Abstract
Donor-matched transplantation of hematopoietic stem cells (HSCs) is widely used to treat hematologic malignancies but is associated with high mortality. The expansion of HSC numbers and their mobilization into the bloodstream could significantly improve therapy. We report here that adult mice conditionally deficient for the transcription Growth factor independence 1b (Gfi1b) show a significant expansion of functional HSCs in the bone marrow and blood. Despite this expansion, Gfi1bko/ko HSCs retain their ability to self-renew and to initiate multilineage differentiation but are no longer quiescent and contain elevated levels of reactive oxygen species. Treatment of Gfi1bko/ko mice with N-acetyl-cystein significantly reduced HSC numbers indicating that increased reactive oxygen species levels are at least partially responsible for the expansion of Gfi1b-deficient HSCs. Moreover, Gfi1b−/− HSCs show decreased expression of CXCR4 and Vascular cell adhesion protein-1, which are required to retain dormant HSCs in the endosteal niche, suggesting that Gfi1b regulates HSC dormancy and pool size without affecting their function. Finally, the additional deletion of the related Gfi1 gene in Gfi1bko/ko HSCs is incompatible with the maintenance of HSCs, suggesting that Gfi1b and Gfi1 have partially overlapping functions but that at least one Gfi gene is essential for the generation of HSCs.
Introduction
Murine hematopoietic stem cells (HSCs) are highly enriched in a bone marrow fraction defined by a combination of markers (Lin−, Sca-1+, c-kit+ [LSK], CD150+, CD48−)1 and are either in a quiescent (dormant) state or undergo cell cycling.2,3 During cell division, one daughter cell retains its stem cell properties, whereas the other daughter cell remains a stem cell or differentiates into multipotential progenitors (MPPs; LSK, CD150+, CD48+ or CD150−, CD48+), which in turn develop into myeloid, lymphoid, and erythroid cells. Expression level of CD34 allows differentiating between dormant and activated stem cells.2,4,5 It has been shown previously that CD34− LSK cells represent long-term stem cells whereas CD34+ LSK cells have only short-term repopulating potential.4 More specifically, CD34+ LSK, CD150+, CD48− HSC are activated and gradually lose their long-term repopulating capacity.2,5,6 In contrast, CD34− LSK, CD150+, CD48− HSCs are quiescent and comprise a pool of stem cells with long-term repopulating capacity.2,5,6
A critical regulatory element for hematopoietic differentiation is provided by the orchestrated activity of transcription factors.6-12 The zinc finger proteins Growth factor independence 1 (Gfi1) or Growth factor independence 1b (Gfi1b) are paradigmatic examples that illustrate the importance of transcriptional regulation during hematopoiesis and in particular for HSCs.13 Previous work has shown that Gfi1b is required for erythroid and megakaryocytic development.14-21 In contrast, Gfi1, the closely related paralogue of Gfi1b, is a key regulator for lymphoid and myeloid development but seems not to be required for erythro- or megakarypoiesis.13,22,23 Expression of Gfi1 and Gfi1b is complementary in many hematopoietic cell types, but in HSCs and some lymphoid subsets both transcription factors are expressed.24-27 Experimental evidence obtained from lymphoid cells demonstrated that the transcription at the Gfi1 and Gfi1b loci is controlled by auto- as well as cross-regulation.24-26
It has been reported by 2 different groups that Gfi1 is required to maintain the ability of HSCs to self-renew and to restrict their proliferation,13,27,28 but the exact mechanisms how Gfi1 exerts this function is unknown. Whether Gfi1b has the same or a different function in adult HSCs could not be analyzed since Gfi1b knockout mice arrest development at midgestation.29,30 Here, we describe the generation of Gfi1b conditional knockout mice, which permits the analysis of the role of Gfi1b in regulating the function and numbers of adult HSCs. Our data point to a pivotal role of Gfi1b in the restriction of HSC proliferation, which is different and more profound than the role of Gfi1, in particular concerning the regulation of HSC dormancy and proliferation.
Methods
Mice
Gfi1bfl/fl mice were generated by homologous recombination in R1 embryonic stem cells. All mice were backcrossed with C57Bl/6 mice, and the C57Bl/6 background was verified by specific satellite polymerase chain reaction. Gfi1fl/fl, Gfi1GFP/WT, and Gfi1bGFP/WT mice were described previously.24,30,31 All mice were housed in specific pathogen–free conditions, and all animal experiments were approved by the institutional animal ethics committee at the Institut de recherches cliniques de Montréal.
Treatment
MxCre tg Gfi1fl/fl or Gfi1bfl/fl mice were injected with polyinosinic-polycytidylic acid (pIpC; Sigma-Aldrich) at a dose of 500 μg per injection every other day for a total of 5 injections. As control, wild-type (wt) or Gfi1bfl/fl mice not carrying the MxCre tg were injected with pIpC. With regard to N-acetyl-cystein (NAC; Sigma-Aldrich) treatment, mice were fed every day with 500 μL of NAC (50 mg/mL).
Flow cytometric analysis, sorting of HSCs, and progenitors
HSCs and progenitors were analyzed with an LSR (BD Biosciences) or CyAn (Beckman Coulter) flow cytometer, and HSCs were sorted with a MoFlo (Cytomation) from adult mouse bone marrow as described previously.1,32 The 5-bromo-2-deoxyuridine (BrdU) experiments and the determination of cell cycle phases by Hoechst staining was done according to described procedures.2 Reactive oxygen species (ROS) were analyzed by staining HSCs with 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA; Invitrogen) for 30 minutes at 37°C. After staining, cells were analyzed by fluorescence-activated cell sorting for level of ROS in HSCs.
Methylcellulose culture
An amount of 20 000 bone marrow cells were seeded on methylcellulose (StemCell Technologies) supplemented with erythropoetin, interleukin-3, interleukin-6, and stem cell factor. After 10 days, the number of colonies was determined. Subsequently cells were resuspended, and 10 000 cells of the suspension were replated on fresh methylcellulose medium.
Transplantation
The number of functional stem cells was determined in vivo using a limiting dilution assay.33 Different amounts of bone marrow cells from pIpC-treated Gfi1bfl/fl and MxCre tg Gfi1bfl/fl mice (both CD45.2+) were transplanted together with 200 000 CD45.1+ bone marrow cells into lethally irradiated CD45.1+ mice. Eighteen weeks after transplantation, the peripheral blood of the recipient mice was analyzed for the contribution of CD45.2+ cells, and a percentage of higher than 1% was considered a positive call. Using the L-Calc software (Version 1.1) from Invitrogen, the frequency of functional stem cells was determined.
Polymerase chain reaction genotyping
Genotyping of Gfi1bfl/fl mice was performed using the following primers:
LP5-3s GGTTTCTACCAGTCTGGCCCTGAACTC; LP3-3r CTCACCTC TCTGTGGCAGTTTCCTATC; LP5-4r TACATTCATGCTTAGAAACTTGAGTC.
The product length of the wt allele is 256 bp, 295 for the floxed allele, and 540 for the deleted allele.
Microarray studies
Microarray data have been deposited in the public database Gene Expression Omnibus (National Center for Biotechnology Information; accession no. GSE20655). Samples were hybridized with Affymetrix Mouse Gene 1.0 ST Arrays. Data were processed using the Affymetrix Expression Console software; algorithm-name: rma-gene-default. Only genes up- or down-regulated more than 2-fold were taken in consideration.
Statistical analysis
The unpaired Student t test was chosen for analyzing the differences in the number of HSCs, common-monocyte progenitors, granulocyte-monocyte progenitors, and platelets. Analysis of variance was used to compare plating efficiency between wt and Gfi1b-deficient bone marrow cells. All P values were calculated 2-sided, and values of P < .05 were considered statistically significant. Statistical analysis was done with GraphPad Prism software (GraphPad Software).
Results
Gfi1b is highly expressed in HSCs and loss of Gfi1b drastically increases HSC numbers
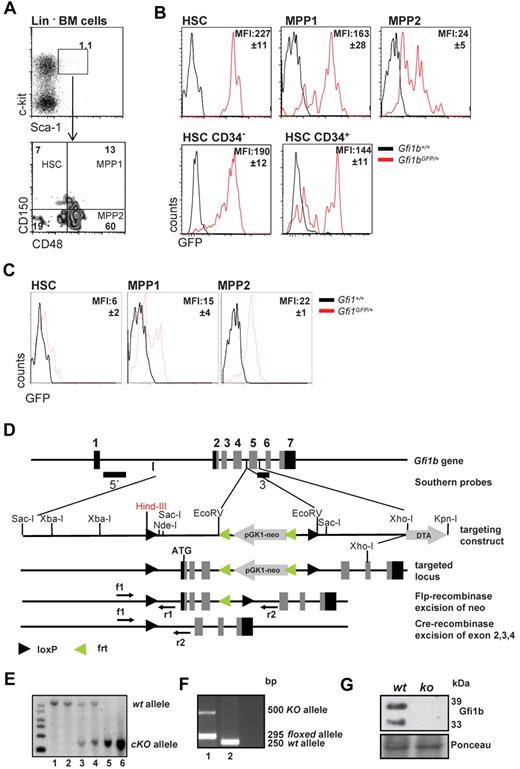
Using previously described Gfi1b:green fluorescent protein (GFP) knock-in mice (Gfi1bGFP/+), in which the level of GFP follows Gfi1b promoter activity and indicates Gfi1b expression levels,30 we observed that Gfi1b is highly expressed in virtually all HSCs (defined as: LSK, CD150+, CD48−) but is significantly down-regulated in the more differentiated MPP subsets (defined as MPP1: LSK, CD150+, CD48+, and MPP2: LSK, CD150−, CD48+; Figure 1A-B). However, the dormant CD34− HSC fraction2,4,6 from Gfi1bGFP/+ mice showed similar mean fluorescence intensities (MFI) as the activated CD34+ HSCs (Figure 1B). In addition, using similar reporter mice for Gfi1 (Gfi1:GFP knock-in mice, Gfi1GFP/+), in which the Gfi1 promoter activity and mRNA levels can be measured by following green flourescence,24,30 we observed that expression levels of Gfi1 and Gfi1b were different in HSC and MPP subsets. In particular, the Gfi1b promoter is highly active in HSCs and down-regulated upon differentiation to the MPPs (Figure 1B-C), whereas Gfi1 shows lowest levels in HSCs and is up-regulated in the MPP fractions. This suggests that both transcription factors are differentially regulated and thus may have different roles in these cells.
Gfi1 and Gfi1b expression in HSCs and MPPs and deletion of Gfi1b by gene targeting. (A) Gating scheme for HSCs and MPPs. Bone marrow cells were stained for the indicated markers and were electronically gated for Lin−, Sca-1+, c-kit+ (LSK) cells. The LSK subset was further analyzed for expression of CD150 and CD48 and was subdivided in HSCs (LSK, CD150+, CD48−), MPP1 (LSK, CD150+, CD48+), and MPP2 (LSK, CD150−, CD48+), according to published procedures. Results are representative for at least 3 independent experiments. (B) Activity of the Gfi1b promoter is followed by green fluorescence in cells isolated from Gfi1b:GFP knock-in mice based on the gating scheme indicated under A. Mean fluorescence intensity of GFP (MFI, representing Gfi1b promoter activity) is indicated. Representative for at least 3 independent experiments. The differences in the MFI of GFP of the different subsets were statistically significant (n = 3; P ≤ .05). (C) Activity of the Gfi1 promoter is followed by green fluorescence in cells isolated from Gfi1:GFP knock-in mice based on the gating scheme indicated under A. Mean fluorescence intensity of GFP (MFI, representing Gfi1 promoter activity) is indicated. Representative for at least 3 independent experiments. The differences in the MFI of GFP of the different subsets were statistically significant (n = 3; P ≤ .05). (D) Scheme of the murine Gfi1b locus and the targeting strategy to generate the conditional Gfi1b mouse allele. Exons 2 (which contains the ATG of Gfi1b), 3, and 4 are flanked by loxP sites. Upon activation of a Cre allele, these exons will be excised, thereby abrogating the expression of the Gfi1b protein. (E) Southern blot of DNA obtained from tails of wt (lane 1-2), Gfi1bfl/+ (lane 3-4), or Gfi1bfl/fl (lane 5-6) mice. DNA samples were restricted with HindIII. Using the 5′ probe described in D, correct recombination of the locus with the targeting vector is demonstrated by appearance of a 6-kb fragment, whereas the wt restriction fragment has a length of 10.5 kb. (F) Polymerase chain reaction genotyping of DNA from tail tip cells of a MxCre tg Gfi1bfl/fl mouse (1) and a wt mouse (2). Mice were injected with pIpC, and the detection of a ko allele is the result of contaminating lymphocytes in the tail. (G) Western blot of Abelson transformed pre-B-cell lines established from bone marrow from pIpC-treated Gfi1bfl/fl and MxCre tg Gfi1bfl/fl mice. Excision of the Gfi1b locus was stimulated with interferon treatment and abrogated the expression of Gfi1b protein in these cell lines. As loading control, Ponceau staining is shown.
Gfi1 and Gfi1b expression in HSCs and MPPs and deletion of Gfi1b by gene targeting. (A) Gating scheme for HSCs and MPPs. Bone marrow cells were stained for the indicated markers and were electronically gated for Lin−, Sca-1+, c-kit+ (LSK) cells. The LSK subset was further analyzed for expression of CD150 and CD48 and was subdivided in HSCs (LSK, CD150+, CD48−), MPP1 (LSK, CD150+, CD48+), and MPP2 (LSK, CD150−, CD48+), according to published procedures. Results are representative for at least 3 independent experiments. (B) Activity of the Gfi1b promoter is followed by green fluorescence in cells isolated from Gfi1b:GFP knock-in mice based on the gating scheme indicated under A. Mean fluorescence intensity of GFP (MFI, representing Gfi1b promoter activity) is indicated. Representative for at least 3 independent experiments. The differences in the MFI of GFP of the different subsets were statistically significant (n = 3; P ≤ .05). (C) Activity of the Gfi1 promoter is followed by green fluorescence in cells isolated from Gfi1:GFP knock-in mice based on the gating scheme indicated under A. Mean fluorescence intensity of GFP (MFI, representing Gfi1 promoter activity) is indicated. Representative for at least 3 independent experiments. The differences in the MFI of GFP of the different subsets were statistically significant (n = 3; P ≤ .05). (D) Scheme of the murine Gfi1b locus and the targeting strategy to generate the conditional Gfi1b mouse allele. Exons 2 (which contains the ATG of Gfi1b), 3, and 4 are flanked by loxP sites. Upon activation of a Cre allele, these exons will be excised, thereby abrogating the expression of the Gfi1b protein. (E) Southern blot of DNA obtained from tails of wt (lane 1-2), Gfi1bfl/+ (lane 3-4), or Gfi1bfl/fl (lane 5-6) mice. DNA samples were restricted with HindIII. Using the 5′ probe described in D, correct recombination of the locus with the targeting vector is demonstrated by appearance of a 6-kb fragment, whereas the wt restriction fragment has a length of 10.5 kb. (F) Polymerase chain reaction genotyping of DNA from tail tip cells of a MxCre tg Gfi1bfl/fl mouse (1) and a wt mouse (2). Mice were injected with pIpC, and the detection of a ko allele is the result of contaminating lymphocytes in the tail. (G) Western blot of Abelson transformed pre-B-cell lines established from bone marrow from pIpC-treated Gfi1bfl/fl and MxCre tg Gfi1bfl/fl mice. Excision of the Gfi1b locus was stimulated with interferon treatment and abrogated the expression of Gfi1b protein in these cell lines. As loading control, Ponceau staining is shown.
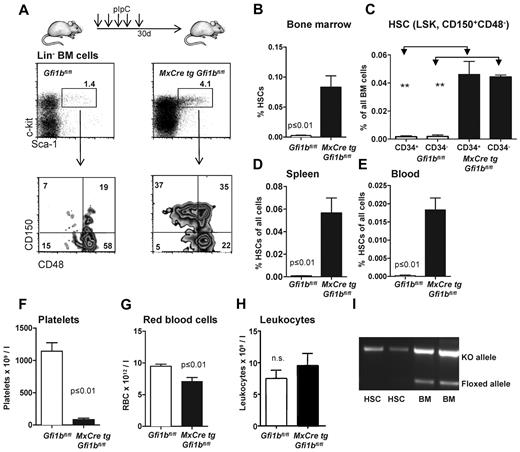
These observations prompted us to further investigate whether Gfi1b plays a particular role in HSCs. Since constitutively Gfi1b-deficient mice die at midgestation29 and thus cannot be used for an analysis of adult hematopoiesis, we generated a Gfi1b-conditional mouse carrying floxed Gfi1b alleles and a MxCre transgene (Figure 1D). In these MxCre tg Gfi1bfl/fl mice, Gfi1b exons 2-4 can be deleted by injection of pIpC, leading to the abrogation of Gfi1b expression (Figure 1E-G). To exclude possible effects of pIpC and Interferon-α on HSCs, MxCre tg Gfi1bfl/fl and Gfi1bfl/fl mice were examined 21 days after the last pIpC injection. It has been shown that this time period is sufficient to wean off effects of pIpC or interferon-α on HSCs.34 When pIpC-injected MxCre tg Gfi1bfl/fl-deficient mice were analyzed, we observed drastically increased frequencies of HSCs in bone marrow, spleen, and in the peripheral blood (between 30- and 100-fold, respectively; Figure 2A-E and Table 1). This is new feature that is not observed in Gfi1-deficient mice (data not shown and as previously reported27,29 ). The expansion affected both short-term (defined as CD34+ LSK, CD150+, CD48−) and long-term (CD34− LSK, CD150+, CD48−) HSCs (Figure 2C and Table 1).
Loss of Gfi1b increases the frequency of hematopoietic stem cells (HSC) in bone marrow, spleen, and blood. (A) Course of pIpC treatment of MxCre tg Gfi1bfl/fl mice and gating strategy to determine HSC and MPP frequencies using the indicated markers to stain bone marrow cells. Loss of Gfi1b significantly enhances the number of HSCs defined as LSK, CD150+,CD48−. Results are representative for at least 3 independent experiments. (B) Frequency of HSCs in the bone marrow (n = 14) of wt and Gfi1b-deficient mice was determined by flow cytometry (P ≤ .001 for both) 30 days after the first (equivalent to 21 days after the last) pIpC injection. (C) Frequency of CD34+ and CD34− HSCs in the bone marrow (n = 4) of wt and Gfi1b-deficient mice was determined by flow cytometry (P ≤ .01) 30 days after the first (equivalent to 21 days after the last) pIpC injection. (D) Frequency of HSCs in the spleen of wt (n = 3) and Gfi1b- (n = 5) deficient mice was determined by flow cytometry (P ≤ .01) 30 days after the first (equivalent to 21 days after the last) pIpC injection. (E) Frequency of HSCs in the peripheral blood (n = 6) of wt and Gfi1b-deficient mice was determined by flow cytometry (P ≤ .01 for both) 30 days after the first (equivalent to 21 days after the last) pIpC injection. (F) Gfi1bfl/fl and MxCre tg Gfi1bfl/fl were treated with pIpC, and 30 days after the first injection, peripheral blood cells were analyzed by an Advia blood analyzer. Loss of Gfi1b decreases platelet numbers (n = 6 for Gfi1bfl/fl and MxCre tg Gfi1bfl/fl) (P ≤ .01). (G) As in F for red blood cells (n = 6; P ≤ .01). (H) As in F for leukocytes. (I) Genotyping of sorted HSC from pIpC-injected MxCre tg Gfi1bfl/fl mice. Excision of the Gfi1b allele was efficient, and nonexcised alleles are below detection limit in HSCs.
Loss of Gfi1b increases the frequency of hematopoietic stem cells (HSC) in bone marrow, spleen, and blood. (A) Course of pIpC treatment of MxCre tg Gfi1bfl/fl mice and gating strategy to determine HSC and MPP frequencies using the indicated markers to stain bone marrow cells. Loss of Gfi1b significantly enhances the number of HSCs defined as LSK, CD150+,CD48−. Results are representative for at least 3 independent experiments. (B) Frequency of HSCs in the bone marrow (n = 14) of wt and Gfi1b-deficient mice was determined by flow cytometry (P ≤ .001 for both) 30 days after the first (equivalent to 21 days after the last) pIpC injection. (C) Frequency of CD34+ and CD34− HSCs in the bone marrow (n = 4) of wt and Gfi1b-deficient mice was determined by flow cytometry (P ≤ .01) 30 days after the first (equivalent to 21 days after the last) pIpC injection. (D) Frequency of HSCs in the spleen of wt (n = 3) and Gfi1b- (n = 5) deficient mice was determined by flow cytometry (P ≤ .01) 30 days after the first (equivalent to 21 days after the last) pIpC injection. (E) Frequency of HSCs in the peripheral blood (n = 6) of wt and Gfi1b-deficient mice was determined by flow cytometry (P ≤ .01 for both) 30 days after the first (equivalent to 21 days after the last) pIpC injection. (F) Gfi1bfl/fl and MxCre tg Gfi1bfl/fl were treated with pIpC, and 30 days after the first injection, peripheral blood cells were analyzed by an Advia blood analyzer. Loss of Gfi1b decreases platelet numbers (n = 6 for Gfi1bfl/fl and MxCre tg Gfi1bfl/fl) (P ≤ .01). (G) As in F for red blood cells (n = 6; P ≤ .01). (H) As in F for leukocytes. (I) Genotyping of sorted HSC from pIpC-injected MxCre tg Gfi1bfl/fl mice. Excision of the Gfi1b allele was efficient, and nonexcised alleles are below detection limit in HSCs.
Change of hematologic compartments and cell populations after Gfi1b deletion
| . | Gfi1bfl/fl . | Gfi1bfl/flMx-Cre tg . | Fold change . | P . |
|---|---|---|---|---|
| Number of BM cells ×106 | 36 ± 9 (n = 28) | 41 ± 13 (n = 27) | 1.13 | .21 |
| Number of splenocytes ×106 | 94 ± 9 (n = 5) | 180 ± 25 (n = 9) | 2 | .04 |
| % Lin− cells in BM | 1.9 ± 0.3 (n = 14) | 3.1 ± 0.4 (n = 14) | 1.63 | .002 |
| Number of Lin− cells ×106 | 0.7 ± 0.1 (n = 14) | 1.5 ± 0.2 (n = 14) | 2 | .01 |
| Number of HSCs in BM | 1000 ± 115 (n = 14) | 39 000 ± 11 000 (n = 14) | 39 | .0001 |
| Number of HSCs in spleen | 442 ± 150 (n = 3) | 51 000 ± 12 800 (n = 5) | 115 | .01 |
| Number of HSCs per 1 mL of blood | 15 ± 13 (n = 6) | 1435 ± 200 (n = 6) | 95 | .01 |
| Number of CD34+ HSC | 1100 ± 500 (n = 3) | 28 000 ± 9000 (n = 3) | 25 | .04 |
| Number of CD34− HSC | 780 ± 280 (n = 3) | 25 500 ± 500 (n = 3) | 32 | .001 |
| . | Gfi1bfl/fl . | Gfi1bfl/flMx-Cre tg . | Fold change . | P . |
|---|---|---|---|---|
| Number of BM cells ×106 | 36 ± 9 (n = 28) | 41 ± 13 (n = 27) | 1.13 | .21 |
| Number of splenocytes ×106 | 94 ± 9 (n = 5) | 180 ± 25 (n = 9) | 2 | .04 |
| % Lin− cells in BM | 1.9 ± 0.3 (n = 14) | 3.1 ± 0.4 (n = 14) | 1.63 | .002 |
| Number of Lin− cells ×106 | 0.7 ± 0.1 (n = 14) | 1.5 ± 0.2 (n = 14) | 2 | .01 |
| Number of HSCs in BM | 1000 ± 115 (n = 14) | 39 000 ± 11 000 (n = 14) | 39 | .0001 |
| Number of HSCs in spleen | 442 ± 150 (n = 3) | 51 000 ± 12 800 (n = 5) | 115 | .01 |
| Number of HSCs per 1 mL of blood | 15 ± 13 (n = 6) | 1435 ± 200 (n = 6) | 95 | .01 |
| Number of CD34+ HSC | 1100 ± 500 (n = 3) | 28 000 ± 9000 (n = 3) | 25 | .04 |
| Number of CD34− HSC | 780 ± 280 (n = 3) | 25 500 ± 500 (n = 3) | 32 | .001 |
The number of bone marrow (BM) cells, splenocytes, and % of Lin− cells was determined in wt and Gfi1b-deficient mice. The increase in number of splenocytes is mostly due to increase of number of erythroid progenitors. HSCs are defined by immunophenotype as Lin−, Sca-1+, Kit+, CD150+, CD48−. Depicted are mean values, SEM, and number of samples. P values are based on unpaired 2-sided t test.
The deletion of Gfi1b increased the number of Lin− cells in the bone marrow but did not significantly alter the overall cellularity of the bone marrow (Table 1). In contrast, there was an increase in the number of splenocytes (Table 1) in Gfi1b-deleted mice, which was mainly the result of an expansion of erythroid progenitors in the spleen (data not shown). Since the total number of bone marrow cells was not altered, the increased frequencies of HSCs correlated well with the increased absolute numbers of HSCs in bone marrow, spleen, and blood, indicating an expansion between 39- and more than 100-fold, respectively (Table 1). We also found that the number of platelets and erythrocytes in the peripheral blood was reduced compared with wt mice, albeit to a different extent, whereas the total number of leukocytes was not changed (Figure 2F-H). This is consistent with the established role of Gfi1b in the erythroid-megakaryocytic lineage.14-21,29 Finally, we also verified whether the excision of the floxed Gfi1b regions was efficient in HSCs after pIpC induction and observed that cells with nonexcised Gfi1b alleles were below detection level (Figure 2I).
HSCs from Gfi1b-deficient mice are less quiescent that wt HSCs and contain more ROS
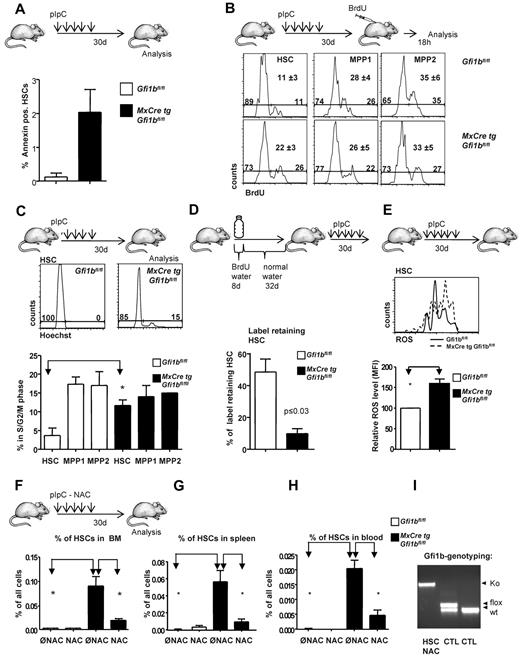
The increased numbers of HSCs in Gfi1b-deficient mice could be the result of a lower rate of spontaneous cell death or more proliferation. Gfi1b-deficient (Gfi1bko/ko) HSCs underwent a slightly higher rate of spontaneous apoptosis than wt HSCs but remained still less than 2.5% (Figure 3A). Using a BrdU pulse chase approach, we found that loss of Gfi1b correlated with significantly increased frequencies of cycling HSCs but had no or little effect on cell cycle progression of cells from the MPP subsets (Figure 3B). Staining with Hoechst showed similarly that a higher percentage of Gfi1bko/ko HSCs was in S and G2/M phases than wt HSCs (Figure 3C) but that cell cycle progression of the MPPs was not altered. These two results suggested that Gfi1b restricts specifically the proliferation of HSCs and hence might control HSCs dormancy but does not affect the rate of cell cycle progression in the different MPP fractions (Figure 3C). In support of this, a label retention assay showed that only 10% of Gfi1bko/ko HSCs were quiescent (ie, did not divide during our observation period; Figure 3D). In contrast, 45% of the pIpC-treated wt HSCs did not undergo a cell division at the end of the same time period (Figure 3D). These findings indicate that a significant proportion of Gfi1bko/ko HSCs is no longer dormant and has entered the cell cycle. It is known that HSCs are kept in a dormant state at the endosteal niche, which provides a hypoxic environment and protects them against oxidative damage by ROS, whereas high ROS are characteristic for activated HSCs and MPPs.35,36 Interestingly, we observed that Gfi1bko/ko HSCs had a significantly increased level of ROS, compared with the wt HSC population (Figure 3E).
Gfi1b restricts proliferation and activation of HSCs. (A) Frequency of apoptosis of HSCs in the bone marrow (n = 3) of wt and Gfi1b-deficient mice was determined by flow cytometry (P ≤ .001 for both) using Annexin staining. (B) Mice were intraperitoneally injected with BrdU 18 hours before analysis. Bone marrow cells were stained for the indicated markers and for BrdU. A representative result from 3 independent examinations is shown. Mean values and SDs of the 3 independent experiments are depicted; P ≤ .05 for difference in cell cycle progression between wt and Gfi1b-deficient HSCs. (C) Bone marrow cells of pIpC-treated Gfi1bfl/fl and MxCre tg Gfi1bfl/fl mice were stained with the specific antibodies to define HSCs, Hoechst 3342 and verapamil according to manufacturer's instruction. Cells were then electronically gated to define HSCs (LSK, CD150+, CD48−), and Hoechst levels were determined. A histogram representative for 3 independent examinations is shown. (Bottom panel) Quantification of 3 independent experiments for HSCs and different MPP fractions; P ≤ .05 for difference in cell cycle progression between wt and Gfi1b-deficient HSCs. Values were obtained 30 days after the first (equivalent to 21 days after the last) pIpC injection. (D) Schematic outline to detect BrdU+ cells following published procedures. Forty percent of wt HSCs were qualified as “label retaining” whereas only 12% of Gfi1bko/ko HSCs still retained the label (BrdU; n = 4 for Gfi1bfl/fl and n = 4 for MxCre tg Gfi1bfl/fl; P ≤ .05). (E) Detection of reactive oxygen species (ROS) in HSCs. (Top panel) A representative result from 3 independent experiments is shown. (Bottom panel) Quantification of ROS levels in HSCs from animals with indicated genotypes (MFI, n = 3). Values were obtained 30 days after the first (equivalent to 21 days after the last) pIpC injection. (F) Frequency of HSCs in the bone marrow of wt (n = 7) and (n = 6) Gfi1b-deficient mice, which received N-Acetylcystein (NAC) or were left untreated (n = 14 for wt and Gfi1b deficient). Frequency of HSCs was determined by flow cytometry (P ≤ .01 between untreated and NAC-treated Gfi1b-deficient HSCs). Values were obtained 30 days after the first (equivalent to 21 days after the last) pIpC injection. (G) Frequency of HSCs in the spleen of wt (n = 3) and Gfi1b- (n = 4) deficient mice, which received NAC or were left untreated (n = 3 for wt and n = 5 Gfi1b-deficient) was determined by flow cytometry (P ≤ .01 between untreated and NAC-treated Gfi1b-deficient HSCs). (H) Frequency of HSCs in the peripheral blood of wt (n = 3) and Gfi1b- (n = 5) deficient mice, which received NAC or were left untreated (n = 6 for both genotypes), was determined by flow cytometry (P ≤ .01 between untreated and NAC-treated Gfi1b-deficient HSCs). (I) Genotyping of Gfi1b-deficient HSCs sorted from NAC and pIpC-treated Gfi1b-deficient mice. HSCs: genotyping of HSCs after treatment with NAC. NAC treatment did not affect excision of floxed Gfi1b exons and nonexcised HCSs were below detection level. CTL: Two controls with 1 sample consisting of cells with a flox/wt constellation and 1 sample consisting of wt cells.
Gfi1b restricts proliferation and activation of HSCs. (A) Frequency of apoptosis of HSCs in the bone marrow (n = 3) of wt and Gfi1b-deficient mice was determined by flow cytometry (P ≤ .001 for both) using Annexin staining. (B) Mice were intraperitoneally injected with BrdU 18 hours before analysis. Bone marrow cells were stained for the indicated markers and for BrdU. A representative result from 3 independent examinations is shown. Mean values and SDs of the 3 independent experiments are depicted; P ≤ .05 for difference in cell cycle progression between wt and Gfi1b-deficient HSCs. (C) Bone marrow cells of pIpC-treated Gfi1bfl/fl and MxCre tg Gfi1bfl/fl mice were stained with the specific antibodies to define HSCs, Hoechst 3342 and verapamil according to manufacturer's instruction. Cells were then electronically gated to define HSCs (LSK, CD150+, CD48−), and Hoechst levels were determined. A histogram representative for 3 independent examinations is shown. (Bottom panel) Quantification of 3 independent experiments for HSCs and different MPP fractions; P ≤ .05 for difference in cell cycle progression between wt and Gfi1b-deficient HSCs. Values were obtained 30 days after the first (equivalent to 21 days after the last) pIpC injection. (D) Schematic outline to detect BrdU+ cells following published procedures. Forty percent of wt HSCs were qualified as “label retaining” whereas only 12% of Gfi1bko/ko HSCs still retained the label (BrdU; n = 4 for Gfi1bfl/fl and n = 4 for MxCre tg Gfi1bfl/fl; P ≤ .05). (E) Detection of reactive oxygen species (ROS) in HSCs. (Top panel) A representative result from 3 independent experiments is shown. (Bottom panel) Quantification of ROS levels in HSCs from animals with indicated genotypes (MFI, n = 3). Values were obtained 30 days after the first (equivalent to 21 days after the last) pIpC injection. (F) Frequency of HSCs in the bone marrow of wt (n = 7) and (n = 6) Gfi1b-deficient mice, which received N-Acetylcystein (NAC) or were left untreated (n = 14 for wt and Gfi1b deficient). Frequency of HSCs was determined by flow cytometry (P ≤ .01 between untreated and NAC-treated Gfi1b-deficient HSCs). Values were obtained 30 days after the first (equivalent to 21 days after the last) pIpC injection. (G) Frequency of HSCs in the spleen of wt (n = 3) and Gfi1b- (n = 4) deficient mice, which received NAC or were left untreated (n = 3 for wt and n = 5 Gfi1b-deficient) was determined by flow cytometry (P ≤ .01 between untreated and NAC-treated Gfi1b-deficient HSCs). (H) Frequency of HSCs in the peripheral blood of wt (n = 3) and Gfi1b- (n = 5) deficient mice, which received NAC or were left untreated (n = 6 for both genotypes), was determined by flow cytometry (P ≤ .01 between untreated and NAC-treated Gfi1b-deficient HSCs). (I) Genotyping of Gfi1b-deficient HSCs sorted from NAC and pIpC-treated Gfi1b-deficient mice. HSCs: genotyping of HSCs after treatment with NAC. NAC treatment did not affect excision of floxed Gfi1b exons and nonexcised HCSs were below detection level. CTL: Two controls with 1 sample consisting of cells with a flox/wt constellation and 1 sample consisting of wt cells.
One possible explanation for the increased number of HSC could be that loss of Gfi1b activates HSCs and that this activation leads to increased level of ROS, which in turn could lead to an expansion of HSCs as previously described.37 To verify this hypothesis, we fed mice NAC, which counteracts the effects of ROS.37 Indeed, we found that administration of NAC significantly limited the expansion of Gfi1bko/ko HSCs in the bone marrow, spleen, and peripheral blood both with regard to frequencies and absolute numbers (Figure 3F-H and Table 2) but did not affect the pIpC-mediated excision of the floxed Gfi1b exons in HSCs (Figure 3I). This suggested that elevated levels of ROS are at least partially responsible for the expansion of Gfi1b-deficient HSCs.
Change of hematologic compartments and cell populations after Gfi1b deletion and NAC injection
| . | Gfi1bfl/fl . | Gfi1bfl/flMx-Cre tg . | Fold change . | P . |
|---|---|---|---|---|
| Number of BM cells ×106 NAC treatment | 45 ± 4 (n = 7) | 44 ± 2 (n = 6) | 1 | .8 |
| Number of splenocytes ×106 NAC treatment | 86 ± 22 (n = 4) | 104 ± 22 (n = 5) | 1.2 | .5 |
| % Lin− cells in BM NAC treatment | 1.5 ± 0.1 (n = 7) | 2.45 ± 0.5 (n = 6) | 1.6 | .15 |
| Number of Lin− cells ×106 NAC treatment | 0.8 ± 0.1 (n = 7) | 1.1 ± 0.4 (n = 6) | 1.4 | .32 |
| Number of HSCs in BM NAC treatment | 1700 ± 700 (n = 7) | 5700 ± 1100 (n = 6) | 3 | .01 |
| Number of HSCs in spleen NAC treatment | 197 ± 77 (n = 3) | 6000 ± 2000 (n = 4) | 30 | .07 |
| Number of HSCs per 1 mL of blood NAC treatment | 11 ± 1 (n = 3) | 307 ± 100 (n = 5) | 28 | .03 |
| . | Gfi1bfl/fl . | Gfi1bfl/flMx-Cre tg . | Fold change . | P . |
|---|---|---|---|---|
| Number of BM cells ×106 NAC treatment | 45 ± 4 (n = 7) | 44 ± 2 (n = 6) | 1 | .8 |
| Number of splenocytes ×106 NAC treatment | 86 ± 22 (n = 4) | 104 ± 22 (n = 5) | 1.2 | .5 |
| % Lin− cells in BM NAC treatment | 1.5 ± 0.1 (n = 7) | 2.45 ± 0.5 (n = 6) | 1.6 | .15 |
| Number of Lin− cells ×106 NAC treatment | 0.8 ± 0.1 (n = 7) | 1.1 ± 0.4 (n = 6) | 1.4 | .32 |
| Number of HSCs in BM NAC treatment | 1700 ± 700 (n = 7) | 5700 ± 1100 (n = 6) | 3 | .01 |
| Number of HSCs in spleen NAC treatment | 197 ± 77 (n = 3) | 6000 ± 2000 (n = 4) | 30 | .07 |
| Number of HSCs per 1 mL of blood NAC treatment | 11 ± 1 (n = 3) | 307 ± 100 (n = 5) | 28 | .03 |
The number of bone marrow (BM) cells, splenocytes, and % of Lin− cells was determined in wt and Gfi1b deficient mice. Mice were fed daily with N-Acetylcystein. HSCs are defined by immunophenotype as Lin−, Sca-1+, Kit+, CD150+, CD48−. Depicted are mean values, SEM, and number of samples. P values are based on unpaired 2-sided t test.
Loss of Gfi1b does not affect multipotency or self-renewal capacity of HSCs
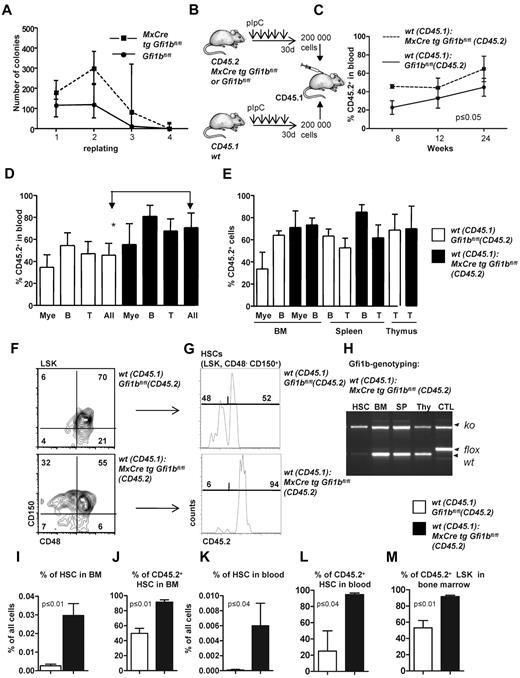
Next, we examined whether loss of Gfi1b might change the self-renewal capacity of HSCs, as was reported in the case of Gfi1 deficiency.27,28 Gfi1bko/ko bone marrow cells generated the same type of colonies (including colony forming unit [CFU[-E, burst-forming unit-E, CFU-G, CFU-M, CFU-GM, CFU-GEMM) as wt cells when seeded in methylcellulose (Figure 4A), and showed initially a higher replating efficiency and generated a higher number of colonies than wt bone marrow (Figure 4A). However, after the fourth replating cycle, Gfi1bko/ko cells exhausted their replating ability similar to wt cells (Figure 4A). We also performed a limiting dilution assay33 to verify the number of functional HSCs in vivo and found an HSC frequency of 1/7000 cells in Gfi1bko/ko mice compared with 1 HSC in 46 000 cells in wt mice (Tables 3–4; P ≤ .03). These findings suggested that Gfi1b deficiency enhances the number of functional HSCs approximately 6-7 times (Table 4).
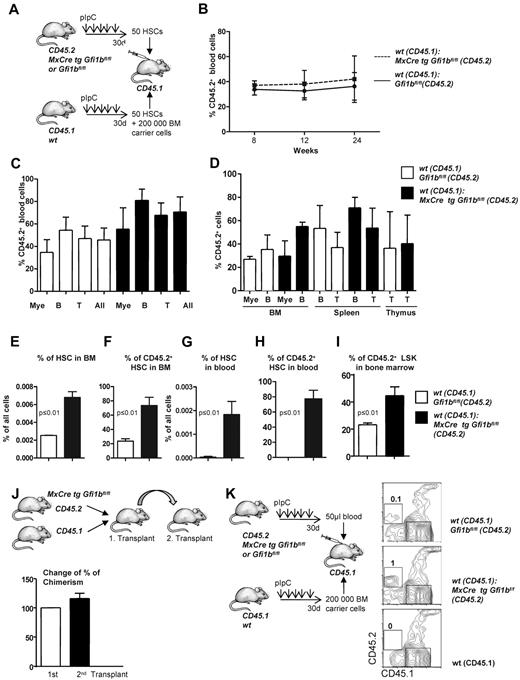
Transplantation of Gfi1b-deficient bone marrow. (A) About 20 000 bone marrow cells of pIpC-treated Gfi1bfl/fl and MxCre tg Gfi1bfl/fl mice were seeded on methylcellulose. After the indicated time periods (10 days), number of colonies was determined, cells were resuspended, and 10 000 cells of the suspension were replated (n = 6). Cell numbers were analyzed at indicated time points. (B) Scheme depicting the transplantation of equal number of bone marrow cells. About 200 000 bone marrow cells from pIpC-treated Gfi1bfl/fl or MxCre tg Gfi1bfl/fl (Gfi1bko/ko; both CD45.2+) mice were transplanted with 200 000 CD45.1+ bone marrow cells into lethally irradiated CD45.1+ mice. (C) Percentage of CD45.2–positive cells (% CD45.2) in the blood after transplantation acquired at indicated time points (n = 4). (D) CD45 chimerism in the blood was determined 24 weeks after transplantation in recipient mice (n = 4) overall (All) and for the indicated lineages. Myeloid (Mac-1), B-lymphoid (B220), T-lymphoid (CD3). The difference is significant (P ≤ .05) for CD45 chimerism between wt and Gfi1b-deficient cells, when all leukocytes are taken into account (All). (E) CD45 chimerism was determined 24 weeks after transplantation in the blood, bone marrow, spleen, and thymus of recipient mice (n = 4). (F) Frequencies of HSCs were determined in mice 24 weeks after transplantation with wt CD45.1 cells and with either wt CD45.2 BM cells or with Gfi1b-deficient CD45.2+ bone marrow cells (n = 4). (G) The relative proportion of HSCs originating from CD45.2 wt or CD45.2 Gfi1b-deficient HSCs was determined and is shown after electronic gating on CD150+CD48− cells depicted in F. (H) HSCs, bone marrow (BM), splenocytes (SP), thymocytes (thy) from mice transplanted with wt CD45.1 and Gfi1b-deficient CD45.2 bone marrow cells were genotyped and tested for the presence of the wt (CD45.1) and Gfi1b flox and Gfi1b ko alleles. (I) The frequencies of HSCs in mice either transplanted with wt CD45.1 and wt CD45.2 bone marrow cells or mice transplanted with wt CD45.1 and Gfi1b-deficient (MxCre tg Gfi1bfl/fl) bone marrow cells was determined (n = 4, P ≤ .01). (J) Quantification of which proportion of HSCs originates from CD45.2 wt or CD45.2 Gfi1b-deficient HSCs in mice transplanted with wt CD45.1 and wt CD45.2 bone marrow cells or mice transplanted with wt CD45.1 and Gfi1b-deficient bone marrow cells (MxCre tg Gfi1bfl/fl; n = 4, P ≤ .01). (K) The frequency of HSCs circulating in the peripheral blood of mice either transplanted with wt CD45.1 and wt CD45.2 bone marrow cells or mice transplanted with wt CD45.1 and CD45.2 Gfi1b-deficient bone marrow cells (MxCre tg Gfi1b fl/fl; n = 4, P ≤ .01). (L) Quantification of which proportion of HSCs circulating in blood originates from CD45.2 wt or CD 45.2 Gfi1b-deficient HSCs in CD45.1 mice transplanted with wt CD45.1 and wt CD45.2 bone marrow cells or CD45.1 mice transplanted with wt CD45.1 and Gfi1b-deficient bone marrow cells (n = 4, P ≤ .01). (M) Quantification of which proportion of Lin−, Sca-1+, c-kit+ (LSK) cells in bone marrow originate from CD45.2 wt or CD45.2 Gfi1b-deficient HSCs in CD45.1 mice transplanted with wt CD45.1 and wt CD45.2 HSCs or CD45.1 mice transplanted with wt CD45.1 and Gfi1b-deficient bone marrow cells (n = 4, P ≤ .01).
Transplantation of Gfi1b-deficient bone marrow. (A) About 20 000 bone marrow cells of pIpC-treated Gfi1bfl/fl and MxCre tg Gfi1bfl/fl mice were seeded on methylcellulose. After the indicated time periods (10 days), number of colonies was determined, cells were resuspended, and 10 000 cells of the suspension were replated (n = 6). Cell numbers were analyzed at indicated time points. (B) Scheme depicting the transplantation of equal number of bone marrow cells. About 200 000 bone marrow cells from pIpC-treated Gfi1bfl/fl or MxCre tg Gfi1bfl/fl (Gfi1bko/ko; both CD45.2+) mice were transplanted with 200 000 CD45.1+ bone marrow cells into lethally irradiated CD45.1+ mice. (C) Percentage of CD45.2–positive cells (% CD45.2) in the blood after transplantation acquired at indicated time points (n = 4). (D) CD45 chimerism in the blood was determined 24 weeks after transplantation in recipient mice (n = 4) overall (All) and for the indicated lineages. Myeloid (Mac-1), B-lymphoid (B220), T-lymphoid (CD3). The difference is significant (P ≤ .05) for CD45 chimerism between wt and Gfi1b-deficient cells, when all leukocytes are taken into account (All). (E) CD45 chimerism was determined 24 weeks after transplantation in the blood, bone marrow, spleen, and thymus of recipient mice (n = 4). (F) Frequencies of HSCs were determined in mice 24 weeks after transplantation with wt CD45.1 cells and with either wt CD45.2 BM cells or with Gfi1b-deficient CD45.2+ bone marrow cells (n = 4). (G) The relative proportion of HSCs originating from CD45.2 wt or CD45.2 Gfi1b-deficient HSCs was determined and is shown after electronic gating on CD150+CD48− cells depicted in F. (H) HSCs, bone marrow (BM), splenocytes (SP), thymocytes (thy) from mice transplanted with wt CD45.1 and Gfi1b-deficient CD45.2 bone marrow cells were genotyped and tested for the presence of the wt (CD45.1) and Gfi1b flox and Gfi1b ko alleles. (I) The frequencies of HSCs in mice either transplanted with wt CD45.1 and wt CD45.2 bone marrow cells or mice transplanted with wt CD45.1 and Gfi1b-deficient (MxCre tg Gfi1bfl/fl) bone marrow cells was determined (n = 4, P ≤ .01). (J) Quantification of which proportion of HSCs originates from CD45.2 wt or CD45.2 Gfi1b-deficient HSCs in mice transplanted with wt CD45.1 and wt CD45.2 bone marrow cells or mice transplanted with wt CD45.1 and Gfi1b-deficient bone marrow cells (MxCre tg Gfi1bfl/fl; n = 4, P ≤ .01). (K) The frequency of HSCs circulating in the peripheral blood of mice either transplanted with wt CD45.1 and wt CD45.2 bone marrow cells or mice transplanted with wt CD45.1 and CD45.2 Gfi1b-deficient bone marrow cells (MxCre tg Gfi1b fl/fl; n = 4, P ≤ .01). (L) Quantification of which proportion of HSCs circulating in blood originates from CD45.2 wt or CD 45.2 Gfi1b-deficient HSCs in CD45.1 mice transplanted with wt CD45.1 and wt CD45.2 bone marrow cells or CD45.1 mice transplanted with wt CD45.1 and Gfi1b-deficient bone marrow cells (n = 4, P ≤ .01). (M) Quantification of which proportion of Lin−, Sca-1+, c-kit+ (LSK) cells in bone marrow originate from CD45.2 wt or CD45.2 Gfi1b-deficient HSCs in CD45.1 mice transplanted with wt CD45.1 and wt CD45.2 HSCs or CD45.1 mice transplanted with wt CD45.1 and Gfi1b-deficient bone marrow cells (n = 4, P ≤ .01).
Determination of functional stem cells by limiting dilution assay
| Genotype /dose, no. of cells . | Positive recipients . |
|---|---|
| wt | |
| 200 000 | 3/3 |
| 100 000 | 3/3 |
| 20 000 | 1/3 |
| 10 000 | 0/3 |
| 5000 | 0/3 |
| Gfi1bko | |
| 200 000 | 3/3 |
| 100 000 | 3/3 |
| 20 000 | 3/3 |
| 5000 | 1/3 |
| Genotype /dose, no. of cells . | Positive recipients . |
|---|---|
| wt | |
| 200 000 | 3/3 |
| 100 000 | 3/3 |
| 20 000 | 1/3 |
| 10 000 | 0/3 |
| 5000 | 0/3 |
| Gfi1bko | |
| 200 000 | 3/3 |
| 100 000 | 3/3 |
| 20 000 | 3/3 |
| 5000 | 1/3 |
The number of functional stem cells was determined in vivo by limiting dilution. Indicated number of pIpC-treated Gfi1bfl/fl and MxCre tg Gfi1bfl/fl (Gfi1bko/ko) (both CD45.2+) bone marrow cells were transplanted with 200 000 CD45.1+ bone marrow cells into lethally irradiated CD45.1+ mice. Approximately 18 weeks after transplantation, peripheral blood was examined for presence of CD45.2+ cells. A percentage higher than 1% was a positive call.
Determination of functional stem cells by limiting dilution assay
| Genotype . | One functional stem cell within . | Upper and lower limit . |
|---|---|---|
| wt | 1:46 000 | 1:20 000-1:200 000 |
| MxCre tg Gfi1bfl/fl | 1:7000* | 1:2000-1:23 000 |
| Genotype . | One functional stem cell within . | Upper and lower limit . |
|---|---|---|
| wt | 1:46 000 | 1:20 000-1:200 000 |
| MxCre tg Gfi1bfl/fl | 1:7000* | 1:2000-1:23 000 |
Based on the results in Table 3 number of functional stem cells was determined.
A statistically significant difference with P ≤ .05.
To further examine whether loss of Gfi1b alters self-renewal and multipotency of HSCs, we first transplanted 200 000 bone marrow cells from either wt or Gfi1b-deficient CD45.2 mice in competition with wt CD45.1 bone marrow cells (Figure 4B). Transplanted Gfi1b-deficient bone marrow cells were able to compete with wt CD45.1 cells with regard to blood, bone marrow, spleen, and thymus repopulation, and recipient mice transplanted with Gfi1b-deficient bone marrow cells even showed a significantly higher level of chimerism (measured as the percentage of CD45.2+ cells) in blood than recipients that received wt CD45.2 cells (Figure 4C-D). However, when frequencies of CD45.2+ myeloid or lymphoid cells were measured in bone marrow, spleen, and thymus, there was no difference between mice that had received wt or Gfi1b-deficient bone marrow (Figure 4E). In addition, we observed a strong and highly significant expansion of transplanted CD45.2+Gfi1b-deficient HSCs in blood and bone marrow (Figure 4F-M). Gf1b-deficient (CD45.2+) HSCs represented almost 90% of all HSCs in the recipient animals (Figure 4F-J). A similar expansion of Gfi1b-deficient HSCs was also detectable in the peripheral blood of recipients that received Gfi1b-deficient bone marrow, indicating that the phenotype of HSC expansion observed in mice lacking Gfi1b is cell autonomous (Figure 4K-L).
The bone marrow of Gfi1b-deficient mice contains approximately 39 times more phenotypically defined stem cells (HSCs; Figure 2B and Table 1). Yet, limiting dilution experiments suggested only 6 time more functional stem cells in Gfi1b-deficient bone marrow (Tables 3–4). One possible explanation for this discrepancy would be that, as a result of activation, Gfi1bko/ko HSCs are at least partially compromised in their stemness and their ability to compete with wt HSCs. To test this, we transplanted a mixture of 50 sorted wt CD45.1+ HSCs (defined as above as LSK, CD48−, CD150+) with either 50 sorted CD45.2+wt HSCs or 50 sorted CD45.2+ HSCs from Gfi1bko/ko mice into syngenic recipient animals (CD45.1+; Figure 5A). We observed that Gfi1bko/ko HSCs could contribute to the same extent to myeloid and lymphoid lineage differentiation in blood and peripheral organs as wt CD45.2+ HSCs (Figure 5B-D). We measured again a significant expansion of Gfi1b-deficient CD45.2+ HSCs and LSK cells in the bone marrow and peripheral blood of recipient animals (Figure 5E-I). This expansion of HSCs is comparable with the result obtained after transplantation of the same number of wt and Gfi1b-deficient bone marrow cells (Figures 4I-L and 5E-I).
Transplantation of Gfi1b-deficient, flow-sorted HSCs. (A) Fifty HSCs originating from either wt (CD45.1) or Gfi1bko/ko (CD45.2) mice were transplanted into lethally irradiated CD45.1+ mice. Twenty-four weeks after transplantation, mice were euthanized and examined for the contribution of Gfi1b-deficient HSCs to the different lineages. (B) Percentage of CD45.2–positive cells (%CD45.2) in the blood at indicated time points after transplantation (n = 3). (C) CD45 chimerism in the blood was determined 24 weeks after transplantation in recipient mice (n = 3) overall (All) and for the indicated lineages. Myeloid (Mac-1), B-lymphoid (B220), T-lymphoid (CD3). The difference is significant (P ≤ .05) for CD45 chimerism between wt and Gfi1b-deficient cells, when all leukocytes are taken into account (All). (D) CD45 chimerism was determined 24 weeks after transplantation in the blood, bone marrow, spleen, and thymus of recipient mice (n = 3). (E) The frequency of bone marrow HSCs in mice either transplanted with wt CD45.1 and wt CD45.2 HSC cells (white) or mice transplanted with wt CD45.1 and Gfi1b-deficient (MxCre tg Gfi1bfl/fl) HSCs (black) was determined (n = 3, P ≤ .01). (F) Quantification which proportion of HSCs originates from CD45.2 wt or CD45.2 Gfi1b-deficient HSCs in mice transplanted with either wt CD45.1 and wt CD45.2 HSCs or mice transplanted with sorted HSC cells from wt CD45.1 and Gfi1b-deficient CD45.2 mice (MxCre tg Gfi1bfl/fl) (n = 3, P ≤ .01). (G) Frequency of HSCs circulating in the peripheral blood of mice either transplanted with wt CD45.1 and wt CD45.2 HSCs or mice transplanted with wt CD45.1 and Gfi1b-deficient CD45.2 HSCs (n = 3, P ≤ .01). (H) Quantification of which proportion of HSCs circulating in blood originates from CD45.2 wt or CD 45.2 Gfi1b-deficient HSCs in CD45.1 mice transplanted with wt CD45.1 and wt CD45.2 HSCs or CD45.1 mice transplanted with wt CD45.1 and Gfi1b-deficient HSCs (n = 3, P ≤ .01). (I) Quantification of which proportion of Lin−, Sca-1+, c-kit+ (LSK) cells in bone marrow originate from CD45.2 wt or CD 45.2 Gfi1b-deficient HSCs in mice transplanted with wt CD45.1 and wt CD45.2 HSCs or CD45.1 mice transplanted with wt CD45.1 and Gfi1b-deficient HSCs (n = 3, P ≤ .01). (J) Serial transplantation. Mice were transplanted with bone marrow from wt CD45.1 and Gfi1b-deficient (CD45.2) mice. After 24 weeks, chimerism in peripheral blood was determined, and 2 million bone marrow of these chimeric mice was transplanted into new lethally irradiated CD45.1 recipient mice. After 16 weeks, chimerism in the blood in these secondary transplanted mice was determined. The percentage of CD45.2 cells in the blood of the secondary transplant recipients was compared with the percentage of CD45.2 cells from the first transplant. The observed chimerism in the first transplant was set to 100% (n = 7 for second transplant, P ≤ .15). (K) Cells from 50 μL of blood were obtained from wt CD45.2 or Gfi1b-deficient CD45.2 mice and were transplanted together with 200 000 bone marrow cells from wt CD45.1 mice. Twelve weeks after transplantation, number of CD45.2 cells (which was set to 1 for CD45.2 Gfi1b-deficient blood cells) within all hematopoietic cells (CD45) in blood was determined. As a control for specificity of the CD45.2 antibody, blood obtained from an untreated CD45.1 mouse was used.
Transplantation of Gfi1b-deficient, flow-sorted HSCs. (A) Fifty HSCs originating from either wt (CD45.1) or Gfi1bko/ko (CD45.2) mice were transplanted into lethally irradiated CD45.1+ mice. Twenty-four weeks after transplantation, mice were euthanized and examined for the contribution of Gfi1b-deficient HSCs to the different lineages. (B) Percentage of CD45.2–positive cells (%CD45.2) in the blood at indicated time points after transplantation (n = 3). (C) CD45 chimerism in the blood was determined 24 weeks after transplantation in recipient mice (n = 3) overall (All) and for the indicated lineages. Myeloid (Mac-1), B-lymphoid (B220), T-lymphoid (CD3). The difference is significant (P ≤ .05) for CD45 chimerism between wt and Gfi1b-deficient cells, when all leukocytes are taken into account (All). (D) CD45 chimerism was determined 24 weeks after transplantation in the blood, bone marrow, spleen, and thymus of recipient mice (n = 3). (E) The frequency of bone marrow HSCs in mice either transplanted with wt CD45.1 and wt CD45.2 HSC cells (white) or mice transplanted with wt CD45.1 and Gfi1b-deficient (MxCre tg Gfi1bfl/fl) HSCs (black) was determined (n = 3, P ≤ .01). (F) Quantification which proportion of HSCs originates from CD45.2 wt or CD45.2 Gfi1b-deficient HSCs in mice transplanted with either wt CD45.1 and wt CD45.2 HSCs or mice transplanted with sorted HSC cells from wt CD45.1 and Gfi1b-deficient CD45.2 mice (MxCre tg Gfi1bfl/fl) (n = 3, P ≤ .01). (G) Frequency of HSCs circulating in the peripheral blood of mice either transplanted with wt CD45.1 and wt CD45.2 HSCs or mice transplanted with wt CD45.1 and Gfi1b-deficient CD45.2 HSCs (n = 3, P ≤ .01). (H) Quantification of which proportion of HSCs circulating in blood originates from CD45.2 wt or CD 45.2 Gfi1b-deficient HSCs in CD45.1 mice transplanted with wt CD45.1 and wt CD45.2 HSCs or CD45.1 mice transplanted with wt CD45.1 and Gfi1b-deficient HSCs (n = 3, P ≤ .01). (I) Quantification of which proportion of Lin−, Sca-1+, c-kit+ (LSK) cells in bone marrow originate from CD45.2 wt or CD 45.2 Gfi1b-deficient HSCs in mice transplanted with wt CD45.1 and wt CD45.2 HSCs or CD45.1 mice transplanted with wt CD45.1 and Gfi1b-deficient HSCs (n = 3, P ≤ .01). (J) Serial transplantation. Mice were transplanted with bone marrow from wt CD45.1 and Gfi1b-deficient (CD45.2) mice. After 24 weeks, chimerism in peripheral blood was determined, and 2 million bone marrow of these chimeric mice was transplanted into new lethally irradiated CD45.1 recipient mice. After 16 weeks, chimerism in the blood in these secondary transplanted mice was determined. The percentage of CD45.2 cells in the blood of the secondary transplant recipients was compared with the percentage of CD45.2 cells from the first transplant. The observed chimerism in the first transplant was set to 100% (n = 7 for second transplant, P ≤ .15). (K) Cells from 50 μL of blood were obtained from wt CD45.2 or Gfi1b-deficient CD45.2 mice and were transplanted together with 200 000 bone marrow cells from wt CD45.1 mice. Twelve weeks after transplantation, number of CD45.2 cells (which was set to 1 for CD45.2 Gfi1b-deficient blood cells) within all hematopoietic cells (CD45) in blood was determined. As a control for specificity of the CD45.2 antibody, blood obtained from an untreated CD45.1 mouse was used.
We next examined whether loss of Gfi1b might exhaust the self-renewal capacity of Gfi1b-deficient HSCs in a serial transplantation assay. We transplanted syngeneic mice (CD45.1) with wt (CD45.1) and CD45.2+Gfi1b-deficient bone marrow and tested the degree of chimerism in the primary and secondary recipient by measuring the percentage of CD45.2+ cells in the blood (Figure 5J). The experiment showed that the degree of chimerism in a secondary transplantation is maintained. The results of these experiments suggest that Gfi1bko/ko HSCs maintain their stemness and multipotency but also their ability to expand in blood and bone marrow beyond wt HSC numbers. It is thus unlikely that the difference between more than 30-fold elevated numbers of phenotypically defined HSCs on one hand and a 6-fold elevated number of functional HSCs (limiting dilution assay) on the other hand is due to a loss of multipotency and self-renewal capacity.
It has been previously shown that HSCs residing in peripheral blood of mice have long-term potential capacity.38 Since we observed a significant expansion of phenotypically defined HSCs in the blood of Gfi1b-deficient mice, we wished to verify whether these blood HSCs represent true functional stem cells. To test this, we transplanted 50 μL of blood originating either from wt or Gfi1bko/ko (both CD45.2+) mice alongside with 200 000 bone marrow cells from wt CD45.1 mice. We observed that Gfi1bko/ko HSCs from peripheral blood were able to give rise to CD45.2+ cells (Figure 5K), indicating that Gfi1bko/ko HSCs found in blood are functionally intact stem cells. Taken together, these data suggest that Gfi1bko/ko HSCs are not compromised in their ability to compete with wt HSCs and maintain their stemness, self-renewal capacity, and multipotency.
Either Gfi1b or Gfi1 are required to maintain HSCs
A direct comparison of both Gfi1- and Gfi1b-deficient mice showed that loss of Gfi1 led to an increase of HSCs, very likely due to higher cell proliferation as previously reported,27,28 but that this increase was by far not as pronounced as in Gfi1b-deficient mice (Figure 6A-B). However, when both Gfi1 and Gfi1b were deleted and mice were examined 15 days after the first pIpC injection, we observed a drastic (> 5-fold) reduction of HSC frequencies compared to wt controls (Figure 6A,C). Genotyping of the few HSCs remaining in these double deficient mice showed repeatedly that one Gfi1b allele was not excised, but both Gfi1 alleles were deleted, indicating a functional Cre recombinase (Figure 6D). We also found that, if double Gfi1/Gfi1b deficient mice were observed for a longer time (40 days after the first pIpC injection), HSCs numbers were restored to wt levels (Figure 6C), but these HSCs showed again only a partial excision of the Gfi1b locus. In addition, we found that loss of Gfi1b leads to an enhanced activity of the Gfi1 promoter in HSCs (Figure 7A) and that HSCs, in which Gfi1b was deleted, up-regulated the expression of Gfi1 mRNA (Figure 7B), confirming previously shown ability of Gfi1b and Gfi1 for cross-regulation.25,26 These data demonstrate that down-regulation of Gfi1b leads to up-regulation of Gfi1 in HSCs and suggest that the complete deletion of both Gfi1 and Gfi1b is incompatible with the generation or maintenance of HSCs.
Loss of both Gfi1 and Gfi1b reduces the number of HSCs. (A) Flow cytometric analysis of bone marrow cells of pIpC-treated wt, MxCre tg Gfi1bfl/fl, MxCre tg Gfi1fl/fl, and MxCre tg Gfi1fl/fl Gfi1bfl/fl mice after electronic gating for LSK cells and for the indicated markers. Results for MxCre tg Gfi1fl/fl Gfi1bfl/fl were obtained 15 days after the first pIpC injection (4 days after the last pIpC injection). (B) As in panel A, with frequencies depicted in percent with regard to total bone marrow (*P ≤ .05; ***P ≤ .001; n = 14 for wt, n = 14 for MxCre tg Gfi1bfl/fl and n = 3 for MxCre tg Gfi1fl/fl). (C) Simultaneous deletion of Gfi1 and Gfi1b reduced the frequency of HSCs in bone marrow by 10-fold approximately 15 days after the first pIpC injection of HSCs (**P ≤ .01). Frequencies of HSCs again reach normal (wt) levels in pIpC-injected MxCre tg Gfi1fl/fl Gfi1bfl/fl mice, when measured 40 days after the first pIpC injection (n = 14 for wt, n = 14 for MxCre tg Gfi1bfl/fl, n = 3 for MxCre tg Gfi1fl/fl, and n = 3 for MxCre tg Gfi1fl/fl Gfi1bfl/fl). (D) Genotyping of sorted HSCs of pIpCinjected MxCre tg Gfi1fl/fl Gfi1bfl/fl mice 15 days after the first pIpC injection. Excision of the Gfi1 allele is complete, showing the presence of a functional Cre recombinase, but excision of the Gfi1b allele is incomplete.
Loss of both Gfi1 and Gfi1b reduces the number of HSCs. (A) Flow cytometric analysis of bone marrow cells of pIpC-treated wt, MxCre tg Gfi1bfl/fl, MxCre tg Gfi1fl/fl, and MxCre tg Gfi1fl/fl Gfi1bfl/fl mice after electronic gating for LSK cells and for the indicated markers. Results for MxCre tg Gfi1fl/fl Gfi1bfl/fl were obtained 15 days after the first pIpC injection (4 days after the last pIpC injection). (B) As in panel A, with frequencies depicted in percent with regard to total bone marrow (*P ≤ .05; ***P ≤ .001; n = 14 for wt, n = 14 for MxCre tg Gfi1bfl/fl and n = 3 for MxCre tg Gfi1fl/fl). (C) Simultaneous deletion of Gfi1 and Gfi1b reduced the frequency of HSCs in bone marrow by 10-fold approximately 15 days after the first pIpC injection of HSCs (**P ≤ .01). Frequencies of HSCs again reach normal (wt) levels in pIpC-injected MxCre tg Gfi1fl/fl Gfi1bfl/fl mice, when measured 40 days after the first pIpC injection (n = 14 for wt, n = 14 for MxCre tg Gfi1bfl/fl, n = 3 for MxCre tg Gfi1fl/fl, and n = 3 for MxCre tg Gfi1fl/fl Gfi1bfl/fl). (D) Genotyping of sorted HSCs of pIpCinjected MxCre tg Gfi1fl/fl Gfi1bfl/fl mice 15 days after the first pIpC injection. Excision of the Gfi1 allele is complete, showing the presence of a functional Cre recombinase, but excision of the Gfi1b allele is incomplete.
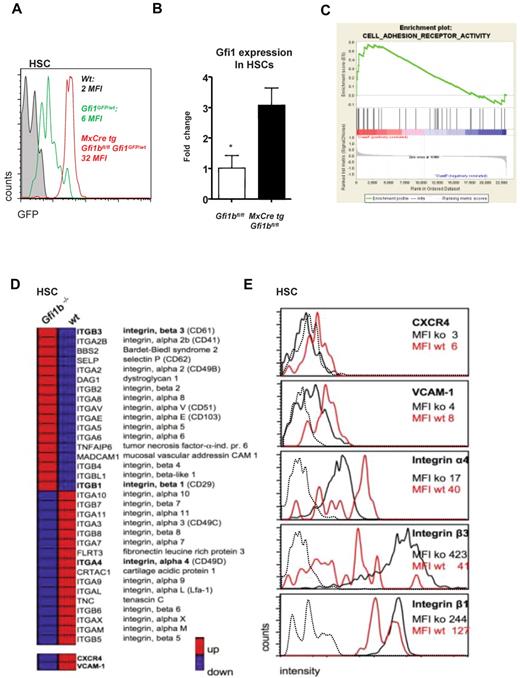
Gfi1b regulates Gfi1 and the expression of surface molecules important for niche organization. (A) MxCre tg Gfi1bfl/fl Gfi1GFP/wt, wt, and Gfi1bfl/fl Gfi1GFP/wt mice were injected with pIpC, and 30 days after the first injection (equivalent to 21 days after the last injection) mice were killed and examined for expression of GFP, which follows the activity of the Gfi1 promoter. Loss of Gfi1b leads to an enhanced activity of the Gfi1 promoter. (B) Real-time polymerase chain reaction analysis of Gfi1 gene expression in HSCs from mice with the indicated genotypes (n = 3). (C) Example of the clustering of Gfi1b effector genes enriched in a specific geneset (Adhesion molecules) generated using the GSEA software (http://www.broad.mit.edu/gsea/).(D) Overview of genes differentially expressed in wt and Gfi1b-deficient HSCs. Red bars represent relatively high and blue bars low expression levels (average fold induction or repression) in Gfi1bko/ko HSCs compared with wt HSCs. CXCR4 and VCAM-1 were not included in the GESA defined adhesion molecule pathway but were also down-regulated on the RNA level. (E) Expression level of different surface adhesion proteins. The expression of these proteins was changed in analogy to the gene expression array results. Mean fluorescence intensities (MFIs) of the respective surface molecules in Gfi1bko/ko (ko) and wt HSCs (wt) are depicted. Dotted line indicates isotype controls.
Gfi1b regulates Gfi1 and the expression of surface molecules important for niche organization. (A) MxCre tg Gfi1bfl/fl Gfi1GFP/wt, wt, and Gfi1bfl/fl Gfi1GFP/wt mice were injected with pIpC, and 30 days after the first injection (equivalent to 21 days after the last injection) mice were killed and examined for expression of GFP, which follows the activity of the Gfi1 promoter. Loss of Gfi1b leads to an enhanced activity of the Gfi1 promoter. (B) Real-time polymerase chain reaction analysis of Gfi1 gene expression in HSCs from mice with the indicated genotypes (n = 3). (C) Example of the clustering of Gfi1b effector genes enriched in a specific geneset (Adhesion molecules) generated using the GSEA software (http://www.broad.mit.edu/gsea/).(D) Overview of genes differentially expressed in wt and Gfi1b-deficient HSCs. Red bars represent relatively high and blue bars low expression levels (average fold induction or repression) in Gfi1bko/ko HSCs compared with wt HSCs. CXCR4 and VCAM-1 were not included in the GESA defined adhesion molecule pathway but were also down-regulated on the RNA level. (E) Expression level of different surface adhesion proteins. The expression of these proteins was changed in analogy to the gene expression array results. Mean fluorescence intensities (MFIs) of the respective surface molecules in Gfi1bko/ko (ko) and wt HSCs (wt) are depicted. Dotted line indicates isotype controls.
Loss of Gfi1b affects expression of surface molecules important for the hematopoietic stem cell niche
To further explore how Gfi1b might function in HSCs and how its function differs from Gfi1, we analyzed relative gene expression levels of wt and Gfi1bko/ko HSCs using Affymetrix gene arrays. We found that genes encoding cell adhesion molecules and integrins were profoundly deregulated in Gfi1bko/ko HSCs (Figure 7C-D). Notably, such proteins as vascular cell adhesion protein 1 (VCAM-1), CXCR4, and integrin α4 that are important to retain HSCs in their endosteal niche1,6,39-42 were expressed at significantly lower levels on Gfi1bko/ko HSCs compared with wt HSCs (Figure 7D-E and Table 5). On the other hand, adhesion molecules such as integrin β1 and β3 that mediate endothelial adhesion43,44 were significantly up-regulated at mRNA and protein level (Figure 7D-E and Table 5), suggesting that loss of Gfi1b directly or indirectly affects expression of cell surface molecules that have a role in niche organization.
Change of expression of different surface proteins on stem cells after deletion of Gfi1b
| Surface protein . | Relative expression level . | P . | |
|---|---|---|---|
| Gfi1bfl/fl . | MxCre tg Gfi1bfl/fl . | ||
| Integrin α4 (CD49d) | 1 | 0.48 ± 0.18 | .02 |
| CXCR4 | 1 | 0.53 ± 0.09 | .01 |
| VCAM-1 | 1 | 0.46 ± 0.07 | .01 |
| Integrin β3 (CD61) | 1 | 13.7 ± 1.9 | .02 |
| Integrin β1 (CD29) | 1 | 1.53 ± 0.2 | .05 |
| Surface protein . | Relative expression level . | P . | |
|---|---|---|---|
| Gfi1bfl/fl . | MxCre tg Gfi1bfl/fl . | ||
| Integrin α4 (CD49d) | 1 | 0.48 ± 0.18 | .02 |
| CXCR4 | 1 | 0.53 ± 0.09 | .01 |
| VCAM-1 | 1 | 0.46 ± 0.07 | .01 |
| Integrin β3 (CD61) | 1 | 13.7 ± 1.9 | .02 |
| Integrin β1 (CD29) | 1 | 1.53 ± 0.2 | .05 |
In 3 independent experiments, expression by mean fluorescence level of the different proteins was measured. To facilitate differences in up- or down-regulation of the different proteins, the expression in the Gfi1bfl/fl was set to 1 (n = 3 for all sets). Depicted are mean values and SEM. P values are based on unpaired 2-sided t test.
Discussion
Gfi1 and Gfi1b are 2 distinct and unique proteins encoded by 2 different genes. Both are nuclear zinc finger proteins sharing their N-terminal SNAG repressor domain and 6 C2H2 C-terminal zinc-coordinating motifs.13 During hematopoiesis, the expression of Gfi1 and Gfi1b is often mutually exclusive,30 but in HSCs both are expressed at the same time, albeit, as we report here, Gfi1 at a much lower levels than Gfi1b. This observation and our observation that Gfi1b expression levels are highest in the quiescent (dormant) CD34− HSC fraction and decrease in subsequent MPP subsets was intriguing and pointed to the possibility that Gfi1b is implicated in regulating the function of HSCs that may be different from the role that its paralogue Gfi1 exerts in stem cells.
The role of Gfi1 in HSCs has been inferred from previously reported experiment, which characterized the LSK bone marrow subset that contains HSCs. Two independent reports suggested that Gfi1 restricts HSCs proliferation and is required to maintain their self-renewal capacity.27,28 Our analysis provides evidence that Gfi1b-deficient HSCs cycle at a higher rate than wt HSCs, which has also been observed, albeit more indirectly for Gfi1 deficient HSCs.27,28 However, Gfi1b-deficient mice show a very strong increase of HSC numbers in the bone marrow, spleen, and peripheral blood, which was never observed in Gfi1-deficient mice. AnnexinV staining, BrdU label experiments, and Hoechst staining showed that loss of Gfi1b does not noticeably affect cell survival but increases specifically cell cycle progression in the HSC fraction but not in the MPP subsets. The absolute difference between these BrdU and Hoechst measurements (4% wt HSC undergoing cell cycle in the Hoechst staining as opposed to 11% for the BrdU staining) is probably due to the fact that application of BrdU itself activates stem cells and promotes their entry into cell cycling.2 Our observations with regard to cell cycle progression and expansion of HSCs suggest that Gfi1b very likely exerts functions in stem cells. These roles and functions of Gfi1b are on one hand similar to the function of Gfi1, as loss of Gfi1 and Gfi1b increases cell cycling. On the other hand, the function of Gfi1b in HSCs is different from Gfi1, as Gfi1b-deficient HSCs do not lose their transplantation capacity, as is the case for Gfi1-deficient HSCs.
According to our label retention assay, the fraction of quiescent HSCs is significantly smaller in Gfi1b-deficient mice than in wt animals. In addition, Gfi1b-deficient HSCs show an increased level of ROS that is not seen in wt HSCs. It is likely that ROS play an important role in the expansion of Gfi1b-deficient HSCs since we can demonstrate that NAC, which counteracts ROS, can significantly limit the expansion of HSCs. The high level of ROS also suggests that Gfi1b-deficient HSCs are metabolically more active than wt HSCs and have a higher mitochondrial activity. However, it is also possible that they are exposed to an environment with higher oxygen levels or that Gfi1b regulates genes involved in the regulation of ROS levels.
We do not have evidence for a genetic regulation of ROS levels, but our observations suggest that Gfi1b-deficient HSCs, in addition to being metabolically more active than wt HSCs, may also be localized in another bone marrow microenvironment that is different from the hypoxic endosteal niche, where they are normally kept in a dormant state. HSCs only leave this niche and enter the cell cycle every 60-100 days or after external activation.2 It is possible that loss of Gfi1b causes a delocalization of HSCs away from the hypoxic endosteal niche to a more oxygen rich environment, which drives them into the cell cycle and causes a significant expansion in bone marrow and blood. Our finding that VCAM-1, integrin α4, and CXCR4, which have been described to play an important role in keeping HSCs in a dormant state in their endosteal niche,1,6,39-42 are all down-regulated in Gfi1bko/ko HSCs, points to this as a possible explanation for the activation of Gfi1b-deficient HSCs. Dysregulation of surface proteins regulating retention of HSCs in the endosteal niche could lead to the release of Gfi1bko/ko HSCs from the hypoxic endosteal niche.45,46 Subsequently, these HSCs would become more exposed to oxygen,35,36 which would result in the appearance of higher ROS triggering entry of these HSCs into the cell cycle.37
Several results reported here would be consistent with this notion as the increased numbers of Gfi1b-deficient HSCs in bone marrow, spleen, and blood, their higher cell cycle rate, their higher levels of ROS, and their lower expression levels of CXCR4, VCAM-1, and integrin α4. However, to definitely show whether Gfi1b-deficient HSCs are indeed localized in different anatomically defined locations in the bone marrow would require future studies with in-depth in vivo imaging as demonstrated elegantly in different recent publications.45,46 In addition, it remains to be shown how precisely Gfi1b regulates the promoters of genes encoding surface and adhesion molecules necessary for niche organization, since we found integrins and adhesion molecules both up- and down-regulated in Gfi1b-deficient HSCs. One exception is CXCR4, which is a Gfi1 target gene47 and might be repressed by Gfi1, since we found Gfi1 mRNA expression to be significantly increased in Gfi1b-deficient HSCs because of cross-regulation between these 2 factors.25,26 A repression of CXCR4 by this mechanism is conceivable and may be implicated in the Gfi1b null phenotype since it has been described that down-regulation of CXCR4 activates HSCs.48
The most dramatic phenotype in Gfi1bko/ko mice remains the strong expansion of HSCs in bone marrow and blood. This feature is clearly cell autonomous since it reappears upon transplantation in recipient mice. It is of interest that such a significant expansion, as seen in Gfi1b adult knockout mice, was not reported for Gfi1b-deficient fetal HSCs29 nor for Gfi1-deficient HSCs.27,28 This underlines that Gfi1b and Gfi1 indeed exert different functions in stem cells and that both also have different roles in adult and fetal stem cells. In contrast to Gfi1, Gfi1b seems to not only restrict HSCs proliferation but also their expansion, and Gfi1bko/ko HSCs do not loose their potential to self-renew nor to initiate multilineage differentiation as was described for Gfi1−/− HSCs. This is supported by our competitive and serial transplantation experiments. Loss of Gfi1b leads to a massive expansion of HSCs in blood and spleen. One attractive hypothesis for this observation would be that Gfi1b regulates mechanisms that govern egress of HSCs from bone marrow to blood. The fact that the expansion of HSCs in spleen is more than 100-fold in the spleen and 95-fold in the blood of Gfi1b-deficient mice compared with wt mice would support such a hypothesis. In addition, it has been shown that antagonizing the binding of stromal cell–derived factor-1 to its receptor CXC motif receptor-4 (CXCR4) can cause the egress of HSCs to the peripheral blood.48 The fact that we see a reduction of CXCR4 on Gfi1bko/ko HSCs and a strong presence of HSCs in the blood and spleen of Gfi1b-deficient mice is certainly consistent with our hypothesis. However, further experiments in particular microscopic analyses need to be done to prove whether Gfi1b indeed regulates HSC egress or blood mobilization. Moreover, it is also possible that the few HSCs that are always present in the blood or spleen expand independently after deletion of Gfi1b following a similar mechanism as their bone marrow counterparts.25,26
Although Gfi1bko/ko mice have a larger pool of both phenotypically and functionally defined HSCs than wt mice, there is a discrepancy between numeric expansion (over 30 times) determined by flow cytometry and functional expansion of HSCs (approximately 6 times) determined by limiting dilution and transplantation. This cannot be explained by the hypothesis that Gfi1bko/ko HSCs may be compromised in their self-renewal capacity, because the transplantation of equal numbers of phenotypically defined wt and Gfi1bko/ko HSCs resulted in an almost identical contribution to the different hematopoietic lineages. In addition, all transplantation and repopulation experiments confirm that Gfi1b-deficient HSCs are functionally intact. However, all these experiments also showed that the population of HSCs itself was always significantly expanded in both bone marrow and blood. One possibility to reconcile these findings would be that only a fraction of the expanded Gfi1bko/ko HSC population is able to give rise to differentiated progeny possibly because the loss of Gfi1b increases the rate of symmetrical over asymmetrical HSC divisions.49 A higher rate of symmetrical divisions could lead to an expansion of the HSCs without increasing the contribution to the different lineages. This could explain why an over 30-fold increased number of HSCs does not translate into an equal higher number of functional HSCs as determined by the limiting dilution assay or into equally increased numbers of differentiated cells after transplantations. Albeit compelling, this can only be proven in future studies by monitoring symmetrical and asymmetrical divisions of HSCs using tools such as the described Notch target reporter mice.49
Since we found that Gfi1 is up regulated in Gfi1b-null HSCs, it is conceivable that the loss of either Gfi1 protein can be compensated by a cross-regulatory mechanism in HSCs as was described for lymphoid cells25,26 and that the knockout of either gene only reveals those parts of the function of both transcription factors that cannot be compensated by the other. It was thus of interest to investigate the consequences of a simultaneous deletion of both Gfi1 and Gfi1b. The experiment showed that a counter-selection occurs for HSCs that lack both Gfi1 and Gfi1b, since the few HSCs that are still found in pIpC-treated MxCre tg Gfi1fl/flGfi1bfl/fl mice always retain one intact Gfi1b allele. This indicates that the deletion of both Gfi1 and Gfi1b is incompatible with the maintenance of an HSC population, and only those HSCs emerge in double KO mice that have escaped Cre-mediated deletion of at least one Gfi1b allele. This also demonstrates that Gfi1b is indispensible in the absence of Gfi1. However, one important conclusion from our data is that in the presence of Gfi1, the modulation of Gfi1b function affects HSC numbers but does not affect their function. This particular feature may render Gfi1b a suitable target for therapeutic intervention in situations where HSC expansion is required,50 either to facilitate HSCs harvesting from donors or to enable faster recovery of patients after bone marrow transplantation. This would protect patients against the lethal effects of the procedure and would improve outcome of leukemia and lymphoma.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are indebted to Mathieu Lapointe and Rachel Bastien for technical assistance; Nancy Laverriere, Marlène Bernier, and Marie-Claude Lavallée for excellent animal care; Eric Massicotte and Julie Lord for fluorescence-activated cell sorting; and Qinzhang Zhu for embryonic stem cell techniques and help generating the conditional mouse strain. We thank the Genome Quebec for performing the arrays and Mark Schlissel and Danae Shultz for providing the Abelson B-cell lines from Gfi1b-deficient bone marrow cells.
This work was supported by grants from the Roche Foundation for Anemia Research and Canadian Institutes of Health Research (MOP-84 238, MOP-94 846; T.M.) and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (J.Z., W.E.P.). T.M. holds a Canada Research Chair (Tier 1), and C.K. is a Cole Foundation fellow.
Authorship
Contribution: C. Khandanpour performed research, analyzed data, and wrote the manuscript; E.S.-A. performed research and analyzed data; L.V. and M.-C.G. performed research, analyzed data, and edited manuscript; J.Z. and W.E.P. provided vital reagents; T.O. assisted in generating the described mouse strain; C. Kosan assisted in generating the described mouse strain and edited the manuscript; and T.M. designed the research, analyzed data, wrote the manuscript, and provided funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tarik Möröy, Institut de recherches cliniques de Montreal, IRCM, 110 avenue des Pins Ouest, H2W 1R7, Montreal, QC, Canada; e-mail: Tarik.Moroy@ircm.qc.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal