23.7%), c.G777A; p.R132H in 6 cases (6.5%), c.C776G; p.R132G in 9 cases (9.7%), and c.C776A; p.R132S in 5 cases (5.4%). All mutations were detected in a “heterozygous” pattern (Figure 1) with a mutation to wild-type ratio of not more than 1, indicating that in all cases one wild-type allele was retained and the mutation should act in a dominant manner.
Electropherograms as generated by Sanger sequencing. Different mutations with their respective frequencies are shown. The respective mutated positions are tagged with a bracket. The base exchange is given in color.
Electropherograms as generated by Sanger sequencing. Different mutations with their respective frequencies are shown. The respective mutated positions are tagged with a bracket. The base exchange is given in color.
IDH1 mutations are numbered relative to the ATG (A = 1) in the Ensembl cDNA sequence ENST00000415913.
Analysis of paired diagnostic and relapse samples
In 12 cases with IDH1 mutation also a sample from relapse and in 2 patients even at second relapse was available. In 11 of these 12 cases (92%) the mutation was stably retained at relapse and in both available cases also at the second relapse. One case, which was mutated for IDH1, NPM1, and also for FLT3-ITD at diagnosis relapsed with both the same NPM1 mutation and FLT3-ITD but lost the IDH1 mutation at relapse. These data suggest that IDH1 mutations are stable in most cases but occasionally can obviously be secondary to other mutations.
Distribution of IDH1 mutations according to WHO/FAB subtypes
Morphology was available in 1180 of 1414 patients. The highest frequency of IDH1 mutations was found in AML without maturation (according to WHO) or AML M1 (according to FAB) with 42 of 274 cases (15.3%), which is significantly more frequent compared with all other subtypes (P < .001). The next most frequently IDH1-mutated groups are AML with maturation/AML M2 (27/432; 6.2%) and AML with minimal differentiation or AML M0 (3/66; 4.5%). IDH1 mutations were rare in acute promyelocytic AML/AML M3(v)/PML-RARA, myelomonocytic AML/AML M4, and acute erythroid leukemia/AML M6 with 2.3%, 2.3%, and 2.0%, respectively, and there was a single case (of 3) with acute megakaryoblastic leukemia or AML M7, which was IDH1-mutated. None of the cases with acute myeloid leukemia with CBFB-MYH11 (n = 57) and of acute monoblastic or monocytic leukemia/AML M5 (n = 44) was found to be IDH1-mutated (in detail see Table 1).
Distribution of IDH1 mutations according to cytogenetics
Karyotype information was available in all 1414 patients. 673/1414 (47.6%) patients had a normal and 741 (52.4%) an aberrant karyotype (in detail see Table 1). IDH1 mutations were significantly more frequent in the normal karyotype AML group (67/673; 10.0%) compared with those with aberrations (26/741; 3.5%; P < .001). In the aberrant karyotypes, there was a higher frequency in trisomy 8 cases (5/30; 16.7%, P = .042), whereas there was a significant underrepresentation of IDH1 mutations in cases with t(15;17)/PML-RARA (2/88; P = .005), inv(16)/CBFB-MYH11 (0/57; P = .029), t(8;21)/RUNX1-RUNX1T1 (0/84; P = .005); and the complex aberrant karyotypes (3/288; P < .001). According to prognostically relevant cytogenetic subgroups there was a significant association of IDH1 mutations to the intermediate risk group (81/779; 10.4%) compared with the favorable (2/229; 0.9%) and the unfavorable group (10/406; 2.5%; P < .001; Table 2).
Distribution in cytogenetically defined prognostic subgroups (according to MRC)49
| Cytogenetic subgroup . | Total cohort (n = 1414) . | IDH1R132 wild-type . | IDH1R132 mutated . |
|---|---|---|---|
| Favorable | 229/16.2% | 227/99.1% | 2/0.9% |
| Intermediate | 779/55.1% | 698/89.6% | 81/10.4% |
| Unfavorable | 406/28.7% | 396/97.5% | 10/2.5% |
| Cytogenetic subgroup . | Total cohort (n = 1414) . | IDH1R132 wild-type . | IDH1R132 mutated . |
|---|---|---|---|
| Favorable | 229/16.2% | 227/99.1% | 2/0.9% |
| Intermediate | 779/55.1% | 698/89.6% | 81/10.4% |
| Unfavorable | 406/28.7% | 396/97.5% | 10/2.5% |
Significant difference (P < .001).
Distribution of IDH1 mutations compared with other cooperating molecular mutations
A further aim of this study was the identification of cooperating mutations in IDH1-mutated AML. The results of this analysis are summarized in Tables 3–4. In the total cohort (Table 3), IDH1 mutations were significantly more frequent in cases with NPM1 mutations (12.5% vs 5.1%; P < .001) or MLL-PTD (15.4% vs 6.5%, P = .020) compared with the respective unmutated status. In addition, there was a trend to a lower frequency of IDH1 mutations in FLT3-TKD–mutated cases (2.2% vs 8.3%, P = .112). Although IDH1R132 mutations were detectable together with all other mutations investigated no differences of IDH1R132 frequency were detected between mutated and unmutated status of FLT3-ITD, NRAS, CEBPA, RUNX1, or JAK2V617F.
Combination of IDH1R132mut with other molecular markers (total cohort)
| Mutation . | cases analyzed . | IDH1 wt . | IDH1 mutated . | Difference compared with respective WT . |
|---|---|---|---|---|
| NPM1wt | 875 | 47 (5.1%) | ||
| NPM1mut | 1251 | 288 | 41 (12.5%) | ↑ P < .001 |
| FLT3-wt | 1020 | 73 (6.7%) | ||
| FLT3-ITD | 1331 | 221 | 17 (7.1%) | n.s. |
| FLT3-TKDwt | 655 | 59 (8.3%) | ||
| FLT3-TKDmut | 759 | 44 | 1 (2.2%) | ↓ P = .112 |
| NRASwt | 447 | 38 (7.8%) | ||
| NRASmut | 544 | 54 | 5 (8.5%) | n.s. |
| MLLwt | 1116 | 78 (6.5%) | ||
| MLL-PTD | 1259 | 55 | 10 (15.4%) | ↑ P = .020 |
| CEBPAwt | 561 | 53 (8.6%) | ||
| CEBPAmut | 660 | 44 | 2 (4.3%) | n.s. |
| RUNX1wt | 215 | 22 (9.3%) | ||
| RUNXmut | 339 | 94 | 8 (7.8%) | n.s. |
| JAK2V617wt | 365 | 23 (5.9%) | ||
| JAK2V617mut | 423 | 31 | 4 (11.4%) | n.s. |
| Mutation . | cases analyzed . | IDH1 wt . | IDH1 mutated . | Difference compared with respective WT . |
|---|---|---|---|---|
| NPM1wt | 875 | 47 (5.1%) | ||
| NPM1mut | 1251 | 288 | 41 (12.5%) | ↑ P < .001 |
| FLT3-wt | 1020 | 73 (6.7%) | ||
| FLT3-ITD | 1331 | 221 | 17 (7.1%) | n.s. |
| FLT3-TKDwt | 655 | 59 (8.3%) | ||
| FLT3-TKDmut | 759 | 44 | 1 (2.2%) | ↓ P = .112 |
| NRASwt | 447 | 38 (7.8%) | ||
| NRASmut | 544 | 54 | 5 (8.5%) | n.s. |
| MLLwt | 1116 | 78 (6.5%) | ||
| MLL-PTD | 1259 | 55 | 10 (15.4%) | ↑ P = .020 |
| CEBPAwt | 561 | 53 (8.6%) | ||
| CEBPAmut | 660 | 44 | 2 (4.3%) | n.s. |
| RUNX1wt | 215 | 22 (9.3%) | ||
| RUNXmut | 339 | 94 | 8 (7.8%) | n.s. |
| JAK2V617wt | 365 | 23 (5.9%) | ||
| JAK2V617mut | 423 | 31 | 4 (11.4%) | n.s. |
wt indicates wild-type, n.s., not significant; ↑, elevated; and ↓, decreased.
Combination of IDH1R132mut with other molecular markers in AML with normal karyotype
| Mutation . | cases analyzed . | IDH1 wt . | IDH1 mutated . | Difference compared with respective WT . |
|---|---|---|---|---|
| NPM1wt | 318 | 28 (8.1%) | ||
| NPM1mut | 651 | 268 | 37 (12.1%) | ↑ P = .090 |
| FLT3-wt | 431 | 50 (10.4%) | ||
| FLT3-ITD | 644 | 149 | 14 (8.6%) | n.s. |
| FLT3-TKDwt | 325 | 40 (11.0%) | ||
| FLT3-TKDmut | 390 | 24 | 1 (4.0%) | n.s. |
| NRASwt | 235 | 24 (9.3%) | ||
| NRASmut | 290 | 29 | 2 (6.5%) | n.s. |
| MLLwt | 551 | 55 (9.1%) | ||
| MLL-PTD | 654 | 39 | 9 (18.8%) | ↑ P = .041 |
| CEBPAwt | 406 | 37 (8.4%) | ||
| CEBPAmut | 478 | 35 | 0 (0%) | ↓ P = .097 |
| RUNX1wt | 124 | 13 (9.5%) | ||
| RUNXmut | 192 | 54 | 1 (1.8%) | ↓ P = .072 |
| JAK2V617wt | 198 | 11 (5.3%) | ||
| JAK2V617mut | 228 | 16 | 3 (15.8%) | n.s. |
| Mutation . | cases analyzed . | IDH1 wt . | IDH1 mutated . | Difference compared with respective WT . |
|---|---|---|---|---|
| NPM1wt | 318 | 28 (8.1%) | ||
| NPM1mut | 651 | 268 | 37 (12.1%) | ↑ P = .090 |
| FLT3-wt | 431 | 50 (10.4%) | ||
| FLT3-ITD | 644 | 149 | 14 (8.6%) | n.s. |
| FLT3-TKDwt | 325 | 40 (11.0%) | ||
| FLT3-TKDmut | 390 | 24 | 1 (4.0%) | n.s. |
| NRASwt | 235 | 24 (9.3%) | ||
| NRASmut | 290 | 29 | 2 (6.5%) | n.s. |
| MLLwt | 551 | 55 (9.1%) | ||
| MLL-PTD | 654 | 39 | 9 (18.8%) | ↑ P = .041 |
| CEBPAwt | 406 | 37 (8.4%) | ||
| CEBPAmut | 478 | 35 | 0 (0%) | ↓ P = .097 |
| RUNX1wt | 124 | 13 (9.5%) | ||
| RUNXmut | 192 | 54 | 1 (1.8%) | ↓ P = .072 |
| JAK2V617wt | 198 | 11 (5.3%) | ||
| JAK2V617mut | 228 | 16 | 3 (15.8%) | n.s. |
wt indicates wild-type, n.s., not significant; ↑, elevated; and ↓, decreased.
Restricting the analyses to the normal karyotype group (Table 4) again a high correlation to MLL-PTD mutations was observed (18.8% vs 9.1% in the MLL-PTD unmutated group, P = .041), whereas there remained only a trend to an association of IDH1mut to the NPM1-mutated group (12.1% in the NPM1-mutated vs 8.1% in the NPM1 unmutated cohort, P = .090). In addition, there was a trend to a lower frequency of IDH1mut in CEBPAmut (no case vs 8.4%, P = .097) and RUNX1mut (1.8% vs 9.5%; .072).
In total, in 25 of 93 IDH1-mutated cases (26.9%), no additional molecular mutation was detected; 49 cases (52.7%) revealed one additional mutation, 18 cases (19.5%) revealed 2 additional mutations; and 1 case revealed 3 additional mutations (in total, 88 additional mutations in 93 cases). Of these additional mutations, the most frequent ones were NPM1 (n = 41; 46.6%), followed by FLT3-ITD (n = 17; 19.3%), MLL-PTD (n = 10; 11.3%), RUNX1 (n = 8; 9.1%), NRAS (n = 5; 5.7%), JAK2 (n = 4; 4.5%), CEBPA (n = 2; 2.3%); and FLT3-TKD (n = 1; 1.1%). In contrast, those cases without molecular mutations in addition to IDH1R132 have significantly higher rates of cytogenetic aberrations than those with additional molecular mutations (11/25; 44.0% vs 15/68; 22%; P = .036).
Correlation to immunophenotype
In 608 patients, immunophenotyping was performed by multiparameter flow cytometry. Cases with IDH1 mutations (n = 48) had a more immature phenotype than those without (n = 560) and expressed monocytic markers to a lower degree. Antigens expressed stronger in IDH1-mutated cases included CD117 (37% vs 27%, P = .015) and MPO (50% vs 39%, P = .012), whereas antigens expressed stronger in IDH1 wild-type cases included CD116 (21% vs 11%, P = .003), CD11b (43% vs 28%, P = .001), CD15 (45% vs 31%, P = .001), CD4 (38% vs 27%, P = .002), CD56 (19% vs 11%, P = .028), CD64 (39% vs 28%, P = .005), CD65 (43% vs 27%, P < .001), and CD7 (30% vs 23%, P = .045). There were no significant differences in the expression of other antigens (CD34, CD13, CD133, CD135, CD14, CD19, CD2, CD33, and human leukocyte antigen DR-1) between IDH1-mutated and unmutated cases. These results are in line with the preponderance of IDH1 mutations in AML M1 and M2 according to FAB or AML without maturation or with maturation according to WHO criteria, respectively.
Correlation of further biologic parameters to IDH1 mutations
There was a high association of IDH1 mutations to female sex (frequency of 8.7% compared with 4.7% in males, P = .003). There was no significant difference in age between the IDH1-mutated cases compared with the unmutated (67.2 vs 65.7 years, P = .134). In addition, there were no further parameters like white blood cell count, platelet count, blast count, or history (de novo, preceding MDS or preceding malignancy), detected to be associated with IDH1R132 mutations.
Prognostic relevance of IDH1 mutations
Clinical follow-up data were available in 769 cases. Median follow-up time was 399 days. Median OS was not reached and OS at 1 year was 79.4%. 339 patients were < 60 years and 430 were ≥ 60 years. Median age of patients from this cohort was 63.0 years (range: 17.1-84.9 years). A total of 287 (37.3%) were included into controlled trials of German study groups. However, all others also received standard AML therapies.
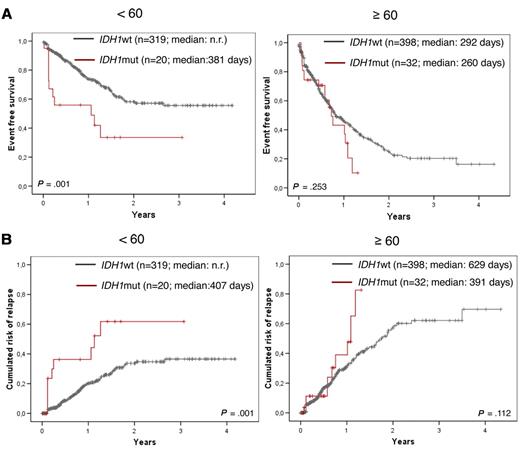
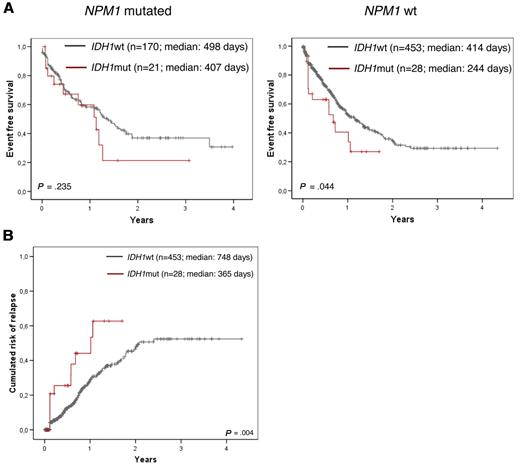
Within the total group there was a significantly worse EFS in the IDH1-mutated compared with the IDH1 wild-type group (median 365 vs 523 days; P = .003; Figure 2A and Table 5). In addition, a trend for shorter OS could be shown (median 533 days vs not reached; P = .110 (Figure 2B). When focusing the analysis to the < 60-year-old and ≥ 60-year-old age groups, the adverse impact on EFS was restricted to the age group < 60 years (median 381 days for IDH1mut vs not reached for IDH1wt, P = .001; Figure 3A and Table 5). Because IDH1 mutations were detected preferentially in NPM1-mutated AML, we did a further differential analysis in NPM1wt compared with NPM1-mutated AML (n = 672 with NPM1 analysis). An adverse impact of IDH1 mutations was detected only in the NPM1wt group (median 244 days for IDH1mut vs 414 days for IDH1wt, P = .044; Figure 4A and Table 5).
Kaplan-Meier plots showing influence of IDH1 mutations in the total cohort. EFS (A), OS (B), and CRR (C) of IDH1-mutated AML (in red) compared with IDH1 wild-type cases (black) in the cohort with outcome data.
Kaplan-Meier plots showing influence of IDH1 mutations in the total cohort. EFS (A), OS (B), and CRR (C) of IDH1-mutated AML (in red) compared with IDH1 wild-type cases (black) in the cohort with outcome data.
Univariate analysis for EFS and CRR
| Subgroup analysis . | Samples total (n) . | IDH1wt (n) . | IDH1mut (n) . | EFS . | CRR . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| wt vs mut (median days) . | P* . | RR . | 95% CI . | wt vs mut (median days) . | P* . | RR . | 95% CI . | ||||
| Total cohort | |||||||||||
| Total | 769 | 717 | 52 | 523 vs 365 | .003 | 1.761 | 1.201-2.582 | 1259 vs 407 | .001 | 2.215 | 1.374-3.570 |
| < 60 y | 339 | 319 | 20 | n.r. vs 381 | .001 | 2.688 | 1.435-5.033 | n.r. vs 407 | .001 | 3.086 | 1.535-6.203 |
| > 60 y | 430 | 398 | 32 | 292 vs 260 | n.s. | n.a. | n.a. | 629 vs 391 | n.s. | n.a. | n.a. |
| NPM1mut | 191 | 170 | 21 | 498 vs 407 | n.s. | n.a. | n.a. | 675 vs 427 | n.s. | n.a. | n.a. |
| NPM1wt | 481 | 453 | 28 | 414 vs 244 | .044 | 1.680 | 1.007-2.802 | 748 vs 365 | .004 | 2.448 | 1.308-4.583 |
| Intermediate risk cytogenetics | |||||||||||
| Total | 450 | 407 | 43 | 372 vs 469 | .103 | 1.500 | 0.961-2.342 | 722 vs 372 | .047 | 1.749 | 0.999-3.062 |
| < 60 y | 180 | 164 | 16 | 375 vs 260 | n.s. | n.a. | n.a. | n.r. vs 455 | n.s. | n.a. | n.a. |
| > 60 y | 270 | 243 | 27 | n.r. vs 455 | n.s. | n.a. | n.a. | 641 vs 391 | n.s. | n.a. | n.a. |
| NPM1mut | 187 | 166 | 21 | 518 vs 407 | n.s. | n.a. | n.a. | 675 vs 427 | n.s. | n.a. | n.a. |
| NPM1wt | 254 | 233 | 21 | 475 vs 244 | n.s. | n.a. | n.a. | 748 vs 365 | .005 | 2.448 | 1.308-4.583 |
| NPM1mut in < 60 y | 91 | 81 | 10 | 568 vs 455 | n.s. | n.a. | n.a. | n.r. vs 455 | n.s. | n.a. | n.a. |
| NPM1wt in < 60 y | 87 | 81 | 6 | n.r. vs 45 | .053 | 1.604 | 0.975-2.642 | n.r. vs 76 | .002 | 4.528 | 1.768-11.660 |
| Subgroup analysis . | Samples total (n) . | IDH1wt (n) . | IDH1mut (n) . | EFS . | CRR . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| wt vs mut (median days) . | P* . | RR . | 95% CI . | wt vs mut (median days) . | P* . | RR . | 95% CI . | ||||
| Total cohort | |||||||||||
| Total | 769 | 717 | 52 | 523 vs 365 | .003 | 1.761 | 1.201-2.582 | 1259 vs 407 | .001 | 2.215 | 1.374-3.570 |
| < 60 y | 339 | 319 | 20 | n.r. vs 381 | .001 | 2.688 | 1.435-5.033 | n.r. vs 407 | .001 | 3.086 | 1.535-6.203 |
| > 60 y | 430 | 398 | 32 | 292 vs 260 | n.s. | n.a. | n.a. | 629 vs 391 | n.s. | n.a. | n.a. |
| NPM1mut | 191 | 170 | 21 | 498 vs 407 | n.s. | n.a. | n.a. | 675 vs 427 | n.s. | n.a. | n.a. |
| NPM1wt | 481 | 453 | 28 | 414 vs 244 | .044 | 1.680 | 1.007-2.802 | 748 vs 365 | .004 | 2.448 | 1.308-4.583 |
| Intermediate risk cytogenetics | |||||||||||
| Total | 450 | 407 | 43 | 372 vs 469 | .103 | 1.500 | 0.961-2.342 | 722 vs 372 | .047 | 1.749 | 0.999-3.062 |
| < 60 y | 180 | 164 | 16 | 375 vs 260 | n.s. | n.a. | n.a. | n.r. vs 455 | n.s. | n.a. | n.a. |
| > 60 y | 270 | 243 | 27 | n.r. vs 455 | n.s. | n.a. | n.a. | 641 vs 391 | n.s. | n.a. | n.a. |
| NPM1mut | 187 | 166 | 21 | 518 vs 407 | n.s. | n.a. | n.a. | 675 vs 427 | n.s. | n.a. | n.a. |
| NPM1wt | 254 | 233 | 21 | 475 vs 244 | n.s. | n.a. | n.a. | 748 vs 365 | .005 | 2.448 | 1.308-4.583 |
| NPM1mut in < 60 y | 91 | 81 | 10 | 568 vs 455 | n.s. | n.a. | n.a. | n.r. vs 455 | n.s. | n.a. | n.a. |
| NPM1wt in < 60 y | 87 | 81 | 6 | n.r. vs 45 | .053 | 1.604 | 0.975-2.642 | n.r. vs 76 | .002 | 4.528 | 1.768-11.660 |
n.r. indicates not reached; n.s., not significant; n.a., not applicable; RR, relative risk; and CI, confidence interval.
P given according to Kaplan-Meier analysis.
Kaplan-Meier plots showing influence of IDH1 mutations according to age. Inferior EFS (A) and CRR (B) of IDH1-mutated AML (in red) compared with IDH1 wild-type cases (black) only in cases < 60 years (left side) but not in those ≥ 60 years (right side).
Kaplan-Meier plots showing influence of IDH1 mutations according to age. Inferior EFS (A) and CRR (B) of IDH1-mutated AML (in red) compared with IDH1 wild-type cases (black) only in cases < 60 years (left side) but not in those ≥ 60 years (right side).
Kaplan-Meier plots showing influence of IDH1 mutations according to NPM1 mutational status. (A) Inferior EFS of IDH1-mutated AML (in red) compared with IDH1 wild-type cases (black) only in cases without NPM1 mutation (right side), but not in the NPM1-mutated cases (left side). (B) CRR is shown for the NPM1wt cohort.
Kaplan-Meier plots showing influence of IDH1 mutations according to NPM1 mutational status. (A) Inferior EFS of IDH1-mutated AML (in red) compared with IDH1 wild-type cases (black) only in cases without NPM1 mutation (right side), but not in the NPM1-mutated cases (left side). (B) CRR is shown for the NPM1wt cohort.
To analyze whether this negative impact of IDH1 in the NPM1wt cohort is due to potentially higher percentage of adverse markers like FLT3-ITD, MLL-PTD, or RUNX1, in this cohort, we did Chi-square tests for these markers. FLT3-ITD and RUNX1 were equally distributed between IDH1mut and IDH1wt cases (7.1% vs 12.7%, P = .558 and 37.5% vs 40.3%, P = 1.0, respectively). MLL-PTD was slightly but nonsignificantly more frequent in the IDH1mut cases (17.9% vs 8.3%, P = .092), and, because this was due to only 5 MLL-PTD cases, this should not account for the overall more unfavorable outcome of IDH1mut cases within the NPM1wt group.
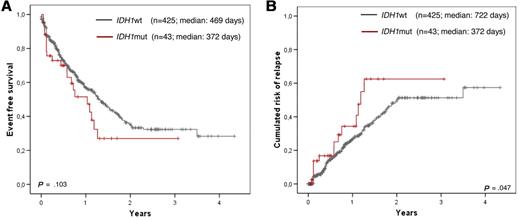
In a last step, we restricted the analysis to the intermediate risk karyotype group (including normal karyotype, which accounted for 86% of all such cases). In this analysis, only a trend for an adverse EFS in the IDH1-mutated group was observed (median, 372 days for IDH1mut vs 469 days for IDH1wt, P = .103; Figure 5A and Table 5). Although numbers are becoming small in further subgroup analysis, a strong trend could also be shown in the < 60-year-old NPM1wt group and an intermediate risk karyotype (median 45 days for IDH1mut vs not reached for IDH1wt, P = .053; Table 5).
Kaplan-Meier plots showing influence of IDH1 mutations in the intermediate risk karyotype. Inferior EFS (A) and CRR (B) of IDH1-mutated AML (in red) compared with IDH1 wild-type cases (black) for the intermediate risk karyotype subgroup.
Kaplan-Meier plots showing influence of IDH1 mutations in the intermediate risk karyotype. Inferior EFS (A) and CRR (B) of IDH1-mutated AML (in red) compared with IDH1 wild-type cases (black) for the intermediate risk karyotype subgroup.
RUNX1 mutations were the only mutations that had an unfavorable impact on EFS (P = .005). All other mutations (FLT3-ITD, FLT3-TKD, MLL-PTD, NPM1, CEBPA, and NRAS) were not found to be prognostically relevant as sole markers. However, the combination of NPM1mut and FLT3wt were found to be favorable (P = .029; Table 6).
Influence of different pretherapeutic parameters on EFS (total cohort of 769 patients)
| . | Cases analyzed . | Univariate . | Multivariate with RUNX1 (n = 256) . | Multivariate without RUNX1 (n = 553) . | |||
|---|---|---|---|---|---|---|---|
| EFS P . | RR (OS) . | EFS P . | RR . | EFS P . | RR . | ||
| Age | 769 | < .001‡ | 1.398* | .013‡ | 0.186* | < .001‡ | 1.351* |
| Sex | 769 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| WBC | 769 | < .001‡ | 1.005 | .041‡ | 1.004† | < .001‡ | 1.005† |
| Blast count in BM | 594 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Blast count in PB | 311 | .060 | 1.005 | n.a. | n.a. | n.a. | n.a. |
| CD34+ | 452 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| FAB subtype | 572 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| History | 755 | .054 | 1.120 | n.a. | n.a. | n.a. | n.a. |
| Aberrant karyotype | 769 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Cytogenetic risk group | 769 | < .001‡ | 2.087 | .111 | 1.429 | .001‡ | 1.624 |
| IDH1 | 769 | .004‡ | 1.761 | .412 | 1.303 | .039‡ | 1.575 |
| NPM1 | 672 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| FLT3-ITD | 735 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| NPM1mut + FLT3wt | 672 | .029‡ | 0.741 | .146 | 0.606 | .174 | 0.790 |
| FLT3-TKD | 561 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| MLL-PTD | 674 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| RUNX1 | 256 | .005‡ | 1.623 | .092 | 1.377 | - | - |
| CEBPA | 393 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| . | Cases analyzed . | Univariate . | Multivariate with RUNX1 (n = 256) . | Multivariate without RUNX1 (n = 553) . | |||
|---|---|---|---|---|---|---|---|
| EFS P . | RR (OS) . | EFS P . | RR . | EFS P . | RR . | ||
| Age | 769 | < .001‡ | 1.398* | .013‡ | 0.186* | < .001‡ | 1.351* |
| Sex | 769 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| WBC | 769 | < .001‡ | 1.005 | .041‡ | 1.004† | < .001‡ | 1.005† |
| Blast count in BM | 594 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Blast count in PB | 311 | .060 | 1.005 | n.a. | n.a. | n.a. | n.a. |
| CD34+ | 452 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| FAB subtype | 572 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| History | 755 | .054 | 1.120 | n.a. | n.a. | n.a. | n.a. |
| Aberrant karyotype | 769 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Cytogenetic risk group | 769 | < .001‡ | 2.087 | .111 | 1.429 | .001‡ | 1.624 |
| IDH1 | 769 | .004‡ | 1.761 | .412 | 1.303 | .039‡ | 1.575 |
| NPM1 | 672 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| FLT3-ITD | 735 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| NPM1mut + FLT3wt | 672 | .029‡ | 0.741 | .146 | 0.606 | .174 | 0.790 |
| FLT3-TKD | 561 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| MLL-PTD | 674 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
| RUNX1 | 256 | .005‡ | 1.623 | .092 | 1.377 | - | - |
| CEBPA | 393 | n.s. | n.a. | n.a. | n.a. | n.a. | n.a. |
RR indicates relative risk.
RR for 10-year intervals, not applicable.
RR for 100-fold increase.
Parameters in the univariate analysis that were included in the multivariate analysis and in the multivariate the independent significant parameters.
In a multivariate analysis of the total cohort of 553 patients (excluding RUNX1 data, as these were available only for a subset of 256 cases) with follow-up data IDH1 mutations were shown to be of independent prognostic relevance for EFS (P = .039; for details, see Table 6). This relevance was also reproducible for cases in the < 60-year-old age group (n = 277; P = .028; Tables 7–8).
Influence of different pretherapeutic parameters on EFS (only patients < 60 years)
| . | Cases analyzed . | Univariate . | Multivariate (n = 277) . | ||
|---|---|---|---|---|---|
| EFS P . | RR . | EFS P . | RR . | ||
| Age | 339 | n.s. | n.a. | - | - |
| Sex | 339 | .063 | 1.451 | - | - |
| WBC* | 277 | < .001‡ | 1.006† | < .001‡ | 1.006 |
| Blast count in BM | 264 | n.s. | n.a. | - | - |
| Blast count in PB | 133 | n.s. | n.a. | - | - |
| CD34+ | 189 | n.s. | n.a. | - | - |
| FAB subtype | 272 | n.s. | n.a. | - | - |
| History | 339 | n.s. | n.a. | - | - |
| Aberrant karyotype | 339 | 0.032 | 0.656 | - | -. |
| Cytogenetic risk group* | 339 | < .001‡ | 1.988 | < .001‡ | 1.947 |
| IDH1* | 339 | .002‡ | 2.688 | .028‡ | 2.166 |
| NPM1 | 280 | .099 | 1.419 | - | - |
| FLT3-ITD | 322 | .080 | 1.472 | - | - |
| NPM1mut + FLT3wt | 339 | n.s. | n.a. | - | - |
| FLT3-TKD | 249 | n.s. | n.a. | - | - |
| MLL-PTD | 277 | n.s. | n.a. | - | - |
| RUNX1 | 77 | .077 | 1.874 | - | - |
| CEBPA | 146 | n.s. | n.a. | - | - |
| . | Cases analyzed . | Univariate . | Multivariate (n = 277) . | ||
|---|---|---|---|---|---|
| EFS P . | RR . | EFS P . | RR . | ||
| Age | 339 | n.s. | n.a. | - | - |
| Sex | 339 | .063 | 1.451 | - | - |
| WBC* | 277 | < .001‡ | 1.006† | < .001‡ | 1.006 |
| Blast count in BM | 264 | n.s. | n.a. | - | - |
| Blast count in PB | 133 | n.s. | n.a. | - | - |
| CD34+ | 189 | n.s. | n.a. | - | - |
| FAB subtype | 272 | n.s. | n.a. | - | - |
| History | 339 | n.s. | n.a. | - | - |
| Aberrant karyotype | 339 | 0.032 | 0.656 | - | -. |
| Cytogenetic risk group* | 339 | < .001‡ | 1.988 | < .001‡ | 1.947 |
| IDH1* | 339 | .002‡ | 2.688 | .028‡ | 2.166 |
| NPM1 | 280 | .099 | 1.419 | - | - |
| FLT3-ITD | 322 | .080 | 1.472 | - | - |
| NPM1mut + FLT3wt | 339 | n.s. | n.a. | - | - |
| FLT3-TKD | 249 | n.s. | n.a. | - | - |
| MLL-PTD | 277 | n.s. | n.a. | - | - |
| RUNX1 | 77 | .077 | 1.874 | - | - |
| CEBPA | 146 | n.s. | n.a. | - | - |
Parameters in the univariate analysis that were included in the multivariate analysis and in the multivariate the independent significant parameters.
RR for 100-fold increase.
See Table 6.
Influence of different pretherapeutic parameters on CRR
| . | Cases analyzed . | Univariate . | Multivariate . | ||
|---|---|---|---|---|---|
| CRR P . | RR . | CRR P . | RR . | ||
| Age* | 696 | < .001† | 1.211 | .016† | 1.161 |
| Sex | 696 | n.s. | n.a. | - | - |
| WBC* | 571 | .001† | 0.005 | .006† | 1.001 |
| Blast count in BM* | 543 | .026† | 1.008 | n.s. | n.a. |
| Blast count in PB | 276 | n.s. | n.a. | - | - |
| CD34+ | 401 | n.s. | n.a. | - | - |
| FAB subtype | 524 | n.s. | n.a. | - | - |
| History | 685 | n.s. | n.a. | - | - |
| Aberrant karyotype | 696 | n.s. | n.a. | - | - |
| Cytogenetic risk group* | 696 | < .001 | 1.865 | < .001† | 1.999 |
| IDH1* | 696 | .001† | 2.215 | .057† | 1.762 |
| NPM1 | 598 | n.s. | n.a. | - | - |
| FLT3-ITD* | 575 | .013† | 1.516 | .017† | 1.716 |
| NPM1mut + FLT3wt | 598 | n.s. | n.a. | - | - |
| FLT3-TKD | 501 | n.s. | n.a. | - | - |
| MLL-PTD | 599 | n.s. | n.a. | - | - |
| RUNX1 | 218 | n.s. | n.a. | - | - |
| CEBPA | 339 | n.s. | n.a. | - | - |
| . | Cases analyzed . | Univariate . | Multivariate . | ||
|---|---|---|---|---|---|
| CRR P . | RR . | CRR P . | RR . | ||
| Age* | 696 | < .001† | 1.211 | .016† | 1.161 |
| Sex | 696 | n.s. | n.a. | - | - |
| WBC* | 571 | .001† | 0.005 | .006† | 1.001 |
| Blast count in BM* | 543 | .026† | 1.008 | n.s. | n.a. |
| Blast count in PB | 276 | n.s. | n.a. | - | - |
| CD34+ | 401 | n.s. | n.a. | - | - |
| FAB subtype | 524 | n.s. | n.a. | - | - |
| History | 685 | n.s. | n.a. | - | - |
| Aberrant karyotype | 696 | n.s. | n.a. | - | - |
| Cytogenetic risk group* | 696 | < .001 | 1.865 | < .001† | 1.999 |
| IDH1* | 696 | .001† | 2.215 | .057† | 1.762 |
| NPM1 | 598 | n.s. | n.a. | - | - |
| FLT3-ITD* | 575 | .013† | 1.516 | .017† | 1.716 |
| NPM1mut + FLT3wt | 598 | n.s. | n.a. | - | - |
| FLT3-TKD | 501 | n.s. | n.a. | - | - |
| MLL-PTD | 599 | n.s. | n.a. | - | - |
| RUNX1 | 218 | n.s. | n.a. | - | - |
| CEBPA | 339 | n.s. | n.a. | - | - |
Parameters in the univariate analysis that were included in the multivariate analysis and in the multivariate the independent significant parameters.
See Table 6.
The adverse impact of IDH1 mutations for outcome could be shown even stronger by the end point of cumulative risk for relapse (summarized in Table 5). This was shorter in the total cohort for IDH1mut compared with IDH1wt (median, 407 vs 1259 days, P = .001; Figure 2C), in the subcohort of patients < 60 years (median, 407 days vs not reached, P = .001; Figure 3B), in the NPM1wt subcohort (median, 365 vs 748 days; P = .004; Figure 4B), in the total intermediate risk karyotype group (median, 372 vs 722 days, P = .047; Figure 5B) and here again for the NPM1wt cases (median, 365 vs 748 days, P = .005) and especially those < 60 years (median, 76 days vs not reached; P = .002). Multivariate analysis also taking into account age, white blood cell count, bone marrow blast count, cytogenetic risk group, IDH1, and FLT3-ITD status shows a strong trend for IDH1 mutation impacting higher cumulative risk for relapse (CRR; P = .057).
Discussion
In a whole-genome sequencing project, IDH1 mutations have recently been identified to be recurrently mutated at position p.R132 in both glioma18 and AML.23 Mardis et al suggested an association of IDH1R132 mutations with normal karyotype AML, especially those with NPM1 mutations, FAB M1, and an unfavorable prognosis in 187 cases analyzed. Although most of these findings could be confirmed recently by others,24,36-38 the impact of IDH1 mutations on prognosis was discussed controversial. Whereas some did not detect an impact of IDH1 mutations on prognosis,24,36 others found them to be unfavorable in distinct subtypes (FLT3wt/NPM1wt)38 or (FLT3wt/NPM1mut).37 In our present study, we were able to further confirm the association of IDH1 mutations to normal karyotype or intermediate risk karyotype AML, the presence of NPM1 mutations, an undifferentiated immunophenotype, and FAB M1 or AML without maturation. In addition, we observed an unfavorable impact of p.R132 mutations to shorter EFS and CRR. This unfavorable effect was most obvious in AML below the age of 60 years and in AML with unmutated NPM1 status. Although the median follow-up of 399 day is relatively short, the negative effect of IDH1 mutations was significant, which may be in part related to the majority of events occurring within the first year after diagnosis. Furthermore, we found a very high prevalence in the female subjects (P = .003) and a high coincidence with MLL-PTD.
We detected 5 different amino acid exchanges at p.R132. Arginine was replaced by cysteine (R132C) in most of the cases (54.8%), followed by leucine (R132L; 23.7%), glycine (R132G; 9.7%), histidine (R132H; 6.5%), and serine (R132S; 5.4%). This pattern is completely different to the mutation pattern in glioma, where R132H was detected in 88% of all cases whereas R132C was detected in only 4.5%.18
In AML, as described for glioma, we also detected a mutation to wild-type ratio of near to but never more than “1,” accounting for retention of a wild-type allele in all cases. This pattern points to a dominant mode of action. This is in accordance with recent data showing that the affinity of the mutated IDH1 protein for the substrate is low. Heterodimerization of the mutant and wild-type proteins then lead to a reduction of the affinity of isocitrate and thus to reduction of the product α-ketoglutarate, which is important for degradation of hypoxia-inducing factor (HIF).21 However, IDH1 mutations enhance the ability of the enzyme to catalyze the NADPH-dependent reduction of α-ketoglutarate to R-2-hydroxyglutarate (2HG).22 Excess accumulation of 2HG has been shown to lead to an elevated risk of malignant brain tumors in patients with inborn errors of 2HG metabolism.39 Similarly, Dang et al22 found markedly elevated levels of 2HG in human malignant gliomas harboring IDH1 mutations. Importantly, very recently it has been shown that 2HG accumulation also leads to malignant progression in AML cells containing IDH1 mutations.40,41
In this study, we observed that IDH1 mutations are frequently found together with other molecular mutations. In more than 73% of all IDH1-mutated cases, at least one additional mutation was detected, and more than 20% have even 2 or more additional mutations. These findings are compatible with a multiple hit model of mutational acquisition in AML. Previous studies have postulated 2 different hits that may lead to acute leukemia,42 one hit leading to loss of differentiation (dominant negative action on transcription factors) and another additional hit leading to enhanced proliferation (mostly defects that act on activation of the STAT and RAS pathway). Because during the last years new mutation types have been described, such as NPM1 mutations, which lead to cellular dislocation of a protein9,43 or MLL-PTD, which leads to increase global hypermethylation,44 a multistep mechanism for AML becomes more probable. Because IDH1 mutations act by accumulation of a metabolite, they seem to represent a completely new type of mechanism in AML leukemogenesis. Although we have shown in this study that IDH1 mutations are most frequently associated with NPM1 mutations, FLT3-ITD and/or MLL-PTD they can be occasionally be associated with any other mutation identified, so far. This supports the hypothesis that IDH1 mutations complements with all these other mutation types.
We detected a striking female preponderance in our study as the IDH1 mutation frequency was twice as often detected in females than in males. This association was not described in 3 previous studies.23,24,45 Because the female/male ratio overall was nearly balanced in our study cohort, we speculate that these differences may be based on diverse ethnical backgrounds (white population in the present study, Asian population in the Chou study). Differences in sex distribution are also known for other malignancies, for example, FIP1L1-PDGFRA in males46 and 5q− syndrome in females.47
Although Mardis and colleagues described a possible adverse effect of IDH1 mutations on survival for normal karyotype and NPM1 wild-type AML, they were not able to demonstrate an independent prognostic impact.23 Also in the cohort described by Chou et al, no influence on prognosis could be shown.24 In contrast, in our study IDH1 mutations were found to have an independent adverse impact on EFS and CCR in AML.
This is in contrast to the data for gliomas, in which IDH1 mutations are prognostically favorable.18 An explanation may be that tumorigenesis is a multi hit scenario and therefore different in different tumors. Different coacting genes and mechanisms have to be anticipated. Although the oncogenic mechanism mediated by neomorphic enzyme activity of mutated IDH1, which leads to conversion of α-ketoglutatate to 2HG has been shown for gliomas and for AML,20,22 the complete mechanism is not fully understood, and 2HG accumulation may be more toxic for AML than for glioma cells. At last, accumulation of HIF has been shown to induce leukemia cell differentiation and growth arrest, whereas in solid tumors HIF seems to enhance tumor growth.48
In conclusion, these data clearly show that IDH1R132 mutations are one of the most frequent mutations in AML and are found in 6.6% of all AML and in 10.0% in the AML with normal karyotype. They are associated with other biologic parameters like FAB M1 morphology, NPM1 mutations and MLL-PTD, and female sex. They seem to be helpful to further specify biologic subgroups of AML. IDH1 mutations have an adverse prognostic effect in AML especially those < 60 years of age and in AML with wild-type NPM1. Thus, it is strongly suggested to implement IDH1 mutation analysis into the diagnostic workup of AML for further stratification in the future. Because accumulation of a metabolite, as it is the case in IDH1-mutated AML for 2HG, is a completely new pathogenetic mechanism in AML, this may open a new possibility for future therapies. Randomized therapeutic trials will show whether there may be specific therapy needed for IDH1-mutated AML.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all the coworkers in our laboratory for their excellent technical assistance and all physicians for referring sample material to our center.
Authorship
Contribution: S.S. was the principal investigator of this study, analyzed data, and wrote the manuscript; M.U. did molecular analysis of the IDH1 mutations; C.H. was responsible for chromosome analysis; W.K. was responsible for immunophenotyping and was involved in the statistical analysis; T.H. was responsible for cytomorphologic analysis; T.A. collected and analyzed clinical data; and all authors read and contributed to the final version of the manuscript.
Conflict-of-interest disclosure: S.S., W.K., C.H., and T.H. in part own the Munich Leukemia Laboratory; and M.U. and T.A. are employed by the Munich Leukemia Laboratory.
Corresponding author: Susanne Schnittger, Munich Leukemia Laboratory, Max-Lebsche-Platz 31, 81377 Munich, Germany; e-mail: susanne.schnittger@mll.com.