Abstract
Gray platelet syndrome (GPS) is an inherited bleeding disorder characterized by macrothrombocytopenia and absence of platelet α-granules resulting in typical gray platelets on peripheral smears. GPS is associated with a bleeding tendency, myelofibrosis, and splenomegaly. Reports on GPS are limited to case presentations. The causative gene and underlying pathophysiology are largely unknown. We present the results of molecular genetic analysis of 116 individuals including 25 GPS patients from 14 independent families as well as novel clinical data on the natural history of the disease. The mode of inheritance was autosomal recessive (AR) in 11 and indeterminate in 3 families. Using genome-wide linkage analysis, we mapped the AR-GPS gene to a 9.4-Mb interval on 3p21.1-3p22.1, containing 197 protein-coding genes. Sequencing of 1423 (69%) of the 2075 exons in the interval did not identify the GPS gene. Long-term follow-up data demonstrated the progressive nature of the thrombocytopenia and myelofibrosis of GPS resulting in fatal hemorrhages in some patients. We identified high serum vitamin B12 as a consistent, novel finding in GPS. Chromosome 3p21.1-3p22.1 has not been previously linked to a platelet disorder; identification of the GPS gene will likely lead to the discovery of novel components of platelet organelle biogenesis. This study is registered at www.clinicaltrials.gov as NCT00069680 and NCT00369421.
Introduction
Gray platelet syndrome (GPS, OMIM #139090) is an inherited platelet disorder characterized by thrombocytopenia and typical gray appearance of platelets by light microscope, due to the absence of α-granules and their constituents.1-3 Since its initial description in 1971, GPS has been reported in various populations.3-21 The diagnosis of GPS requires demonstration of the absence or marked reduction of α-granules in platelets observed by electron microscopy (EM).2,22,23 Megakaryocytes also show decreased α-granules.24 Platelet dense bodies and lysosomes are unaffected.22,25 Alpha granules, the most abundant vesicles in platelets, store proteins that promote platelet adhesiveness and wound healing when secreted during platelet activation.26,27 Some α-granule proteins (eg, platelet factor 4 and β-thromboglobulin) are synthesized in megakaryocytes and packed into the vesicles, whereas others are either passively (eg, immunoglobulins and albumin) or actively (eg, fibrinogen) taken up from the plasma by receptor-mediated endocytosis.26,27 Proteins synthesized in megakaryocytes are markedly reduced in GPS, whereas other α-granule constituents are less affected.25,27 Studies of granule membrane-specific proteins have shown that platelets and megakaryocytes of GPS patients have rudimentary α-granule precursors.7,28,29 Therefore, the basic defect in GPS is thought to be the inability of megakaryocytes to pack endogeneously synthesized secretory proteins into developing α-granules.
Most GPS patients have macrothrombocytopenia.3,5 The number of megakaryocytes in the bone marrow is normal, but platelet survival is reduced.6 The shortened survival of α-granule-deficient platelets and the inability of defective megakaryocytes to mature normally are thought to contribute to the thrombocytopenia of GPS. Myelofibrosis3,10,26 and splenomegaly1,3,5,14,26 occur in GPS. Although rare patients with severe hemorrhage have been reported,8 the bleeding tendency in GPS is thought to be mild to moderate.3,5,10,26,27 In vitro platelet aggregation tests give variable results.3 Treatment is nonspecific and includes platelet transfusions before surgeries, DDAVP (1-desamino-8-D-arginine vasopressin), and splenectomy.3,10,26
Most GPS families involve a single case or multiple affected siblings born to unaffected, often consanguineous parents, suggesting autosomal recessive (AR) inheritance. Rare families with autosomal dominant (AD)30 and X-linked19,31 variants of GPS have been reported. X-linked GPS-like disease, caused by mutations in GATA-1, results in generalized hypogranularity of platelets and erythroid abnormalities.19,31 The genetic cause of the most common, AR type of GPS remains unknown. HZF was suggested as a candidate gene because of similarities of the Hzf-null mouse model to GPS; however, no mutations were identified in the human HZF gene in 5 patients from 3 GPS families.32
Here, we present the clinical and molecular characterization of 25 GPS patients from 14 unrelated families from various populations including Bedouin, Turkish, Mennonite, French, German, Somalian, African American, and mixed Northern and Southern European backgrounds. We demonstrate the progressive nature of thrombocytopenia and myelofibrosis in GPS and emphasize the fatal nature of the disorder in some cases. In addition, using genome-wide linkage analysis and homozygosity mapping we localized the GPS gene to a 9.4-Mb interval on 3p21.1-3p22.1 including 197 protein-coding genes and excluded 1423 (69%) of the 2075 exons in the interval.
Methods
Patients
Between January 2000 and January 2010, a total of 116 individuals (25 patients) from 14 unrelated families were evaluated. All patients or their parents gave written informed consent in accordance with the Declaration of Helsinki. Fifty-nine individuals from 14 unrelated GPS families were enrolled in the protocol, “Genetic Analysis of Gray Platelet Syndrome” (www.clinicaltrials.gov, NCT00069680), approved by the National Human Genome Research Institute (NHGRI) Institutional Review Board. Sixty individuals from the Bedouin family were enrolled in the “Clinical and Genetic Analysis of Gray Platelet Syndrome” study approved by the Israeli Supreme Helsinki Committee of the Israeli Ministry of Health. In addition, 25 individuals including 10 patients from 7 families (Figure 1, families 1, 2, 4, 5, 11, 13, and 14) who were clinically evaluated at the National Institutes of Health (NIH) Clinical Center, were also enrolled in the protocol “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism” (www.clinicaltrials.gov, NCT00369421).
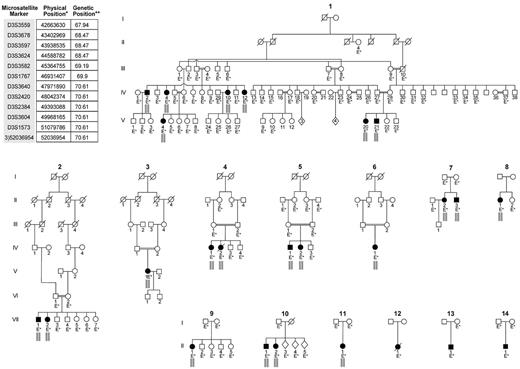
Pedigrees of 14 GPS families included in the study and the microsatellite markers used to establish linkage. The microsatellite markers listed in the top left corner of the figure are the set of markers located within the homozygosity region that is indicated by the small gray bars placed under affected individuals in the pedigrees. These gray bars indicate that, for this segment of chromosome 3, these patients inherited the same set of microsatellite genotypes from their mother and father; hence they are homozygous by descent. *Physical map positions are based on UCSC genome browser version hg18. **Genetic map positions in centiMorgans are based on Marshfield map. Patients in families 12, 13, and 14 did not display a homozygous haplotype in the linked region.
Pedigrees of 14 GPS families included in the study and the microsatellite markers used to establish linkage. The microsatellite markers listed in the top left corner of the figure are the set of markers located within the homozygosity region that is indicated by the small gray bars placed under affected individuals in the pedigrees. These gray bars indicate that, for this segment of chromosome 3, these patients inherited the same set of microsatellite genotypes from their mother and father; hence they are homozygous by descent. *Physical map positions are based on UCSC genome browser version hg18. **Genetic map positions in centiMorgans are based on Marshfield map. Patients in families 12, 13, and 14 did not display a homozygous haplotype in the linked region.
The inclusion criterion was platelet EM characteristic for GPS. Clinical investigations included medical and family history and physical examination. Pedigrees were drawn for each family. All patients had abdominal ultrasonography. Several patients had bone marrow (BM) evaluations. The degree of bleeding was classified as “severe” if blood transfusion was required, “moderate” if no blood transfusion but iron supplementation was required, and “mild” if bleeding was only minor.
Laboratory evaluations
Hematologic and biochemical analysis.
Complete blood count and peripheral blood smears were performed on all patients, available parents, and unaffected siblings. Additional evaluations included prothrombin time (PT), partial thromboplastin time (PTT), serum iron, transferrin, transferrin saturation, ferritin, vitamin B12, folate, liver and kidney function tests, serum protein electrophoresis, quantitative immunoglubulins, sedimentation rate, and TSH and T4 measurements using routine methods.
Bone marrow analysis.
Posterior iliac crest bone marrow aspirate smears were stained with Wright–Giemsa. B-Plus fixed, paraffin-embedded biopsies were stained with hematoxylin and eosin. For assessment of fibrosis, bone marrow biopsy sections were stained with Gomori silver impregnation. Images were obtained via Olympus BX-51 digital microscope (Olympus America) equipped with a UPlanFL 2×/0.50 numeric aperture objective and Olympus DP70 digital camera system. Imaging software was Adobe Photoshop CS3 (Adobe Systems).
Electron microscopy.
For platelet EM, blood samples were mixed immediately with citrate-dextrose, pH 6.5, in a ratio of 9:1 blood to anticoagulant. Platelet-rich plasma (PRP) was separated by centrifugation at 100g for 10 minutes at room temperature. Samples of PRP were fixed, embedded, sectioned, and stained for EM as described.33,34 For white blood cell (WBC) EM, citrate PRP was prepared by centrifugating blood at 100g for 20 minutes at room temperature. Buffy coats were obtained by high speed centrifugation and further processed as described.35
Platelet studies
Platelet aggregation studies.
PRP samples were studied in a lumiaggregometer (Chrono-Log) using 1800 U/mL luciferase and 16mM adenosine diphosphate, 22mM epinephrine, 4.4 mg/mL equine skin collagen, and 540 mg/mL arachidonic acid.
Analysis of platelet lysates.
Detergent lysates of washed platelets were prepared as described36 and analyzed for platelet factor 4 (PF4) and fibronectin using commercial enzyme-linked immunosorbent assay (ELISA; Asserachrom PF4, Diagnostica Stago; Imuclone Fibronectin ELISA Kit, American Diagnostica,) and for thrombospondin-1 (TSP-1) using Western blot analysis. The activity of von Willebrand factor (ristocetin cofactor) activity, antigen, and multimeric pattern were analyzed using published methods.36 Lysate fibrinogen was measured with an ELISA using an immunopurified, rabbit polyclonal immunoglobulin G (Calbiochem).37
Platelet flow cytometry.
The membrane surface expression of P-selectin and fibrinogen receptor proteins as well as the rebinding of platelet TSP-1 in resting and activated platelets was studied by flow cytometry. Immediately after collection, citrated whole blood (450 mL) was mixed for 5 minutes with either 50 mL phosphate-buffered saline, 50 mL 200mM lyophilized adenosine-5′-diphosphate solution (Bio/Data Corp), or 50 mL 100mM thrombin receptor-activating peptide from Biosearch and further processed and evaluated as described.17,28 Thrombin stimulation of platelets in citrated venous whole blood and labeling with anti-CD62P or anti-TSP-1 antibodies was performed as described.38
Genome-wide linkage analysis
Seventy-four individuals, including 14 patients from 6 families (Figure 1, families 1, 4, 5, 8, 9, and 13) were included in the genome-wide analysis (GWA) performed in 2 groups.
GWA of group 1, consisting of 60 individuals including 7 patients from the Bedouin family (Family 1), was performed using the ABI PRISM Linkage Mapping Set using 400 fluorescently labeled polymerase chain reaction (PCR) primer pairs with 10-centiMorgan (cM) intervals. Genotypes were determined using the ABI Prism 3100 DNA analyzer (Applied Biosystems) and GENE SCAN 3.1, GENOTYPER 3.1, and GENEMAPPER 1.1 software. GWA of group 2, consisting of 26 individuals including 7 patients from 5 families (families 4, 5, 8, 9, and 13), was performed using 1000 microsatellite markers at 5-cM intervals (DeCODE). The data were evaluated for Mendelian inheritance of marker alleles by 2-point and multipoint analysis and haplotype analysis programs, using the Superlink Online software Version 1.5.39 Two-point and multipoint logarithm of the odds (LOD) score analyses were performed based on the compatibility of the pedigrees with AR inheritance with full penetrance. In addition, homozygosity mapping40 was carried out on the data provided by the GWA using the algorithm in the software package MAPMAKER/MAPHOMOZ (web/ftp://ftp-genome.wi.mit.edu/distribution/software/homoz), version: May 1995.
Fine mapping
Fine mapping was initially performed on 102 individuals, including 21 patients from 11 families (Figure 1, families 1, 4, 5, 7, 8, 9, 10, 11, 12, 13, and 14), using 40 microsatellite markers (between D3S1277 at position 34630714 and D3S3717 at position 55156784 on University of California Santa Cruz [UCSC] hg18) localized at approximately 500-kb intervals (DeCODE). A second round of fine mapping was performed on 37 individuals, including 16 patients from 11 families (No 1, 2, 4, 5, 7, 9, 10, 11, 12, 13, and 14), using 63 markers (between D3S3559 at position 42663630 and D3S1588 at position 54072888 on UCSC hg18) with intervals < 200 kb. Custom-made markers were developed by locating tandem repeats using DNA sequences from UCSC Genome Bioinformatics (http://www.genome.ucsc.edu/ Build 36.1; hg18) and Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html).41 Primer sequences were identified using Primer3 (http://primer3.sourceforge.net) and checked with the UCSC Genome Bioinformatics In-Silico PCR program (http://www.genome.ucsc.edu/cgi-bin/hgPcr?command = start). Custom-made markers were named as “chr #)starting position of the primer” such as 3)52036954; primers are available upon request. The PCR products were pooled based on fluorescent labels and expected allele size. Fragments were separated by capillary electrophoresis on a Genetic Analyzer 3100 and analyzed using GENESCAN 3.7 and GENOTYPER 2.5 software (Applied Biosystems). All genotyping was done including Centre d'Etude du Polymorphisme Humain control individual #1347-02 (Applied Biosystems).
SNP analysis
Whole genome single-nucleotide polymorphism (SNP) analysis was performed using the Illumina Human 1M–Duo SNP array and Illumina's standard protocol on 8 GPS patients: 1(V-20), 2(VII-2), 3(V-1), 6(IV-1), 10(II-1), 12(II-1), 13(II-1), and 14(II-1). The region of homozygosity was identified based on the “B allele frequency” plots of the Genome Studio software Illumina Version: 2010.1.
Haplotype analysis
Haplotypes were constructed for 25 affected individuals and their parents using the fine mapping data.
Sequence analysis
Data regarding position, coding region sequence, and exon-intron boundaries were obtained from Ensemble Genome Browser (http://www.ensembl.org/), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/mapview/), and UCSC (http://genome.ucsc.edu/), and used to designate regions of interest. Bidirectional sequencing of the PCR-amplified products was analyzed as described.42 A first-pass ranking of missense variants by sequence conservation, in conjunction with the identification of nonsense, splice-site, and frame-shift variants, was used to create prioritized listings of sequence variants; further analysis established the likelihood of disease relevance and/or correlation with inheritance pattern.
Results
Clinical manifestations
Table 1 presents the clinical features of the 21 GPS patients (13 females, 8 males) from 14 families; 10 of the 21 patients were not previously published. Duration of follow up, available for 16 patients, ranged from 2 to 32 years (13.3 ± 9.4 years).
Clinical and laboratory characteristics of GPS patients
| Family no. (patient no.) . | Sex . | Inheritance pattern . | Ethnicity . | Age at onset (y) . | Age at diagnosis (y) . | Age at most recent evaluation (y) . | Age at previous report (y)ref . | Duration of follow-up (y) . | Presenting symptoms . | Types of hemorrhage . | Severity of hemorrhage . | Splenomegaly . | Myelofibrosis age/grade* . | PLT count (K/μL) . | Hb (g/dL) . | WBC (K/μL) . | Vitamin B12 (pg/mL)† . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (IV-2) | M | AR, First Cousins | Muslim Bedouins | NA | 46 | 57 | Not reported | 9 | Low PLT count identified incidentally | No symptoms | Mild | Yes | NE | 37 | 14.3 | 3.6 | 1378 |
| 1 (V-20) | F | AR, First Cousins | Muslim Bedouins | 0.7 | 6 | 17 | 813 | 9 | Low PLT count identified incidentally | Epistaxis, menometrorrhagia | Moderate | Yes | 8/grade 1-2 | 59-225 | 11.3 | 3.3 | 5699 |
| 1 (V-21) | M | AR, First Cousins | Muslim Bedouins | 3 | 3 | 14 | 513 | 9 | Low PLT count identified incidentally | No symptoms | Mild | Yes | NE | 68- 351 | 12.3 | 3.7 | 3627 |
| 2 (VII-1) | M | AR, 3rd cousins once removed | Mennonite | 4 | 8 | 10 | Not reported | 2 | Sister with GPS | Easy bruising, epistaxis | Mild | Yes | NE | 84-97 | 13 | 5.6 | 1852 |
| 2 (VII-2) | F | AR, 3rd cousins once removed | Mennonite | 0.6 | 6 | 9 | Not reported | 3 | Major bruise after minor injury | Easy bruising, epistaxis, intracranial hemorrhage | Severe | Yes | NE | 88-97 | 12.7 | 6.1 | 1859 |
| 3 (V-1) | F | AR, Second Cousins | Caucasian, (French) | Childhood | Childhood | 64 | 5517 | 9 | Mild epistaxis, easy bruising | Epistaxis, menometrorrhagia, hemorrhage during surgery and after deliveries, gastric hemorrhage without lesion | Moderate | Yes | NE | 18-70 | NA | NA | NE |
| 4 (IV-1) | F | AR, First Cousins | Caucasian, (Turkish) | 2.5 | 15 | 29 | Not reported | 14 | Easy bruising, anemia | Easy bruising, epistaxis, menometrorrhagia | Severe | Yes | 2/no fibrosis, 23/grade 3 | 28-46 | 8.6 | 3.3 | 1441 |
| 4 (IV-2) | F | AR, First Cousins | Caucasian, (Turkish) | 6 | 10 | Died at 21 | Not reported | 11 | Prolonged bleeding after dental work | Easy bruising, epistaxis, menometrorrhagia | Severe | Yes | NE | 5-25 | 10.9 | 4.1 | 2139 |
| 5 (IV-1) | M | AR, First Cousins | Caucasian, (Turkish) | 3 | 12 | 21 | Not reported | 9 | Splenomegaly, low PLT count | Easy bruising, epistaxis | Mild | Yes | NE | 65 | 16‡ | 4.7 | 927 |
| 5 (IV-2) | F | AR, First Cousins | Caucasian, (Turkish) | 6 | 6 | 13 | Not reported | 6 | Low PLT count identified incidentally | Easy bruising | Mild | No | NE | 23-105 | 12.5 | 4.8 | NE |
| 6 (IV-1) | F | AR, First Cousins | Caucasian, (Turkish) | 6 | 33 | 33 | Not reported | 0 | Low PLT count identified incidentally | Epistaxis, menometrorrhagia | Mild | Yes | 33/grade 3-4 | 16 | 8.3 | 1.7 | 1078 |
| 7 (II-2) | F | AR | Caucasian (German) | Early childhood | 17 | 17 | 176 | 0 | Easy bruising | Easy bruising, epistaxis, hemorrhage after tonsillectomy, menometrorrhagia | Severe | NA | 17/grade 3-4 | 33-150 | NA | NA | NE |
| 7 (II-3) | M | AR | Caucasian (German) | 4 | 12 | 12 | 126 | 0 | Easy bruising | Hemorrhage after tonsillectomy and tooth extraction | Severe | NA | NE | 69-125 | NA | NA | NE |
| 8 (II-1) | F | Simplex | Caucasian, (mixed Northern European) | 0 | 3 | 30 | 48 | 26 | Easy bruising | Subgaleal hemorrhage, nasopharyngeal hemorrhage, menometrorrhagia | Severe | Yes | 10/grade 1, 29/grade 2-3 | 31 | 10 | 6 | NE |
| 9 (II-1) | F | Simplex | Caucasian, (Turkish) | 0.8 | 5 | 24 | Not reported | 19 | Petechiae, epistaxis | Severe epistaxis, hematemesis, melena | Severe | Yes | NE | 20- 72 | 10.8 | 3.3 | 1524 |
| 10 (II-1) | F | Simplex | African American, (Somalian) | NA | 16 | 16 | 1616 | 0 | NA | NA | NA | NA | NE | NA | NA | NA | NE |
| 10 (II-2) | M | Simplex | African American, (Somalian) | NA | 14 | 14 | 1416 | 0 | NA | NA | NA | NA | NE | 95 | NA | NA | NE |
| 11 (II-1) | F | Simplex | Caucasian, (German, mixed Northern European) | 0.8 | 2 | 36 | 434 | 32 | Major bruise after minor injury | Easy bruising, epistaxis, menometrorrhagia | Moderate | No | 8/grade 1 | 15-56 | 13.7 | 4.7 | 5135 |
| 12 (II-1) | F | Simplex | Caucasian, (French, Scottish, Irish) | 4 | 4 | Died at 20 | Not reported | 16 | Excessive bleeding after tonsillectomy | Menometrorrhagia, hemorrhage after dental work | Severe | Yes | 4/no fibrosis, 20/no fibrosis | 40-150 | 12 | 3.6 | NE |
| 13 (II-1) | M | Simplex | Caucasian, (Russian, Yugoslavian, Irish, German) | 0 | 11 | 54 | 111 2234 | 32 | Easy bruising | Easy bruising, epistaxis | Moderate | Yes, Splenectomy | 11/no fibrosis, 47/grade 3 | 25-300 | 13 | 7.2 | 1555 |
| 14 (II-1) | M | Simplex | African American | 0.8 | 6 | 13 | Not reported | 7 | Easy bruising | Easy bruising | Mild | Yes | NE | 71 | 13.2 | 3.2 | 4008 |
| Family no. (patient no.) . | Sex . | Inheritance pattern . | Ethnicity . | Age at onset (y) . | Age at diagnosis (y) . | Age at most recent evaluation (y) . | Age at previous report (y)ref . | Duration of follow-up (y) . | Presenting symptoms . | Types of hemorrhage . | Severity of hemorrhage . | Splenomegaly . | Myelofibrosis age/grade* . | PLT count (K/μL) . | Hb (g/dL) . | WBC (K/μL) . | Vitamin B12 (pg/mL)† . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (IV-2) | M | AR, First Cousins | Muslim Bedouins | NA | 46 | 57 | Not reported | 9 | Low PLT count identified incidentally | No symptoms | Mild | Yes | NE | 37 | 14.3 | 3.6 | 1378 |
| 1 (V-20) | F | AR, First Cousins | Muslim Bedouins | 0.7 | 6 | 17 | 813 | 9 | Low PLT count identified incidentally | Epistaxis, menometrorrhagia | Moderate | Yes | 8/grade 1-2 | 59-225 | 11.3 | 3.3 | 5699 |
| 1 (V-21) | M | AR, First Cousins | Muslim Bedouins | 3 | 3 | 14 | 513 | 9 | Low PLT count identified incidentally | No symptoms | Mild | Yes | NE | 68- 351 | 12.3 | 3.7 | 3627 |
| 2 (VII-1) | M | AR, 3rd cousins once removed | Mennonite | 4 | 8 | 10 | Not reported | 2 | Sister with GPS | Easy bruising, epistaxis | Mild | Yes | NE | 84-97 | 13 | 5.6 | 1852 |
| 2 (VII-2) | F | AR, 3rd cousins once removed | Mennonite | 0.6 | 6 | 9 | Not reported | 3 | Major bruise after minor injury | Easy bruising, epistaxis, intracranial hemorrhage | Severe | Yes | NE | 88-97 | 12.7 | 6.1 | 1859 |
| 3 (V-1) | F | AR, Second Cousins | Caucasian, (French) | Childhood | Childhood | 64 | 5517 | 9 | Mild epistaxis, easy bruising | Epistaxis, menometrorrhagia, hemorrhage during surgery and after deliveries, gastric hemorrhage without lesion | Moderate | Yes | NE | 18-70 | NA | NA | NE |
| 4 (IV-1) | F | AR, First Cousins | Caucasian, (Turkish) | 2.5 | 15 | 29 | Not reported | 14 | Easy bruising, anemia | Easy bruising, epistaxis, menometrorrhagia | Severe | Yes | 2/no fibrosis, 23/grade 3 | 28-46 | 8.6 | 3.3 | 1441 |
| 4 (IV-2) | F | AR, First Cousins | Caucasian, (Turkish) | 6 | 10 | Died at 21 | Not reported | 11 | Prolonged bleeding after dental work | Easy bruising, epistaxis, menometrorrhagia | Severe | Yes | NE | 5-25 | 10.9 | 4.1 | 2139 |
| 5 (IV-1) | M | AR, First Cousins | Caucasian, (Turkish) | 3 | 12 | 21 | Not reported | 9 | Splenomegaly, low PLT count | Easy bruising, epistaxis | Mild | Yes | NE | 65 | 16‡ | 4.7 | 927 |
| 5 (IV-2) | F | AR, First Cousins | Caucasian, (Turkish) | 6 | 6 | 13 | Not reported | 6 | Low PLT count identified incidentally | Easy bruising | Mild | No | NE | 23-105 | 12.5 | 4.8 | NE |
| 6 (IV-1) | F | AR, First Cousins | Caucasian, (Turkish) | 6 | 33 | 33 | Not reported | 0 | Low PLT count identified incidentally | Epistaxis, menometrorrhagia | Mild | Yes | 33/grade 3-4 | 16 | 8.3 | 1.7 | 1078 |
| 7 (II-2) | F | AR | Caucasian (German) | Early childhood | 17 | 17 | 176 | 0 | Easy bruising | Easy bruising, epistaxis, hemorrhage after tonsillectomy, menometrorrhagia | Severe | NA | 17/grade 3-4 | 33-150 | NA | NA | NE |
| 7 (II-3) | M | AR | Caucasian (German) | 4 | 12 | 12 | 126 | 0 | Easy bruising | Hemorrhage after tonsillectomy and tooth extraction | Severe | NA | NE | 69-125 | NA | NA | NE |
| 8 (II-1) | F | Simplex | Caucasian, (mixed Northern European) | 0 | 3 | 30 | 48 | 26 | Easy bruising | Subgaleal hemorrhage, nasopharyngeal hemorrhage, menometrorrhagia | Severe | Yes | 10/grade 1, 29/grade 2-3 | 31 | 10 | 6 | NE |
| 9 (II-1) | F | Simplex | Caucasian, (Turkish) | 0.8 | 5 | 24 | Not reported | 19 | Petechiae, epistaxis | Severe epistaxis, hematemesis, melena | Severe | Yes | NE | 20- 72 | 10.8 | 3.3 | 1524 |
| 10 (II-1) | F | Simplex | African American, (Somalian) | NA | 16 | 16 | 1616 | 0 | NA | NA | NA | NA | NE | NA | NA | NA | NE |
| 10 (II-2) | M | Simplex | African American, (Somalian) | NA | 14 | 14 | 1416 | 0 | NA | NA | NA | NA | NE | 95 | NA | NA | NE |
| 11 (II-1) | F | Simplex | Caucasian, (German, mixed Northern European) | 0.8 | 2 | 36 | 434 | 32 | Major bruise after minor injury | Easy bruising, epistaxis, menometrorrhagia | Moderate | No | 8/grade 1 | 15-56 | 13.7 | 4.7 | 5135 |
| 12 (II-1) | F | Simplex | Caucasian, (French, Scottish, Irish) | 4 | 4 | Died at 20 | Not reported | 16 | Excessive bleeding after tonsillectomy | Menometrorrhagia, hemorrhage after dental work | Severe | Yes | 4/no fibrosis, 20/no fibrosis | 40-150 | 12 | 3.6 | NE |
| 13 (II-1) | M | Simplex | Caucasian, (Russian, Yugoslavian, Irish, German) | 0 | 11 | 54 | 111 2234 | 32 | Easy bruising | Easy bruising, epistaxis | Moderate | Yes, Splenectomy | 11/no fibrosis, 47/grade 3 | 25-300 | 13 | 7.2 | 1555 |
| 14 (II-1) | M | Simplex | African American | 0.8 | 6 | 13 | Not reported | 7 | Easy bruising | Easy bruising | Mild | Yes | NE | 71 | 13.2 | 3.2 | 4008 |
Age at bone marrow evaluation in years and grade of myelofibrosis using a scale of 1-4.
Normal: 220-960.
This patient had a Hb variant with increased affinity for oxygen.
No. indicates number; M, male; F, female; AR, autosomal recessive; NE, not evaluated; NA, not available; and PLT, platelet.
Diagnosis
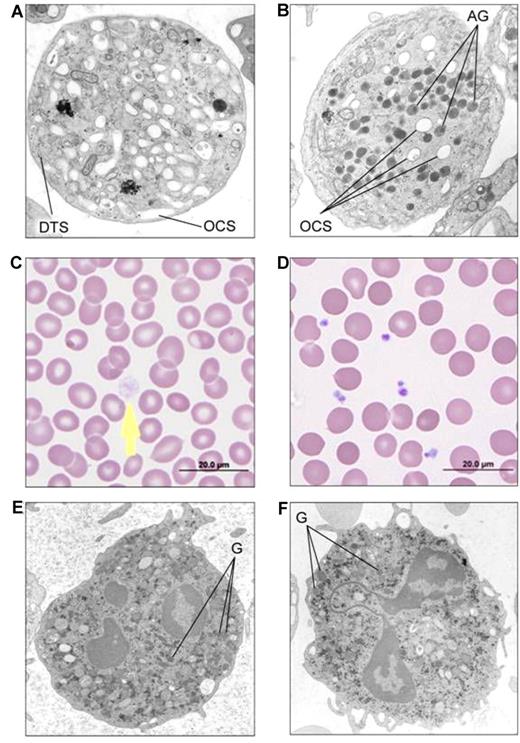
Diagnosis was based on EM studies that revealed lack of α-granules, with normal dense bodies and other platelet organelles (Figure 2A). Peripheral smears showed typical large, pale gray platelets (Figure 2B). Granularity of neutrophils on peripheral smears and EM of white cells were normal (Figure 2C). All 20 parents available, including those with single cases, had normal platelet counts and peripheral smears, and all 12 parents evaluated had normal platelet EM supporting AR inheritance. In 6 families, the parents were consanguineous; in 4 of these families and in 2 without consanguinity, multiple siblings were affected (Figure 1).
Platelet and neutrophil images of GPS patients in comparison with controls. (A) EM of thin sections of a platelet from patient 1(V-20) showing absence of α-granules and abundant channels of the open canalicular system (labeled as OCS) in comparison with EM of a control platelet (B) showing normal alpha granules (labeled as AG). DTS: Dense tubule system. (C) Light microscopy of the peripheral smear of GPS patient 1(V-20) showing a large pale gray platelet (arrow) without the cytoplasmic color of α-granules, in comparison with a normal smear (D). (E) EM of a neutrophil of GPS patient 11(II-1) showing normal cytoplasmic granules (labeled as G) similar to control neutrophil from a normal volunteer (F); Phillips 301 Electrone microscope.
Platelet and neutrophil images of GPS patients in comparison with controls. (A) EM of thin sections of a platelet from patient 1(V-20) showing absence of α-granules and abundant channels of the open canalicular system (labeled as OCS) in comparison with EM of a control platelet (B) showing normal alpha granules (labeled as AG). DTS: Dense tubule system. (C) Light microscopy of the peripheral smear of GPS patient 1(V-20) showing a large pale gray platelet (arrow) without the cytoplasmic color of α-granules, in comparison with a normal smear (D). (E) EM of a neutrophil of GPS patient 11(II-1) showing normal cytoplasmic granules (labeled as G) similar to control neutrophil from a normal volunteer (F); Phillips 301 Electrone microscope.
Most patients had bleeding symptoms from infancy. The average age of onset of symptoms was 2.6 ± 2.2 standard deviation years, ranging from birth to 6 years; diagnosis occurred between 2 and 46 years (11.8 ± 10.7 years). Age at diagnosis was similar in females (10.3 ± 8.8 years) and males (14.0 ± 13.4 years). Idiopathic thrombocytopenic purpura (ITP) was the initial diagnosis in 8 of 15 patients (53%) for whom this information was available. Age at our evaluation ranged from 9 to 64 years (25.0 ± 15.9 years). Splenomegaly was detected in 88% (15/17) of patients; one underwent splenectomy.
Platelet aggregation studies in response to most agents were normal in those patients tested. (Table 2). PF4 and fibrinogen were reduced in platelet lysates of those patients tested (Table 2). Expression of p-selectin and TSP-1 was decreased on plasma membranes of stimulated platelets in those patients evaluated (Table 2). Expression of fibrinogen receptor after stimulation was decreased in 1 of the 2 patients tested.
Platelet-related laboratory results
| . | Platelet aggregation . | Platelet lysate . | Plasma . | Platelet plasma membrane at rest . | Platelet plasma membrane after thrombin receptor-mediated stimulation . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family no. (patient no.) . | ADP . | Epinephrine . | Arachidonic acid . | Collagen . | Thrombin . | PF4 . | Fibrinogen . | Fibronectin . | Platelet vWf antigen (%) . | Platelet vWf activity (%) . | vWf antigen (IU/dL) . | vWf activity (IU/dL) . | P-selectin (%)* . | Fibrinogen receptor (%)* . | TSP-1 (%)* . | P-selectin (%)* . | Fibrinogen receptor (%)* . | TSP-1 (%)* . |
| 1 (V-20) | Normal | Normal | Normal | Normal | Normal | - | 45 ng/106 cells (Normal:70) | 0.34 ng/106 cells | < 3 | < 3 | - | - | - | - | - | - | - | - |
| 1 (V-21) | Normal | Normal | Normal | Normal | - | 2.3 ng/106 cells (Normal:17-23) | 33 ng/106 cells (Normal:70) | - | < 3 | < 3 | - | - | - | - | - | - | - | - |
| 2 (VII-1) | Normal | - | - | Normal | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 2 (VII-2) | Normal | - | - | Normal | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 3 (IV-1) | Normal17 | No aggregation17 | Normal | No aggregation17 | - | - | 1.25 μg/L08 cells17 (Normal:28) | - | 417 | - | - | - | - | - | - | 50 | - | - |
| 4 (IV-1) | Reduced | Reduced | - | Reduced | - | - | - | - | 4 | < 3 | 116 | 123 | - | - | - | - | - | - |
| 4 (IV-2) | Reduced | - | - | Reduced | - | - | - | - | 4 | < 3 | 96 | 99 | - | - | - | - | - | - |
| 7 (II-2) | Normal6 | Normal6 | - | Reduced to ∼30%6 | - | 66 ng/108 cells6 (Normal: 2040 ± 320) | - | - | - | - | - | - | 121 | - | 105 | 12 | - | 7 |
| 7 (II-3) | - | - | - | - | - | 145 ng/108 cells6 (Normal: 2040 ± 320) | - | - | - | - | - | - | 213 | - | 144 | 17 | - | 7 |
| 8 (II-1) | Normal8 | Normal8 | - | Normal8 | - | 17 ng/108 cells8 (Normal: 1200-3000) | Undetectable8 | - | - | - | - | - | - | - | - | - | - | - |
| 11 (II-1) | Normal | Normal | Normal | Normal | Normal | - | - | - | - | - | - | - | 173 | 160 | - | 68 | 76 | - |
| 13 (II-1) | Normal | Normal | Normal | Normal | Normal | - | - | - | - | - | - | - | 72 | 128 | - | 60 | 173 | - |
| 14 (II-1) | Normal | Normal | Normal | Normal | Normal | - | - | - | < 3 | < 3 | 80 | 63 | - | - | - | - | - | - |
| Normal | Normal | Normal | Normal | Normal | Normal | Varies as above | Varies as above | 1.4-1.8 | 16-55 | 44-115 | 41-132 | 48-144 | 100 | 100 | 100 | 100 | 100 | 100 |
| . | Platelet aggregation . | Platelet lysate . | Plasma . | Platelet plasma membrane at rest . | Platelet plasma membrane after thrombin receptor-mediated stimulation . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family no. (patient no.) . | ADP . | Epinephrine . | Arachidonic acid . | Collagen . | Thrombin . | PF4 . | Fibrinogen . | Fibronectin . | Platelet vWf antigen (%) . | Platelet vWf activity (%) . | vWf antigen (IU/dL) . | vWf activity (IU/dL) . | P-selectin (%)* . | Fibrinogen receptor (%)* . | TSP-1 (%)* . | P-selectin (%)* . | Fibrinogen receptor (%)* . | TSP-1 (%)* . |
| 1 (V-20) | Normal | Normal | Normal | Normal | Normal | - | 45 ng/106 cells (Normal:70) | 0.34 ng/106 cells | < 3 | < 3 | - | - | - | - | - | - | - | - |
| 1 (V-21) | Normal | Normal | Normal | Normal | - | 2.3 ng/106 cells (Normal:17-23) | 33 ng/106 cells (Normal:70) | - | < 3 | < 3 | - | - | - | - | - | - | - | - |
| 2 (VII-1) | Normal | - | - | Normal | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 2 (VII-2) | Normal | - | - | Normal | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 3 (IV-1) | Normal17 | No aggregation17 | Normal | No aggregation17 | - | - | 1.25 μg/L08 cells17 (Normal:28) | - | 417 | - | - | - | - | - | - | 50 | - | - |
| 4 (IV-1) | Reduced | Reduced | - | Reduced | - | - | - | - | 4 | < 3 | 116 | 123 | - | - | - | - | - | - |
| 4 (IV-2) | Reduced | - | - | Reduced | - | - | - | - | 4 | < 3 | 96 | 99 | - | - | - | - | - | - |
| 7 (II-2) | Normal6 | Normal6 | - | Reduced to ∼30%6 | - | 66 ng/108 cells6 (Normal: 2040 ± 320) | - | - | - | - | - | - | 121 | - | 105 | 12 | - | 7 |
| 7 (II-3) | - | - | - | - | - | 145 ng/108 cells6 (Normal: 2040 ± 320) | - | - | - | - | - | - | 213 | - | 144 | 17 | - | 7 |
| 8 (II-1) | Normal8 | Normal8 | - | Normal8 | - | 17 ng/108 cells8 (Normal: 1200-3000) | Undetectable8 | - | - | - | - | - | - | - | - | - | - | - |
| 11 (II-1) | Normal | Normal | Normal | Normal | Normal | - | - | - | - | - | - | - | 173 | 160 | - | 68 | 76 | - |
| 13 (II-1) | Normal | Normal | Normal | Normal | Normal | - | - | - | - | - | - | - | 72 | 128 | - | 60 | 173 | - |
| 14 (II-1) | Normal | Normal | Normal | Normal | Normal | - | - | - | < 3 | < 3 | 80 | 63 | - | - | - | - | - | - |
| Normal | Normal | Normal | Normal | Normal | Normal | Varies as above | Varies as above | 1.4-1.8 | 16-55 | 44-115 | 41-132 | 48-144 | 100 | 100 | 100 | 100 | 100 | 100 |
Percentage of geometric fluorescence mean where 100% is the value obtained for the normal control subject.
ADP indicates adenosine diphosphate; PF4, platelet factor 4; vWf, von Willebrand factor; TSP-1, thrombospondin-1; and -, not tested.
Bleeding tendency
Of 19 patients, 37% (7/19) of patients had mild, 21% (4/19) had moderate, and 42% (8/19) had severe bleeding (Table 1). Of the 8 patients with severe bleeding, 7 were females in whom menometrorrhagia and resultant severe anemia required repeated blood transfusions, and led to the death of 2. Two males had no bleeding symptoms. Five patients had mild symptoms in the form of bruising and epistaxis, and 4 had moderate menometrorrhagia. Most patients required supplemental iron, perioperative and peripartum DDVAP, or platelet transfusions.
Progressive nature of GPS
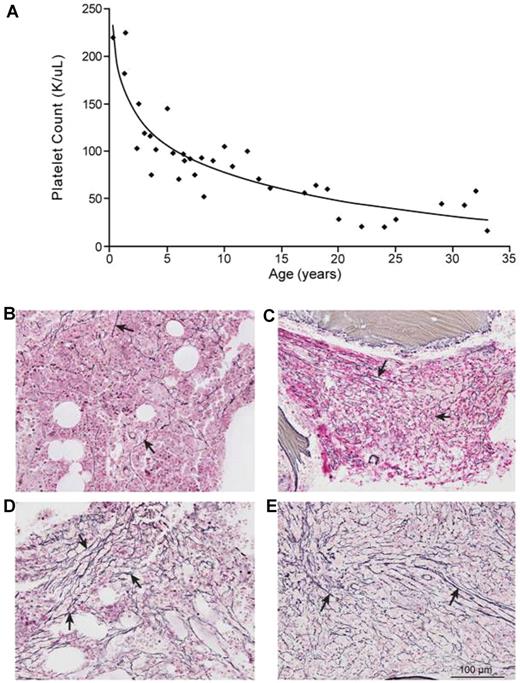
Thrombocytopenia of variable severity was present in all patients. Platelet counts were inversely correlated to age (R2:0.81; Figure 3A). Patients (8) from 8 families underwent a BM evaluation; 7 (88%) had variable degrees of myelofibrosis (Table 1 and Figure 3B-E). In patient 8 (II-1), BM at age 10 years showed mild (grade 1 of 4) reticulin fibrosis (Figure 3B); repeat BM at age 29 showed significant (grade 2-3) fibrosis (Figure 3C). Patient 13 (II-1) had no myelofibrosis at age 11, but had marked (grade 3) reticulin fibrosis at age 47 (Figure 3D). Patient 4 (IV-1)'s BM at age 2 years was normal; repeat BM at age 23 showed significant (grade 3) reticulin fibrosis (Figure 3E).
GPS is a progressive disorder. (A) Platelet counts of 14 GPS patients plotted against their age showing a significant negative correlation (y = -42.04 ln(x) + 174.14, R2 = 0.805). Patient 13(II-1), who had splenectomy, is not included. (B) Bone marrow of patient 8 (II-1) at age 10 showing mild (grade 1) reticulin fibrosis (arrows). (C) Bone marrow of patient 8 (II-1) at age 29 showing grade 2–3 reticulin fibrosis (arrows). (D) Bone marrow of patient 13(II-1) at age 47 showing grade 3 reticulin fibrosis (arrows). (E) Bone marrow of patient 4(IV-2) showing grade 3 reticulin fibrosis (arrows). Olympus BX-51 microscope (Olympus America, Melville, NY) equipped with a UPlanFL 20×/0.50 numeric aperture objective. Images were captured using an Olympus DP70 digital camera system. Imaging software was Adobe Photoshop CS3 (Adobe Systems); magnification 200×, all images.
GPS is a progressive disorder. (A) Platelet counts of 14 GPS patients plotted against their age showing a significant negative correlation (y = -42.04 ln(x) + 174.14, R2 = 0.805). Patient 13(II-1), who had splenectomy, is not included. (B) Bone marrow of patient 8 (II-1) at age 10 showing mild (grade 1) reticulin fibrosis (arrows). (C) Bone marrow of patient 8 (II-1) at age 29 showing grade 2–3 reticulin fibrosis (arrows). (D) Bone marrow of patient 13(II-1) at age 47 showing grade 3 reticulin fibrosis (arrows). (E) Bone marrow of patient 4(IV-2) showing grade 3 reticulin fibrosis (arrows). Olympus BX-51 microscope (Olympus America, Melville, NY) equipped with a UPlanFL 20×/0.50 numeric aperture objective. Images were captured using an Olympus DP70 digital camera system. Imaging software was Adobe Photoshop CS3 (Adobe Systems); magnification 200×, all images.
Hemoglobin concentrations of affected males ranged from 12.3 to 14.3 g/dL (13.2 ± 0.7), excluding patient 5 (IV-1) with increased affinity to oxygen. Values for females ranged from 8.3 to 13.7 g/dL (11.1 ± 1.7 g/dL). All 7 postpubertal females had a history of iron deficiency anemia and received iron supplementation. Iron levels of the 7 male patients were normal. PT, PTT, liver and kidney function, thyroid function tests, and urine analysis were normal in all patients.
Vitamin B12
We found markedly elevated serum vitamin B12 concentrations in 12 of 13 patients tested from 9 unrelated families. Ranging from 1078 pg/mL to 5699 pg/mL with a mean ± standard deviation value of 2608 ± 1590 pg/mL. Repeat measurements confirmed consistently high values. Patient 5 (IV-1) had a high normal vitamin B12 level at 927 pg/mL whereas both his parents were vitamin B12 deficient.
Illustrative cases
Below, we provide brief summaries of 5 GPS patients, one selected because he is the oldest patient alive [13(II-1)], 2 because they suffered fatal hemorrhages [4(IV-2)] and [12(II-1)], one to emphasize the severity of menometrorrhagia and the progressive nature of myelofibrosis [8(II-1)], and one to emphasize the mild nature of bleeding tendency [5(IV-2)].
Case 13(II-1)
This 54-year-old male is the original GPS patient described in 1971 when he was 11 years old1 and again at age 22.34 We evaluated him at the NIH Clinical Center at ages 47, 49, and 50. He had easy bruising in infancy and suffered recurrent knee pain. Platelet counts within the first 10 years ranged between 150 and 25 K/μL. He was initially diagnosed with ITP, treated with steroids and underwent splenectomy at 11 years when platelets decreased to 25 K/μL. After splenectomy, platelet counts increased to 239 to 300 K/μL. BM at age 11 showed no fibrosis, normal number of megakaryocytes with decreased granularity, and normal granulocytic and erythroid series. At age 47, his platelet count was 107 K/uL and hemoglobin was 13.9 g/dL. An elevated total protein (9.2 g/dL) prompted measurement of quantitative immunoglobulins; IgM was elevated at 3560 mg/dL. Serum protein electrophoresis identified excessive IgM kappa; urine protein electrophoresis revealed IgM mu and kappa chains. BM showed grade 2-3 reticulin fibrosis (Figure 3D) and increased plasma cells or plasmocytoid lymphocytes expressing CD38/CD138. Flow cytometry of peripheral blood showed no immunophenotypic evidence of a lymphoproliferative disorder. The diagnosis of Waldenstrom macroglobulinemia was made. At age 49, BM continued to show increased plasma cells. IgM was increased to 6130 mg/dL with a serum viscosity of 2.5. He remained free of hyperviscosity-related symptoms. At age 52, he underwent hip replacement surgery with prophylactic platelet infusions. Although no major bleeding occurred immediately after surgery, he required transfusion the following week. At age 54, IgM was 6550 mg/dL, serum viscosity was 3.6, and platelet count was 88 K/μL. He had occasional dizziness and headaches and specific treatment for Waldenstrom macroglobulinemia was under consideration.
Case 12(II-1)
This female patient was asymptomatic in her first 4 years of life. She was diagnosed with GPS following a 4-day massive hemorrhage after tonsillectomy and adenoidectomy. Throughout childhood, her platelet counts were in the low hundreds. She had an enlarged spleen at age 13, when she was evaluated for severe anemia caused by menometrorrhagia and required long term iron supplementation. At age 20, her car was hit from behind and her abdomen was strained by the seatbelt. Although she was asymptomatic immediately after the accident, 2 days later she collapsed at home and expired despite multiple transfusions. Autopsy revealed hemoperitoneum and an enlarged (940 g) spleen with a 4-cm laceration in the anteriomedial aspect. There was no fibrosis of the bone marrow. Platelet count 3 weeks before she died was 31 K/μL.
Case 8(II-1)
This 30-year-old female had easy bruising since birth and was diagnosed with GPS at age 3 years. At age 3.5, she experienced prolonged epistaxis, with a decrease in hemoglobin to 6.8 g/dL; at age 4, she presented with a massive subgaleal hemorrhage and left periorbital ecchymosis after minor head trauma.8 Platelet count was 94 K/μL. Between ages 5 and 10 years, she presented with recurrent spontaneous retropharyngeal hemorrhages and required platelet transfusions. At age 10, she developed massive hepatosplenomegaly due to CMV infection and dropped her platelets to 10 K/μL, requiring multiple platelet and blood transfusions during the following year. BM at age 10 showed grade 1 reticulin staining (Figure 3B). After menarche, she developed severe anemia (hemoglobin 5 g/dL) and required depo-progesterone injections. At ages 23 and 29, she had 2 additional episodes of severe anemia (hemoglobin of 4 g/dL) after prolonged vaginal bleeding and required blood transfusions, intravenous iron, and estrogen. Platelet count at 29 was 32 K/μL. BM at age 29 showed grade 2-3 reticulin fibrosis (Figure 3C), and nearly absent iron staining. Cytogenetic analysis of BM was normal and fluorescent in situ hybridization (FISH) for common genetic aberrations indicating myelodysplastic disease was negative.
Case 4(IV-2)
This female was diagnosed with GPS at age 10, when screened after the diagnosis of GPS in her older sister [4 (IV-1)]. She had prolonged bleeding after dental work at age 6 and later suffered from severe menometrorrhagia requiring blood transfusions and iron supplementation. Platelet counts were 105 K/μL, 45 K/μL, and 25 K/μL at ages 10, 13, and 20, respectively. At age 21, she developed diffuse petechia without any other symptoms. Platelet count was 5 K/μL. She was started on platelet infusions every 2 days. After 6 days, she developed sudden projectile vomiting, was admitted to intensive care with intracranial hemorrhage, and died later that day.
Case 5(IV-2)
Before she was diagnosed with GPS, this female patient underwent brain surgery at the age of 4.5 years for removal of a meduloblastoma, without major hemorrhage. She was diagnosed with GPS when thrombocytopenia persisted several months after cessation of chemotherapy. Currently, at age 13, she is free of bleeding symptoms. Her platelet counts at ages 7 and 13 were 105 K/μL, and 102 K/μL, respectively.
Genetic analyses
Inheritance pattern.
The facts that 6 families are consanguineous (1, 2, 3, 4, 5, and 6; Figure 1) and 6 families have multiple affected individuals with unaffected parents (1, 2, 4, 5, 7, and 10) support AR inheritance. Bedouin kindred from Israel (Figure 1, family 1) with multiple consanguineous marriages contained 7 GPS individuals. Segregation analysis performed in this family showed that AR inheritance was 5 orders of magnitude more likely than AD inheritance.
Genome-wide linkage analyses.
Genome-wide linkage analysis the 60 members, including 7 affected, of the Israeli Bedouin kindred (Figure 1, family 1), provided maximal LOD scores of 7.2, 7, and 7.2 at 3 consecutive markers: D3S3563, D3S3685, and D3S3559, determining the GPS-linked interval between markers D3S3521 and D3S1029 (Table 3). In addition, homozygosity mapping was performed on group 2, that consisted of 26 individuals including 7 patients from 5 GPS families (Figure 1, families 4, 5, 8, 9, and 13). Two consanguineous Turkish families with 2 patients in each (Families 4 and 5) provided significant linkage (LOD = 4.14) to locate the linked region between markers D3S1612 and D3S3532 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These 2 intervals overlap, supporting the linkage of GPS gene to 3p21.1-3p22.1 with a combined LOD score of 11.3.
Microsatellite markers and recombination fractions documenting linkage of group 1
| Marker name . | Recombination fraction . | ||||||
|---|---|---|---|---|---|---|---|
| 0 . | 0.01 . | 0.05 . | 0.1 . | 0.2 . | 0.3 . | 0.4 . | |
| D3S1266 | −22.82 | −1.03 | 0.17 | 0.5 | 0.45 | 0.18 | 0.01 |
| D3S3567 | 1.49 | 1.48 | 1.43 | 1.33 | 0.97 | 0.51 | 0.12 |
| D3S1277 | 3.54 | 3.45 | 3.09 | 2.64 | 1.74 | 0.89 | 0.25 |
| D3S3521 | −9.8 | 3.47 | 3.64 | 3.26 | 2.17 | 0.99 | 0.15 |
| AFM206ZF2 | 5.13 | 5 | 4.48 | 3.82 | 2.5 | 1.23 | 0.27 |
| D3S3522 | 5.3 | 5.17 | 4.64 | 3.96 | 2.58 | 1.24 | 0.24 |
| D3S2407 | −5.52 | 0 | 0.57 | 0.69 | 0.58 | 0.34 | 0.13 |
| D3S3563 | 7.21 | 7.06 | 6.47 | 5.7 | 4.1 | 2.45 | 0.94 |
| D3S3685 | 6.99 | 6.85 | 6.26 | 5.51 | 3.94 | 2.34 | 0.9 |
| D3S3559 | 7.22 | 7.07 | 6.48 | 5.71 | 4.11 | 2.46 | 0.94 |
| D3S2304 | 3.08 | 3 | 2.71 | 2.34 | 1.57 | 0.86 | 0.3 |
| D3S1449 | 2.46 | 2.4 | 2.13 | 1.79 | 1.15 | 0.58 | 0.18 |
| D3S3678 | 4.96 | 4.87 | 4.49 | 4 | 2.95 | 1.84 | 0.77 |
| D3S966 | 2.54 | 2.49 | 2.28 | 2 | 1.4 | 0.78 | 0.27 |
| D3S1029 | −9.24 | −3.26 | −0.86 | 0.04 | 0.57 | 0.5 | 0.22 |
| D3S3624 | 2.8 | 2.74 | 2.48 | 2.14 | 1.46 | 0.82 | 0.31 |
| D3S1289 | 1.36 | 1.8 | 2.04 | 1.88 | 1.28 | 0.62 | 0.14 |
| Marker name . | Recombination fraction . | ||||||
|---|---|---|---|---|---|---|---|
| 0 . | 0.01 . | 0.05 . | 0.1 . | 0.2 . | 0.3 . | 0.4 . | |
| D3S1266 | −22.82 | −1.03 | 0.17 | 0.5 | 0.45 | 0.18 | 0.01 |
| D3S3567 | 1.49 | 1.48 | 1.43 | 1.33 | 0.97 | 0.51 | 0.12 |
| D3S1277 | 3.54 | 3.45 | 3.09 | 2.64 | 1.74 | 0.89 | 0.25 |
| D3S3521 | −9.8 | 3.47 | 3.64 | 3.26 | 2.17 | 0.99 | 0.15 |
| AFM206ZF2 | 5.13 | 5 | 4.48 | 3.82 | 2.5 | 1.23 | 0.27 |
| D3S3522 | 5.3 | 5.17 | 4.64 | 3.96 | 2.58 | 1.24 | 0.24 |
| D3S2407 | −5.52 | 0 | 0.57 | 0.69 | 0.58 | 0.34 | 0.13 |
| D3S3563 | 7.21 | 7.06 | 6.47 | 5.7 | 4.1 | 2.45 | 0.94 |
| D3S3685 | 6.99 | 6.85 | 6.26 | 5.51 | 3.94 | 2.34 | 0.9 |
| D3S3559 | 7.22 | 7.07 | 6.48 | 5.71 | 4.11 | 2.46 | 0.94 |
| D3S2304 | 3.08 | 3 | 2.71 | 2.34 | 1.57 | 0.86 | 0.3 |
| D3S1449 | 2.46 | 2.4 | 2.13 | 1.79 | 1.15 | 0.58 | 0.18 |
| D3S3678 | 4.96 | 4.87 | 4.49 | 4 | 2.95 | 1.84 | 0.77 |
| D3S966 | 2.54 | 2.49 | 2.28 | 2 | 1.4 | 0.78 | 0.27 |
| D3S1029 | −9.24 | −3.26 | −0.86 | 0.04 | 0.57 | 0.5 | 0.22 |
| D3S3624 | 2.8 | 2.74 | 2.48 | 2.14 | 1.46 | 0.82 | 0.31 |
| D3S1289 | 1.36 | 1.8 | 2.04 | 1.88 | 1.28 | 0.62 | 0.14 |
Maximum LOD scores of 6.99 to 7.22 were achieved along three consecutive markers D3S3563, D3S3685, and D3S3559, supporting strong linkage of GPS to the interval between D3S1029 and D3S3521.
Fine mapping, haplotype, and SNP analyses.
Two rounds of fine mapping of the linked interval on a total of 111 individuals including 23 patients from 12 families (Figure 1, families 1, 2, 4, 5, 7, 8, 9, 10, 11, 12, 13, and 14), refined the GPS region to a 9.4-Mb interval between markers D3S3559 (position 42663630 at UCSC hg18) and NIH custom-made marker 3)52036954 (position 52036954 at UCSC hg18; Figure 1). Two-point analysis of 6 families (1, 4, 5, 7, 9, and 10) revealed a combined maximum LOD score of 12.05 for this region (Table 4). Multipoint analysis supported these results (data not shown).
Results of 2-point analysis for families 1, 4, 5, 7, 9, and 10
| Marker name . | Recombination fraction . | ||||||
|---|---|---|---|---|---|---|---|
| 0 . | 0.01 . | 0.05 . | 0.1 . | 0.2 . | 0.3 . | 0.4 . | |
| D3S1277 | −0.57 | 4.60 | 5.55 | 5.53 | 4.55 | 3.07 | 1.45 |
| D3S3718 | 0.12 | 2.81 | 3.46 | 3.38 | 2.61 | 1.62 | 0.70 |
| D3S1561 | 0.74 | 5.35 | 5.90 | 5.74 | 4.54 | 2.85 | 1.14 |
| D3S1611 | 5.66 | 5.63 | 5.23 | 4.51 | 2.97 | 1.61 | 0.59 |
| D3S3623 | 11.84 | 11.56 | 10.39 | 8.93 | 6.13 | 3.62 | 1.47 |
| D3S1260 | 3.99 | 4.02 | 3.86 | 3.46 | 2.46 | 1.46 | 0.62 |
| D3S3521 | 10.52 | 10.39 | 9.62 | 8.43 | 5.97 | 3.63 | 1.47 |
| D3S3593 | 6.86 | 6.70 | 6.02 | 5.17 | 3.53 | 2.03 | 0.82 |
| D3S3527 | 9.27 | 9.17 | 8.48 | 7.40 | 5.15 | 3.06 | 1.21 |
| D3S3522 | 9.26 | 9.06 | 8.22 | 7.10 | 4.74 | 2.42 | 0.53 |
| D3S3559 | 11.00 | 10.86 | 10.04 | 8.80 | 6.25 | 3.85 | 1.65 |
| D3S3678 | 3.00 | 5.55 | 5.93 | 5.57 | 4.31 | 2.80 | 1.29 |
| D3S3647 | 6.94 | 6.76 | 6.02 | 5.11 | 3.42 | 1.99 | 0.85 |
| D3S3597 | 4.24 | 4.18 | 3.89 | 3.47 | 2.56 | 1.63 | 0.76 |
| D3S3624 | 9.95 | 9.73 | 8.84 | 7.70 | 5.37 | 3.15 | 1.30 |
| D3S3582 | 6.13 | 5.97 | 5.33 | 4.52 | 2.97 | 1.68 | 0.71 |
| D3S1767 | 6.67 | 6.48 | 5.73 | 4.82 | 3.22 | 1.88 | 0.76 |
| DG3S758 | 9.72 | 9.46 | 8.44 | 7.16 | 4.66 | 2.42 | 0.69 |
| D3S3640 | 11.90 | 11.71 | 10.81 | 9.54 | 6.82 | 4.09 | 1.71 |
| D3S2420 | 7.72 | 7.53 | 6.78 | 5.82 | 4.00 | 2.40 | 0.98 |
| D3S3560 | 8.09 | 7.89 | 7.08 | 6.07 | 4.16 | 2.46 | 0.99 |
| D3S2384 | 5.01 | 4.84 | 4.18 | 3.37 | 1.98 | 1.03 | 0.40 |
| DG13S126 | 12.05 | 11.78 | 10.69 | 9.31 | 6.54 | 3.87 | 1.57 |
| D3S3629 | 11.24 | 10.98 | 9.95 | 8.65 | 6.05 | 3.57 | 1.40 |
| D3S3604 | 7.92 | 7.73 | 7.00 | 6.08 | 4.29 | 2.63 | 1.14 |
| D3S3667 | 7.56 | 7.39 | 6.69 | 5.81 | 4.10 | 2.52 | 1.11 |
| D3S1573 | 10.43 | 10.17 | 9.13 | 7.81 | 5.21 | 2.82 | 0.91 |
| D3S3026 | 10.74 | 10.48 | 9.43 | 8.11 | 5.54 | 3.18 | 1.21 |
| D3S3561 | 8.05 | 7.84 | 6.98 | 5.92 | 3.92 | 2.20 | 0.86 |
| D3S1578 | 4.34 | 6.49 | 6.52 | 5.94 | 4.44 | 2.81 | 1.22 |
| D3S1588 | 7.85 | 7.81 | 7.34 | 6.50 | 4.60 | 2.71 | 1.06 |
| D3S1289 | 0.11 | 3.56 | 4.55 | 4.45 | 3.39 | 2.03 | 0.78 |
| D3S3672 | −4.05 | 3.53 | 4.24 | 4.12 | 3.18 | 2.03 | 0.88 |
| DG3S1126 | −7.44 | 2.70 | 4.15 | 4.23 | 3.27 | 2.00 | 0.88 |
| DG3S1017 | 6.99 | 7.09 | 6.89 | 6.21 | 4.52 | 2.84 | 1.30 |
| DG3S1039 | 9.17 | 8.96 | 8.11 | 7.05 | 4.96 | 3.03 | 1.33 |
| D3S3660 | −5.27 | 7.58 | 7.56 | 6.80 | 4.87 | 2.91 | 1.21 |
| DG3S1047 | 5.76 | 5.85 | 5.68 | 5.07 | 3.56 | 2.09 | 0.87 |
| DG3S1048 | −2.89 | 0.25 | 2.02 | 2.31 | 1.77 | 0.95 | 0.26 |
| D3S3717 | −3.48 | 2.19 | 3.87 | 4.05 | 3.31 | 2.16 | 1.00 |
| Marker name . | Recombination fraction . | ||||||
|---|---|---|---|---|---|---|---|
| 0 . | 0.01 . | 0.05 . | 0.1 . | 0.2 . | 0.3 . | 0.4 . | |
| D3S1277 | −0.57 | 4.60 | 5.55 | 5.53 | 4.55 | 3.07 | 1.45 |
| D3S3718 | 0.12 | 2.81 | 3.46 | 3.38 | 2.61 | 1.62 | 0.70 |
| D3S1561 | 0.74 | 5.35 | 5.90 | 5.74 | 4.54 | 2.85 | 1.14 |
| D3S1611 | 5.66 | 5.63 | 5.23 | 4.51 | 2.97 | 1.61 | 0.59 |
| D3S3623 | 11.84 | 11.56 | 10.39 | 8.93 | 6.13 | 3.62 | 1.47 |
| D3S1260 | 3.99 | 4.02 | 3.86 | 3.46 | 2.46 | 1.46 | 0.62 |
| D3S3521 | 10.52 | 10.39 | 9.62 | 8.43 | 5.97 | 3.63 | 1.47 |
| D3S3593 | 6.86 | 6.70 | 6.02 | 5.17 | 3.53 | 2.03 | 0.82 |
| D3S3527 | 9.27 | 9.17 | 8.48 | 7.40 | 5.15 | 3.06 | 1.21 |
| D3S3522 | 9.26 | 9.06 | 8.22 | 7.10 | 4.74 | 2.42 | 0.53 |
| D3S3559 | 11.00 | 10.86 | 10.04 | 8.80 | 6.25 | 3.85 | 1.65 |
| D3S3678 | 3.00 | 5.55 | 5.93 | 5.57 | 4.31 | 2.80 | 1.29 |
| D3S3647 | 6.94 | 6.76 | 6.02 | 5.11 | 3.42 | 1.99 | 0.85 |
| D3S3597 | 4.24 | 4.18 | 3.89 | 3.47 | 2.56 | 1.63 | 0.76 |
| D3S3624 | 9.95 | 9.73 | 8.84 | 7.70 | 5.37 | 3.15 | 1.30 |
| D3S3582 | 6.13 | 5.97 | 5.33 | 4.52 | 2.97 | 1.68 | 0.71 |
| D3S1767 | 6.67 | 6.48 | 5.73 | 4.82 | 3.22 | 1.88 | 0.76 |
| DG3S758 | 9.72 | 9.46 | 8.44 | 7.16 | 4.66 | 2.42 | 0.69 |
| D3S3640 | 11.90 | 11.71 | 10.81 | 9.54 | 6.82 | 4.09 | 1.71 |
| D3S2420 | 7.72 | 7.53 | 6.78 | 5.82 | 4.00 | 2.40 | 0.98 |
| D3S3560 | 8.09 | 7.89 | 7.08 | 6.07 | 4.16 | 2.46 | 0.99 |
| D3S2384 | 5.01 | 4.84 | 4.18 | 3.37 | 1.98 | 1.03 | 0.40 |
| DG13S126 | 12.05 | 11.78 | 10.69 | 9.31 | 6.54 | 3.87 | 1.57 |
| D3S3629 | 11.24 | 10.98 | 9.95 | 8.65 | 6.05 | 3.57 | 1.40 |
| D3S3604 | 7.92 | 7.73 | 7.00 | 6.08 | 4.29 | 2.63 | 1.14 |
| D3S3667 | 7.56 | 7.39 | 6.69 | 5.81 | 4.10 | 2.52 | 1.11 |
| D3S1573 | 10.43 | 10.17 | 9.13 | 7.81 | 5.21 | 2.82 | 0.91 |
| D3S3026 | 10.74 | 10.48 | 9.43 | 8.11 | 5.54 | 3.18 | 1.21 |
| D3S3561 | 8.05 | 7.84 | 6.98 | 5.92 | 3.92 | 2.20 | 0.86 |
| D3S1578 | 4.34 | 6.49 | 6.52 | 5.94 | 4.44 | 2.81 | 1.22 |
| D3S1588 | 7.85 | 7.81 | 7.34 | 6.50 | 4.60 | 2.71 | 1.06 |
| D3S1289 | 0.11 | 3.56 | 4.55 | 4.45 | 3.39 | 2.03 | 0.78 |
| D3S3672 | −4.05 | 3.53 | 4.24 | 4.12 | 3.18 | 2.03 | 0.88 |
| DG3S1126 | −7.44 | 2.70 | 4.15 | 4.23 | 3.27 | 2.00 | 0.88 |
| DG3S1017 | 6.99 | 7.09 | 6.89 | 6.21 | 4.52 | 2.84 | 1.30 |
| DG3S1039 | 9.17 | 8.96 | 8.11 | 7.05 | 4.96 | 3.03 | 1.33 |
| D3S3660 | −5.27 | 7.58 | 7.56 | 6.80 | 4.87 | 2.91 | 1.21 |
| DG3S1047 | 5.76 | 5.85 | 5.68 | 5.07 | 3.56 | 2.09 | 0.87 |
| DG3S1048 | −2.89 | 0.25 | 2.02 | 2.31 | 1.77 | 0.95 | 0.26 |
| D3S3717 | −3.48 | 2.19 | 3.87 | 4.05 | 3.31 | 2.16 | 1.00 |
Combined maximum LOD score of 12.05 was achieved (marker DG13S126).
Genome-Wide SNP analysis using 1M–Duo SNP array, performed on 8 patients [1(V-20), 2(VII-2), 3(V-1), 6(IV-1), 10(II-1), 12(II-1), 13(II-1), and 14(II-1)] excluded copy number variants such as large deletions/duplications along the linked interval.
Affected individuals in 11 of the 14 families (except for families 12, 13, and 14) revealed a homozygous haplotype (Figure 1; indicated with gray bars) that segregated with the disease and was unique to each family. This included 3 families (3, 6, and 8) for which parental DNA was not available, but the patients had homozygous markers along the identical interval. In 3 additional families (12, 13, and 14), affected individuals did not display a homozygous region of microsatellite markers in this interval and SNP analysis of the affected individuals in these families did not uncover any small (< 200-kb) region of homozygosity. SNP analysis did not show any heterozygous SNPs at the boundaries of the homozygous region to enable narrowing from the edges.
Sequencing of genes
The GPS-linked interval contained a total of 197 protein-coding genes. After exclusion of genes already assigned to well-described diseases, we initially sequenced 20 selected candidate genes, finding no pathogenic mutations. Subsequently, we sequenced all 2075 exons and flanking splice junctions of 165 candidate genes (supplemental Table 1) using genomic DNA; 78 genes were sequenced in 4 independent patients [1(V-21), 4(IV-2), 10(II-1), 11(II-1)] and 87 genes were sequenced in 6 independent patients [1(V-21), 2(VII-2), 3(II-2), 8(II-1), 10(II-1), 11(II-1)], along with 4 controls (2 mixed Northern European and Turkish). We successfully sequenced 1423 (69%) of the 2075 exons in the linked interval without identifying homozygous or compound heterozygous pathogenic mutations. This excluded mutations in all the coding exons of 37 genes; in an additional 38 genes only one difficult exon could not be excluded. The remaining 652 exons spread throughout the region could not be amplified or sequenced despite several attempts.
In addition, we sequenced the X-linked GATA1 gene in patients 13(II-1) and 14(II-1) who were males without a homozygous haplotype in the linked region, and identified no pathogenic mutations.
Discussion
The pedigree structures and homozygous haplotypes confirmed AR inheritance in all but 3 families and indicated that the GPS gene resides in a 9.4-Mb interval on chromosome 3p. For the other 3 families (12, 13, and 14), the inheritance pattern could be AR due to compound heterozygous mutations of a gene in the linked interval on chromosome 3, AR due to another gene located elsewhere, AD due to new mutations, or X-linked in the case of families 13 and 14 with male patients.
Although we enrolled almost all known GPS families from around the world to achieve the highest possible number of meiosis, we did not identify any recombination events to further decrease the size of the candidate region. This was probably influenced by the relatively poor meiotic recombination rates in this region of chromosome 3.43 Although the physical distance of the linked region is 9.4 Mb, its genetic distance based on the sex-averaged recombination rates on the Marshfield map43 is 2.7 cM, suggesting a significantly lower recombination rate (approximately 0.28% for 1 Mb) in this region in comparison to the genome-wide average (approximately 1% for 1 Mb).
By performing fine mapping with microsatellite markers, located densely at every 200 Kb, and by SNP analysis, we exhausted the available molecular genetics methods to refine the region at the breakpoints. We also excluded the possibility of a small (< 200 Kb) region of homozygosity in the 3 patients (Figure 1, families 12, 13, and 14) who did not display the 9.4 Mb homozygous haplotype present in the other 22 GPS patients. Finally, we successfully sequenced 1423 (69%) of the 2075 exons of 165 genes. A next generation sequencing approach is likely to increase the success rate in the remaining 652 difficult exons that are spread throughout the region.
No previous study has described the natural history of GPS. Long-term follow-up, including serial bone marrow evaluations in several patients, demonstrated the progressive nature of GPS with respect to thrombocytopenia and myelofibrosis, and showed that thrombocytopenia in GPS worsens with age (Figure 3A). The platelet count was negatively correlated with age (R2:0.81); platelet counts of patients older than 12 years (46 ± 18 K/μL) were significantly lower than platelet counts of patients 12 and younger (113 ± 45 K/μL; P < .0001). Patient 13(II-1), who was splenectomized, maintained a platelet count of 88 K/μL at age 54, significantly higher than the average platelet count of GPS patients over age 20 (33 K/μL) who were not splenectomized. The etiology of the progressive nature of thrombocytopenia in GPS remains to be determined (worsening myelofibrosis and splenic enlargement with more platelet consumption).
Our data show that myelofibrosis affects the majority (88%) of GPS patients. Serial BM evaluations demonstrate the progressive nature of myelofibrosis in GPS (Figure 3B-E). In several patients, myelofibrosis was not present in childhood but developed in adulthood [4(IV-1), 13(II-1)] and its severity progressed as patients aged [8(II-1)]. The youngest age at which myelofibrosis was demonstrated was 8 years, in patients 1(V-20) and 11(II-1). Premature release of platelet-derived growth factor and other profibrotic substances from the megakaryocytes into the BM is thought to cause the myelofibrosis in GPS.3 Microarray studies on fibroblasts of patient 1(V-20) had shown increased expression of cytoskeleton-related proteins, especially fibronectin, further supporting this hypothesis.44
Another novel finding in this study is the markedly high serum vitamin B12 levels in 11 of the 12 GPS patients tested. Although the underlying mechanism is not completely understood, elevated serum vitamin B12 is observed in various conditions including liver diseases, myelogeneous and promyelocytic leukemia, polycythemia vera, hypereosinophilic syndrome, and reactive macrophage activation syndrome; it serves as a diagnostic criterion in the latter 3 diseases.45,46 In most patients, the diagnosis of GPS was not an immediate consideration and there was a delay of several years (6.2 ± 6.7 years) between the onset of symptoms and the diagnosis. The fact that platelet aggregation studies are generally normal in GPS probably complicates the initial diagnosis. Platelet EM, the “gold standard” for the diagnosis of GPS, is not widely available. Elevated vitamin B12 can be used as a biochemical marker in patients with suspected GPS to reinforce the indication for platelet EM.
Hematologic malignancies have been reported in several families with other platelet storage pool deficiencies,47,48 but not in GPS. It is intriguing that patient 13(II-1) developed Waldenstrom macroglobulinemia. We are aware of only 2 other GPS patients who are of similar age or older; patient 3(V-1) did not have a BM evaluation, but an Ashkenazi-Jewish GPS patient had a BM at age 72 that showed no increase in plasma cells.9 Longer follow-up may answer the question of whether or not GPS predisposes to Waldenstrom macroglobulinemia. Similarly, whether the medullablastoma observed in patient 5(IV-2) is causally related to GPS remains to be determined.
Consistent with previous reports,3,4,16,18 bleeding symptoms in the presented patients were mostly mucosal (Table 1). Although the degree of bleeding tendency was variable, a significant proportion of patients (42%) had severe bleeding. The results of platelet aggregation studies did not correlate with the severity of bleeding. 3 of the 5 GPS patients with severe bleeding who had platelet aggregation studies performed, had normal [2 (VII-2), 7 (II-2), 8 (II-1)] or only partially impaired [2 (VII-2), 7 (II-2), 8 (II-1)] platelet aggregation. Patient 3 (V-1) who had moderate bleeding showed no aggregation in response to epinephrine and collagen (Tables 1–2). The majority of the severe cases were postpubertal females with menometrorrhagia. The platelet counts of the 2 females who had fatal hemorrhages were between 25 and 30 K/μL during the months before their deaths. The value of splenectomy in such cases remains an open question.
Understanding the molecular basis of organelle biogenesis in megakaryocytes is of significant importance. Identification of the genes of organelle formation and trafficking disorders such as Griscelli, Hermansky-Pudlak and Chediak-Higashi Syndromes49,50 have contributed to our understanding of fundamental aspects of cell function. Similarly, identification of the GPS gene is likely to teach us about organelle biosynthesis, contribute to our understanding of myelofibrosis, and potentially help design targeted novel therapies.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all GPS patients and families for numerous interviews, physical examinations, laboratory and imaging testing and for providing samples. We thank Isa Bernardini, and Roxanne Fischer, funded by National Human Genome Research Institute and Khanh Nghiem, funded by NIH Clinical Center and Marcy Krumwiede for handling and processing specimens.
This study was supported by the Intramural Research Programs of the National Human Genome Research Institute,and the NIH Clinical Center, and by the Israeli Ministry of Justice; Izvonot Fund, grants 84/2004, 85/2004, and 9090-25/2007 to T.F.-Z.
National Institutes of Health
Authorship
Contribution: M.G.-A. was principal investigator of GPS clinical trial NCT00069680, designed research, recruited patients, performed and supervised genotyping and haplotype analysis, performed DNA sequencing, analyzed clinical and molecular data, and wrote the paper; Y.Z.-E. performed GWS, DNA sequencing, analyzed linkage data, and participated in preparing the manuscript; F.G. recruited and provided clinical data on families 4, 5 and 9, performed research, performed DNA sequencing, and participated in preparing the manuscript; D.G. analyzed linkage data and participated in preparing the manuscript; M.C. recruited and provided clinical data on families 4, 5 and 9, performed research, performed DNA sequencing, and participated in preparing the manuscript; M.K. participated in recruiting members family 1, participated in DNA sequencing, and participated in preparing the manuscript; R.K. recruited family 7, established collaborations, sequenced genes, coordinated and assisted in linkage analyses, and participated in preparing the manuscript; N.K. participated in recruiting family members form family 1, and participated in preparing the manuscript; Y.A assisted in linkage analyses, established collaborations, and participated in preparing the manuscript; J.C. performed hematologic profile of members from family 1 and participated in preparing the manuscript; M.A.-B. performed homozygosity mapping, assisted in linkage analyses, and participated in preparing the manuscript; A.S. participated in follow up of patients from family 1 and participated in preparing the manuscript; H.S. performed linkage analyses and participated in preparing the manuscript; J.M. performed hematologic work up of family 1 and participated in preparing the manuscript; M.A. recruited and provided clinical data on family 6, performed research, and participated in preparing the manuscript; H.E. performed DNA sequencing; A.S.F. recruited and provided clinical data on families 2 and 14, performed research, and participated in preparing the manuscript; P.S.H. performed genotyping of microsatellite markers, assisted in linkage and haplotype analyses, and participated in preparing the manuscript; L.C.R., K.P., and P.T. performed DNA sequencing; T.M. analyzed SNP data; M.H. assisted in DNA sequencing and participated in preparing the manuscript; I.M. evaluated the bone marrows and participated in preparing the manuscript; M.H. evaluated platelet studies, recruited and provided clinical data on family 8, performed clinical evaluation of patients, and participated in preparing the manuscript; B.E.K. provided clinical data on family 7, analyzed platelet proteins and function, and participated in preparing the manuscript; K.J. provided DNA samples of family 7, performed platelet function experiments, analyzed platelet proteins, and participated in preparing the manuscript; N.F.H., P.F.C., M.J., P.C., and J.C.M. performed DNA sequencing, participated in preparing the manuscript; A.N. recruited and provided clinical data on family 3 and participated in preparing the manuscript; J.G.W. performed EM of platelets and WBCs, recruited and provided clinical data on families 10, 11, and 13, and participated in preparing the manuscript; W.A.G. was principal investigator of clinical trial NCT00369421, accountable investigator of clinical trial NCT00069680, designed research, supervised research, and participated in preparing the manuscript; and T.F.-Z. was principal investigator of the Israeli protocol “Clinical and Genetic Analysis of Gray Platelet Syndrome,” designed research, recruited patients, performed and supervised research, analyzed clinical and molecular data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Meral Gunay-Aygun, National Human Genome Research Institute, NIH, 10 Center Dr Bldg 10, Rm 10C103, Bethesda, MD; e-mail: mgaygun@mail.nih.gov.