Within the healthy population, there is substantial, heritable, and interindividual variability in the platelet response. We explored whether a proportion of this variability could be accounted for by interindividual variation in gene expression. Through a correlative analysis of genome-wide platelet RNA expression data from 37 subjects representing the normal range of platelet responsiveness within a cohort of 500 subjects, we identified 63 genes in which transcript levels correlated with variation in the platelet response to adenosine diphosphate and/or the collagen-mimetic peptide, cross-linked collagen-related peptide. Many of these encode proteins with no reported function in platelets. An association study of 6 of the 63 genes in 4235 cases and 6379 controls showed a putative association with myocardial infarction for COMMD7 (COMM domain-containing protein 7) and a major deviation from the null hypo thesis for LRRFIP1 [leucine-rich repeat (in FLII) interacting protein 1]. Morpholino-based silencing in Danio rerio identified a modest role for commd7 and a significant effect for lrrfip1 as positive regulators of thrombus formation. Proteomic analysis of human platelet LRRFIP1-interacting proteins indicated that LRRFIP1 functions as a component of the platelet cytoskeleton, where it interacts with the actin-remodeling proteins Flightless-1 and Drebrin. Taken together, these data reveal novel proteins regulating the platelet response.
Introduction
Platelets are anucleate cell fragments derived from megakaryocytes (MKs) that play a pivotal role in the regulation of normal hemostasis. These cells are also intimately involved in the pathological processes associated with atherothrombotic diseases, in particular, the thrombotic response to plaque rupture seen in myocardial infarction (MI). The platelet response to agonist stimulation shows a wide interindividual variation1,2 that is reproducible over time1 and shows substantial heritability,3 suggesting regulation through genetic variation.
We have successfully used genomics approaches to identify novel regulators of platelet function4,–6 and, in addition, identified 32 quantitative trait loci (QTLs) for platelet count, volume,7,–9 and function.1,10 An association study showed that one of the QTLs for platelet count on chromosome 12q24 is also a risk gene for MI and other diseases.8 In addition to having identified a novel risk gene for MI, these studies have also provided new insights in the processes of megakaryopoiesis, proplatelet formation, and the regulation of the platelet functional response to activatory signals. Here, we expanded on our previous studies and investigated whether other genes, hitherto unknown to play a role in the platelet, can be identified by correlating transcript levels in platelets with the functional response.
Genome-wide expression (GWE) studies of platelets, and from MKs derived by culture from CD34+ hematopoietic progenitor cells, have shown that approximately half of the ∼ 10 000 transcripts present in MKs can also be detected in platelets.11,–13 For the present study, we used samples from the previously described Platelet Function Cohort (PFC),1,10 comprising 500 healthy subjects in whom the platelet response to 2 key agonists, adenosine diphosphate (ADP) and a collagen mimetic, cross-linked collagen-related peptide (CRP-XL), was quantified by measuring the exposure of P-selectin (a marker of α-granule secretion) and the binding of fibrinogen (a marker of the activation of α-IIb-β3 and therefore a surrogate marker of aggregation). Transcript levels in platelet RNA from 37 subjects, selected from the PFC to represent the full spectrum of the platelet response, were determined, and a correlative analysis identified 63 transcripts that showed an association between abundance and at least 1 of the 4 functional readouts measured in the PFC. Six of the corresponding genes were selected for an association study in more than 11 500 DNA samples from cases and controls to identify putative, novel risk genes for MI. The 2 genes with the lowest P value of association with MI were selected for a functional study in a laser-induced vessel wall damage model in Danio rerio.5 This showed that the silencing of both commd7 and lrrfip1 by morpholino (MO) injection resulted in reduced thrombus formation. To elucidate the underlying molecular mechanism of the strongest effect, we characterized the protein-protein interaction network of LRRFIP1 in resting and activated human platelets, and the results suggest that this protein plays a critical role in the regulation of the platelet cytoskeleton.
Methods
Whole-genome expression studies with platelet RNA
For selection of representative subjects from the PFC for whole-genome expression studies, the measurement of the platelet response in the 500 subjects in the PFC has been described previously.1 Briefly, the platelet response to a standardized, midrange concentration of ADP (10−7M), or CRP-XL (0.1 μg/mL), was measured using flow cytometry to analyze 2 quantitative readouts for each agonist; P-selectin expression (P) and fibrinogen binding to the activated α-IIb-β3 integrin (F). This provides 4 separate measurements of platelet function, which are represented in figures and tables by PA, PC, and FA and FC, where A and C denote ADP and CRP-XL, respectively. The functional responses observed were normalized by logit transformation and adjusted for confounders, and the standardized residuals for each measure were used to select a group of 46 subjects representative of the full range of platelet response, as previously described.1 At the time of recall, their platelet response was retested.1
Whole-genome expression profiling of platelet RNA from the PFC: Leukocyte-depleted platelet concentrates were obtained by apheresis from the recalled subjects from the PFC as described previously.12,13 RNA quality and purity were assessed by nonreducing agarose gel electrophoresis and duplex reverse-transcription polymerase chain reaction (RT-PCR) to determine the levels of the pan-leukocyte marker, CD45 (PTPRC), also as previously described.12,13 Nine samples, evenly distributed across the functional spectrum, did not meet the quality criteria and were removed from the study (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Total RNA from the remaining 37 samples was further purified using the RNeasy MinElute Cleanup kit (QIAGEN). RNA (100 ng) was amplified and labeled using the Illumina TotalPrep RNA Amplification kit (Applera). Biotinylated cRNA (1500 ng per sample) was applied to Illumina HumanWG-6 v1 Expression BeadChips and hybridized overnight at 58°C. This array contains more than 46 000 probes bound to more than 1.2 million beads corresponding to genes annotated in the National Center for Biotechnology Information. RefSeq and UniGene databases. Arrays were washed, detected and scanned according to the manufacturer's instructions. WGE data are available at ArrayExpress at the European Bioinformatics Institute (EBI) at http://www.ebi.ac.uk/microarray-as/ae under the accession number E-MTAB-374.
Identification of transcripts correlating with platelet function: The platelet WGE data were analyzed using R and Bioconductor (www.bioconductor.org) to identify transcripts whose levels correlated with 1 or more readout of platelet function.14 Probes were only considered positive if they had a detection score more than 0.95 in 19 or more samples. Data were normalized using quantile normalization to remove nonbiological variation between arrays,15 the signal intensity (SI) of each probe was plotted against the logit transformed function data,1 and the Pearson correlation (r-value) and associated P value were calculated. The probability of a significant correlation was calculated for each probe by comparing the observed distribution of r-values to the null distribution of r-values observed if the null hypothesis of no correlation between values was true. After this, an additional filtering step was applied to remove all probes that had a low signal (average log2 SI < 7.64), which resulted in a list of 63 genes for further study.
Selection of genes for further study
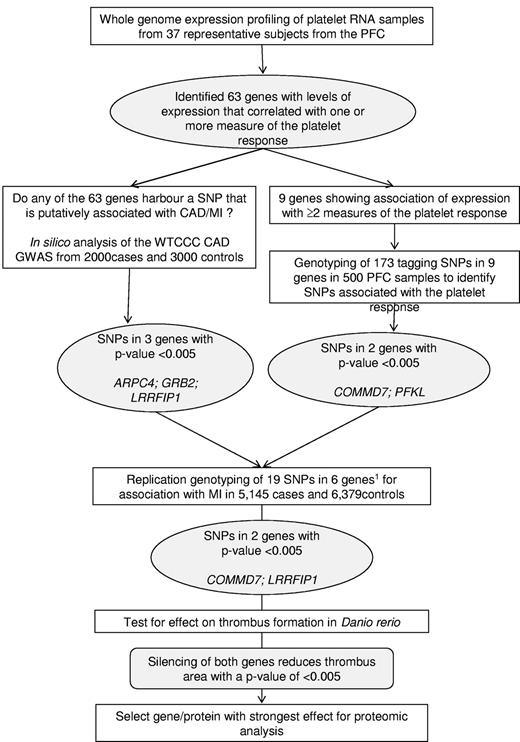
Within this group of 63 genes, we then identified genes that harbored single-nucleotide polymorphisms (SNPs) that showed a putative association with MI by in silico analysis of the association results for coronary artery disease (CAD)/MI of the Wellcome Trust Case Control Consortium (WTCCC) genome-wide association study of 2000 CAD/MI cases and 3000 controls16 (subsequently referred to as WTCCC CAD/MI GWAS), or SNPs associated with one or more measure of platelet response (by genotyping tagging SNPs in the PFC). Replication genotyping of these selected SNPs was then carried out in a larger cohort of more than 11 500 MI/CAD cases and controls to identify genes associated with MI for further study in vivo and in vitro. The strategy for this selection is illustrated in Figure 1.
Schematic summarizing the overall study design. A stepwise approach was followed to select 6 genes for an association study in cases of MI and controls. The results of the association study prompted a study to determine the effect of the silencing of 2 genes on thrombus formation in D rerio. The protein encoded by the gene with the strongest effect on thrombus formation was selected for proteomics analysis in human platelets. PFC, Platelet Function Cohort; CAD, coronary artery disease; WTCCC, Wellcome Trust Case Control Consortium; GWAS, genome-wide association study; SNP, single-nucleotide polymorphism. Inclusion of the 6th gene (GTF2A2) was based on association of expression with all 4 measures of the platelet response and association.
Schematic summarizing the overall study design. A stepwise approach was followed to select 6 genes for an association study in cases of MI and controls. The results of the association study prompted a study to determine the effect of the silencing of 2 genes on thrombus formation in D rerio. The protein encoded by the gene with the strongest effect on thrombus formation was selected for proteomics analysis in human platelets. PFC, Platelet Function Cohort; CAD, coronary artery disease; WTCCC, Wellcome Trust Case Control Consortium; GWAS, genome-wide association study; SNP, single-nucleotide polymorphism. Inclusion of the 6th gene (GTF2A2) was based on association of expression with all 4 measures of the platelet response and association.
In silico analysis of WTCCC CAD/MI GWAS data identified 3 of the 63 genes (ARPC4, GRB2, and LRRFIP1) that harbored a SNP nominally associated with the risk of CAD/MI at a relaxed P ≤ .005. These SNPs were subsequently genotyped in the PFC DNA samples by a Sequenom genotyping assay.
For genotyping in subjects of the PFC, to further identify SNPs associated with platelet function, the 500 DNA samples from the PFC were genotyped using a GoldenGate 1536 Custom array (Illumina) that included 173 SNPs in 9 of the 63 genes encoding the transcripts that showed an association with at least 2 measures of platelet function in the PFC (ALDOA, COMMD7, GTF2A2, NFE2L2, PFKL, PPP2CA, PRKACB, TACC3, and TBPL1). Genotyping of this cohort of 500 healthy blood donors, of European ancestry, from the Cambridge BioResource at the Cambridge BioMedical Research Center and National Health Service Blood and Transplant (NHSBT) Cambridge has been described previously.10
Genotyping of MI/CAD cases and controls
A total of 19 tagging SNPs in 6 genes were selected for genotyping by Sequenom in 5145 MI/CAD cases and 6379 controls. The tagging SNPs were selected on the basis of the following criteria: (1) the SNP with the lowest P value of association for platelet function in the PFC, (2) the SNP with the lowest P value of association with the risk of CAD/MI in the WTCCC-GWAS, and (3) nonsynonymous (ns)SNPs that alter the amino acid sequence of the corresponding protein. For the latter, nsSNPs from HapMap and the Wellcome Trust Sanger Institute Exome sequencing project, known at the time of the design of the experiment, were included. For all SNPs in categories (1) and (2), technical back-up SNPs that were in linkage disequilibrium (LD) of r2 = 1 were included.
The cohort comprised 5145 samples from MI/CAD cases from 3 studies: the British Heart Foundation Family Study (BHF-FHS; n = 2207); the Associazione per lo Studio della Trombosi in Cardiologia-Premature MI cohort (ASTC-PMI; n = 1958); and the Amsterdam Premature Atherosclerosis Study (AMC-PAS; n = 980). The controls comprised 6379 DNA samples from the following: the UK Blood Services Common Controls (UKBS-CC) Panels 1 and 2 (n = 1528 and 1644, respectively); ASTC-PMI (n = 1976); and the Sanquin Common Controls (SANQUIN-CC; n = 1231). Eligibility criteria and clinical baseline figures for the 3 case collections have been reported elsewhere17,–19 and are provided in supplemental Table 1A and B, which include their demographic and classical risk factors (eg, smoking, hypertension, and diabetes). Controls for the BHF-FHS and AMC-PAS cohorts were collected from healthy blood donors from either the United Kingdom16,20 or the Dutch Blood Services, respectively (UKBS-CC and Sanquin-CC). For the ASTI-PMI cohort, controls were recruited locally, then matched to the cases for age and sex.18 Processing and quality control of the DNA samples is described in the supplemental data. All DNA samples were collected with informed consent and written approval, and all studies were approved by the relevant ethical research committees. All investigations were conducted in accordance with the principles of the Declaration of Helsinki.
Laser-induced thrombus formation in D rerio
We have previously reported the development of a MO based gene knockdown model in D rerio (zebrafish) for identifying genes with a role in thrombus formation.5 This model was used to assess the function of the fish orthologs of COMMD7 and LRRFIP1, herein named commd7 and lrrfip1, and of the control gene itga2b. Orthologs were first identified by reciprocal BLAST analysis using Ensembl (http://www.ensembl.org) and showed 61%, 39%, and 42% sequence homology at the amino acid level, respectively (supplemental Table 2). Fish handling, MO injection, and laser injury were conducted as described previously.5 For the latter, injury of the caudal artery endothelium was by a nitrogen-pulse ablating laser delivered for 1-5 seconds at 3 pulses/sec. Each larva was injured once. Images acquired with a charge-coupled device camera were analyzed with Metamorph software (version 6.0; Universal Imaging Corporation). Time to attachment (TTA) corresponded to the first cell adhering to the vessel after injury (t = 0). Thrombus area (TA) was calculated in square pixels at 1-minute intervals after injury by measuring thrombus surface area using ImageJ Version 1.38 software (National Institutes of Health).5 For each experiment, typically up to 15 MO-injected larvae were screened per MO with a similar number of matched sibling controls tested before and afterward, with the total experiment lasting less than 4 hours. Each experiment was repeated at least once using offspring of different parentage.
Statistical analysis In this study, TTA of the first cell was modeled using Cox regression, stratifying by the experimental day. The severity of the injury did not significantly affect the model and so was not adjusted for. Although the TA is highly skewed, log10(TA) is approximately normally distributed. In all cases, a substantial thrombus had formed at the site of injury within 1 minute, but, in some instances, this thrombus had detached by later time points, in which case the TA value was set to “not available” (NA). Log10(area) was fitted with a linear mixed model with fixed effects for MO type (nonspecific or MO directed to ATG site or splice site), time after laser injury (1, 2, 3, or 4 minutes), and random effects for experimental day and fish within experimental day. The estimated coefficients are antilogged to transform back to the original scale (ie, square pixels).
LRRFIP1 interactome in platelets
The antibodies used in proteomic studies were as follows: anti-LRRFIP1 (clone 32; BD Biosciences), anti-FLI1 (Flightless 1, clone 116.40; Abcam), anti-DBN1 (Drebrin 1, clone M2F6; Abcam), and normal mouse immunoglobulin G (IgG; Santa Cruz Biotechnology). Washed platelets were prepared, and control and thrombin receptor activation peptide (TRAP or SFLLRN, 5μM)-activated samples were prepared as previously described21 : Platelets were lysed in a 2× lysis buffer (100mM Tris [tris(hydroxymethyl)aminomethane], pH 7.4, 300mM NaCl; 2% [vol/vol] Nonidet P-40 [NP-40], complete protease inhibitors, 2 mM sodium orthovanadate, 20 mM 2-glycerophosphate, and 50mM sodium fluoride), and anti-LRRFIP1 and IgG control immunoprecipitations were performed from resting and activated platelets as described previously.21 After extensive washing, bound proteins were eluted from the Protein A/G beads (Pierce) by boiling in Laemmli buffer. Gel-based liquid chromatography–tandem mass spectrometry (GeLC-MS/MS) was performed as previously described.22 Spectra were searched using both the X!TANDEM and SEQUEST algorithms, against the International Protein Index (IPI) database (http://www.ebi.ac.uk/IPI/) V3.38 as described before.23 The probability-based evaluation program, Protein Prophet, was used to extract identifications greater or equal to a score of 0.9.24 The list of proteins was further refined manually to remove one-peptide hits, keratin, and IgG contamination, and to exclude nonspecific interactions (defined as any protein that had one or more peptide(s) found in the IgG control samples).23
Results
Association between platelet transcript levels and platelet function
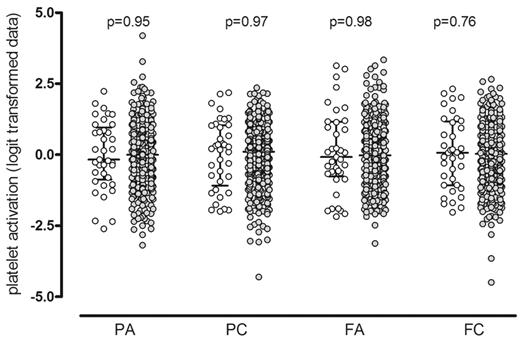
The wide distribution of the functional responses of the 500 PFC subjects to a midrange concentration of ADP (10−7M) and CRP-XL (0.1 μg/mL) has been previously described.1,10 They are shown in Figure 2 to illustrate that the platelet responses of the 37 selected subjects for the WGE study were distributed across the full spectrum of platelet responses (P > .75 for all). The response on recall was repeated in the same functional assay and was consistent with the original response for all subjects and for all measures of the platelet response, as previously described1 (P > .28 for all; data not shown).
Platelet functional response in the Platelet Function Cohort. Comparison of the platelet response to ADP and CRP-XL in the 37 selected subjects (○) with the 500 subjects in the whole PFC (•). Platelet activation was measured by flow cytometry, and data are shown as the standardized residuals of logit transformed data, after adjustment for confounding variables, as described by Jones et al.1 PA, P-selectin expression in response to ADP; PC, P-selectin expression in response to CRP-XL; FA, fibrinogen binding in response to ADP; FC, fibrinogen binding in response CRP-XL. Error bars show the median and interquartile ranges.
Platelet functional response in the Platelet Function Cohort. Comparison of the platelet response to ADP and CRP-XL in the 37 selected subjects (○) with the 500 subjects in the whole PFC (•). Platelet activation was measured by flow cytometry, and data are shown as the standardized residuals of logit transformed data, after adjustment for confounding variables, as described by Jones et al.1 PA, P-selectin expression in response to ADP; PC, P-selectin expression in response to CRP-XL; FA, fibrinogen binding in response to ADP; FC, fibrinogen binding in response CRP-XL. Error bars show the median and interquartile ranges.
cRNA from the 37 RNA samples was applied to Illumina HumanWG-6 v1 Expression BeadChips. A total of 5325 probes were detected as being present in more than 50% of the 37 microarrays, which represents 5169 unique genes. After calculation of the Pearson correlation between the normalized SI of these 5325 probes, and each of the 4 logit transformed functional responses, correlations with a P ≥ .95 were considered significant. At this probability level, the signal generated by 262 probes were correlated with the platelet response; 61 with PA and 112, 46, and 43 with the PC, FA, and FC responses, respectively. From these 262 probes, 63 were selected by excluding those with low abundance (log2 SI < 7.64), except those probes that showed a ratio exceeding 3 between highest and the lowest SI (GTF2A2, LPAR4, NFE2L2, NM1, and SORD) in the 37 samples. All 63 probes showed a wide range of SIs across the 37 samples (supplemental Table 3), with RPS26 having the highest ratio (mean log2 SI, 9.35; range, 7.97-10.32) and FUT6 the lowest (mean log2 SI, 8.26; range, 8.25-9.70).
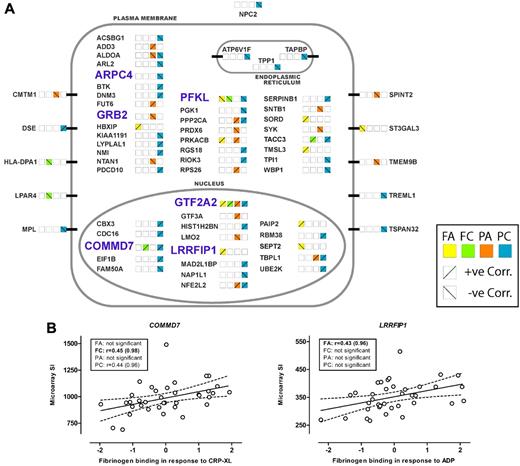
The cellular location of the genes, based on gene ontology and refined by manual curation, is illustrated in Figure 3A, with approximately half (31/63) identified as having a cytoplasmic location, 10 transmembrane proteins, and the remaining third having either a nuclear (18/63), endoplasmic reticulum (3/63), or lysosomal/secreted (1/63) location. Figure 3A also illustrates that there were both positive and negative correlations between the level of transcript and the functional response(s). The results of the correlation analysis for the 2 transcripts, COMMD7 and LRRFIP1, selected for further study, are shown in Figure 3B.
Genes showing association of transcript abundance in platelets with platelet response. (A) Sixty-three transcripts in platelets that show a correlation between microarray signal intensity (SI) and functional responses. Genes are grouped alphabetically and by cellular location of the encoded proteins. Platelet activation was measured as fibrinogen binding (F) and P-selectin expression (P) in response to ADP (A) or the collagen mimetic, CRP-XL (C). Correlation with functional responses for each transcript is indicated by the colored squares, with positive and negative correlations indicated by the diagonal lines. The 6 genes selected for genotyping in the cases of myocardial infarction (MI) and controls are highlighted in light blue and bold upper case. (B) Correlation between platelet transcript level and platelet function, in 37 subjects selected for the whole-genome expression study, for the 2 genes showing association with MI. SI was measured with Illumina probes GI_29789284-S and GI_4758689-S for COMMD7 and LRRFIP1, respectively. Platelet activation is shown as the standardized residuals of logit transformed flow-cytometric measurement of the percentage of platelets positive for specific markers of activation, as previously described.1 Graphs display the correlation with the highest significance, and boxes show Pearson correlation coefficients (r) and probability values (in parentheses) for all 4 functional parameters, with the highest significance shown in bold.
Genes showing association of transcript abundance in platelets with platelet response. (A) Sixty-three transcripts in platelets that show a correlation between microarray signal intensity (SI) and functional responses. Genes are grouped alphabetically and by cellular location of the encoded proteins. Platelet activation was measured as fibrinogen binding (F) and P-selectin expression (P) in response to ADP (A) or the collagen mimetic, CRP-XL (C). Correlation with functional responses for each transcript is indicated by the colored squares, with positive and negative correlations indicated by the diagonal lines. The 6 genes selected for genotyping in the cases of myocardial infarction (MI) and controls are highlighted in light blue and bold upper case. (B) Correlation between platelet transcript level and platelet function, in 37 subjects selected for the whole-genome expression study, for the 2 genes showing association with MI. SI was measured with Illumina probes GI_29789284-S and GI_4758689-S for COMMD7 and LRRFIP1, respectively. Platelet activation is shown as the standardized residuals of logit transformed flow-cytometric measurement of the percentage of platelets positive for specific markers of activation, as previously described.1 Graphs display the correlation with the highest significance, and boxes show Pearson correlation coefficients (r) and probability values (in parentheses) for all 4 functional parameters, with the highest significance shown in bold.
Interrogation of the HaemAtlas25 showed that all but 2 of the 63 transcripts (DNM3 and MPL) had pleiomorphic expression in the 6 main blood-cell types, erythroblasts, and MKs. The log2 SIs for the 63 transcripts varied widely in MKs (mean log2, 12.01; range, 7.27-14.19) and platelets (mean log2, 10.52; range, 6.17-14.02). On average, the level of the 63 transcripts was higher in cells of the myeloid lineage, compared with the 4 lymphoid cell types.
Selection of 6 genes for genotyping in a case-control study
We then investigated whether the transcripts associated with platelet function are encoded by genes that harbor SNPs that confer a risk for MI. For this, we performed a genotyping study in 11 524 DNA samples from 3 European Caucasoid MI cohorts and geographically matched controls (supplemental Table 1).
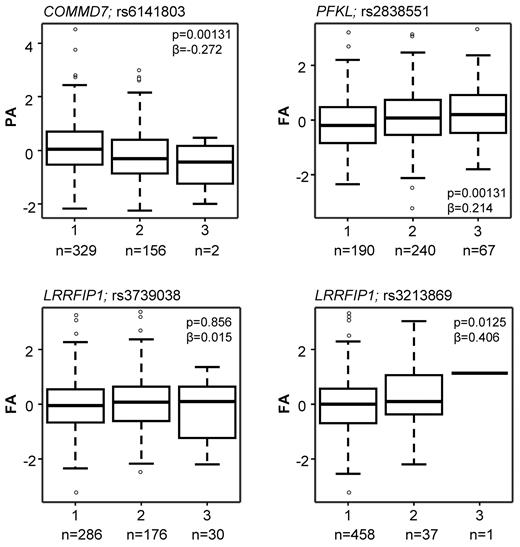
As illustrated in Figure 1, 6 the 63 genes (highlighted in pale blue bold font in Figure 3A and detailed in Table 1) were selected for this association study based on one or more of the following criteria: (1) correlations between transcript level and at least 3 of the 4 functional responses (GTF2A2, PFKL); (2) presence of an SNP nominally associated with the risk of CAD/MI at a P < .005 in the WTCCC GWAS (ARPC4, GRB2, and LRRFIP1); or (3) presence of an SNP with a P value of association < .005 for platelet function in the PFC (COMMD7, PFKL). The effect of the SNPs in COMMD7 and PFKL on the platelet response is illustrated in Figure 4, together with the effect of the 2 SNPs in LRRFIP1 that also showed the lowest P value for association with MI in the WTCCC CAD/MI GWAS data (rs3213869) and the replication cohort (rs3739038; see below), respectively.
The 6 genes selected for genotyping in cases with MI and controls
| HGNC Gene IDa . | Description . | Microarray analysis . | Genotyping in PFCb (lowest P) . | Association with CAD, Pj,k . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACc . | MSId . | SDe . | Ratiof . | No. SNP typedg . | rs no. of SNP . | MAFh . | Pi . | |||
| ARPC4 | Actin-related protein 2/3 complex, subunit 4, 20 kDa | −0.48 | 577 | 94 | 2.0 | 2 | rs3806667 | 0.098 | 1.70 × 10−1 | 2.6 × 10−3 |
| COMMD7 | COMM domain containing protein 7 | 0.45 | 987 | 144 | 2.2 | 10 | rs6141803 | 0.181 | 1.27 × 10−3 | ns |
| GRB2 | Growth factor receptor-bound protein 2 | −0.44 | 364 | 45 | 1.7 | 3 | rs41282071 | 0.065 | 5.56 × 10−2 | 8.3 × 10−4 |
| GTF2A2 | General transcription factor IIA, 2, 12 kDa | 0.46 | 185 | 65 | 3.7 | 17 | rs1057058 | 0.050 | 1.13 × 10−2 | ns |
| LRRFIP1 | Leucine-rich repeat (in FLII) interacting protein 1 | 0.43 | 352 | 54 | 1.9 | 4 | rs3213869 | 0.038 | 1.26 × 10−2 | 2.5 × 10−3 |
| PFKL | Phosphofructokinase, liver | −0.47 | 271 | 47 | 2.2 | 31 | rs2838551 | 0.376 | 1.27 × 10−3 | ns |
| HGNC Gene IDa . | Description . | Microarray analysis . | Genotyping in PFCb (lowest P) . | Association with CAD, Pj,k . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACc . | MSId . | SDe . | Ratiof . | No. SNP typedg . | rs no. of SNP . | MAFh . | Pi . | |||
| ARPC4 | Actin-related protein 2/3 complex, subunit 4, 20 kDa | −0.48 | 577 | 94 | 2.0 | 2 | rs3806667 | 0.098 | 1.70 × 10−1 | 2.6 × 10−3 |
| COMMD7 | COMM domain containing protein 7 | 0.45 | 987 | 144 | 2.2 | 10 | rs6141803 | 0.181 | 1.27 × 10−3 | ns |
| GRB2 | Growth factor receptor-bound protein 2 | −0.44 | 364 | 45 | 1.7 | 3 | rs41282071 | 0.065 | 5.56 × 10−2 | 8.3 × 10−4 |
| GTF2A2 | General transcription factor IIA, 2, 12 kDa | 0.46 | 185 | 65 | 3.7 | 17 | rs1057058 | 0.050 | 1.13 × 10−2 | ns |
| LRRFIP1 | Leucine-rich repeat (in FLII) interacting protein 1 | 0.43 | 352 | 54 | 1.9 | 4 | rs3213869 | 0.038 | 1.26 × 10−2 | 2.5 × 10−3 |
| PFKL | Phosphofructokinase, liver | −0.47 | 271 | 47 | 2.2 | 31 | rs2838551 | 0.376 | 1.27 × 10−3 | ns |
Gene identifier (ID) is by the HUGO Gene Nomenclature Committee (HGNC).
PFC, Platelet Function Cohort.
AC, average correlation r value.
MSI, mean signal intensity obtained with the 37 platelet RNA samples for the corresponding feature on the Illumina platform.
SD, standard deviation.
Ratio = maximum signal intensity/minimum signal intensity.
Number (no.) of single-nucleotide polymorphisms (SNPs) typed in the 500 DNA samples of the PFC (COMMD7 and PFKL were genotyped on a custom Illumina 1536 platform; ARPC4, GRB2, and LRRFIP1 were genotyped by Sequenom).
MAF, minor allele frequency.
Lowest P value: the association between genotype and platelet function was tested for each SNP against the 4 functional responses, and the lowest P value is presented.
Lowest P value: the association between genotype and CAD/MI in the WTCCC GWAS cohort.
In silico analysis from WTCCC CAD cases and controls; ns, not significant, and P values in bold reached the predefined level of significance.
Box plots showing the effect of sequence variation in the COMMD7, LRRFIP1, and PFKL genes on the platelet response. Data are shown for the SNPs that showed the lowest P value in the PFC and the lowest P value in the replication MI/CAD cohort. The association between sequence variation and the platelet response is shown as the mean and interquartile range. Platelet activation was measured by flow cytometry, and data are shown as the standardized residuals of logit transformed data, after adjustment for confounding variables, as described by Jones et al.1 PA, P-selectin expression in response to ADP; FA, fibrinogen binding in response to ADP. Numbers (n) below graphs indicate the numbers of subjects homozygous for the major allele (1), heterozygous (2), or homozygous for the minor allele (3).
Box plots showing the effect of sequence variation in the COMMD7, LRRFIP1, and PFKL genes on the platelet response. Data are shown for the SNPs that showed the lowest P value in the PFC and the lowest P value in the replication MI/CAD cohort. The association between sequence variation and the platelet response is shown as the mean and interquartile range. Platelet activation was measured by flow cytometry, and data are shown as the standardized residuals of logit transformed data, after adjustment for confounding variables, as described by Jones et al.1 PA, P-selectin expression in response to ADP; FA, fibrinogen binding in response to ADP. Numbers (n) below graphs indicate the numbers of subjects homozygous for the major allele (1), heterozygous (2), or homozygous for the minor allele (3).
Genotyping in MI cases and controls
The 19 tagging SNPs in the 6 genes selected for replication genotyping (see supplemental Table 4) were all (apart from rs3743119 in the GTF2A2 gene) in Hardy-Weinberg equilibrium and typed correctly. The association between each SNP and MI was first tested in the 3 separate populations using the Cochrane-Armitage trend test (British and Dutch studies) and the Cochrane-Mantel-Haenzsel test (Italian study of matched case-control pairs; supplemental Table 4). Having observed the effect of the SNPs on MI in separate populations, the 3 collections were combined, and a Cochrane-Mantel-Haenzsel test was performed, controlling for the 3 population clusters. A P value threshold of 1 × 10−4 (after Bonferroni correction) was considered significant. The combined analysis revealed one SNP rs6141803 in COMMD7 that exceeded this threshold (P = 5.3 × 10−6) and another, SNP rs3739038 in LRRFIP1, which showed a major deviation from the null hypothesis (P = 7.9 × 10−4). These results suggest a possible association with MI, although the P values of association did not reach genome-wide significance.
Gene silencing of COMMD7 and LRRFIP1 in D rerio shows modification of thrombosis
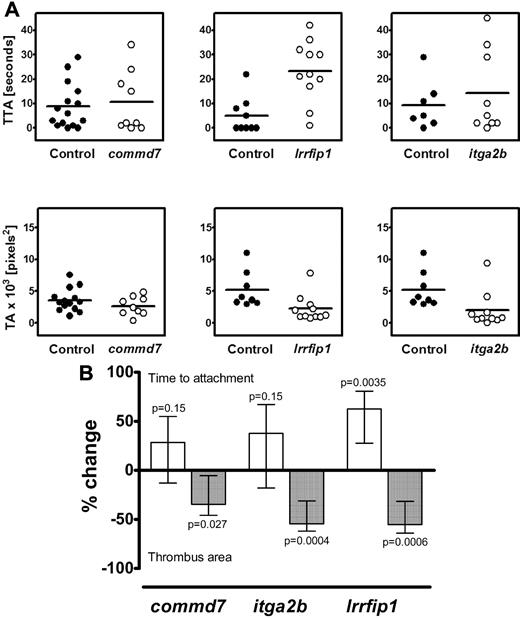
The observations that the transcript levels of both COMMD7 and LRRFIP1 are correlated with platelet function (Figure 3), and that the genes harbor SNPs that are associated with platelet function in the PFC (Table 1; Figure 4), or showed a putative association with the risk for MI (supplemental Table 4), prompted functional studies in D rerio. The effect of gene silencing by MO injection on thrombus formation after laser injury of the caudal artery was determined using a recently validated experimental approach.5,26 Figure 5 a shows representative data of the TTA and TA from a single experiment for commd7 and lrrfip1, compared with the positive control, itga2b, that encodes the α-chain of the platelet α-IIb-β3 integrin. Figure 5B shows the combined analysis of the results of all experiments, expressed as the percentage change, compared with the wild-type controls. Knockdown of commd7 produced a modest, nonsignificant increase in TTA, of the same order as that seen when itga2b was silenced, accompanied by a significant decrease in TA (P = .027). For lrrfip1, the TTA was significantly increased (P = .0035), which was linked to a significant, strong reduction in the thrombus area (P = .0006), comparable with that seen in the itga2b knockdown (P = .0004). The strong effect of the lrrfip1 silencing on both parameters of thrombus formation therefore warranted further studies of this protein in human platelets to delineate the molecular pathway by which LRRFIP1 may affect thrombus formation.
The effect of antisense knockdown of commd7, lrrfip1, and itga2b in D. rerio on thrombus kinetics. (A) Examples of results for the knockdown of commd7, lrrfip1, and itga2b (positive control) from 1 experiment showing the time to attachment of the first cell (TTA) in the top panels and the thrombus area (TA) in the bottom panels in control (•) and morpholino (MO)–injected (○) embryos. (B) Combined data for all experiments showing data as the percentage difference between control and the gene of interest (± confidence intervals) in either TTA (□) or TA (■). Injection of complementary MO resulted in a nonsignificant increase in TTA in the commd7 and itga2b morphants and a significant increase (P = .0035) in the lrrfip1 morphants. Knockdown of all 3 genes resulted in a reduction in TA of 30% (P = .027), 54% (P = .0004), and 55% (P = .0006) respectively. The effect of knockdown on TTA was estimated using Cox regression, and stratifying by experimental day and the effect on TA was estimated using a linear mixed-effects model as described in “Methods.”
The effect of antisense knockdown of commd7, lrrfip1, and itga2b in D. rerio on thrombus kinetics. (A) Examples of results for the knockdown of commd7, lrrfip1, and itga2b (positive control) from 1 experiment showing the time to attachment of the first cell (TTA) in the top panels and the thrombus area (TA) in the bottom panels in control (•) and morpholino (MO)–injected (○) embryos. (B) Combined data for all experiments showing data as the percentage difference between control and the gene of interest (± confidence intervals) in either TTA (□) or TA (■). Injection of complementary MO resulted in a nonsignificant increase in TTA in the commd7 and itga2b morphants and a significant increase (P = .0035) in the lrrfip1 morphants. Knockdown of all 3 genes resulted in a reduction in TA of 30% (P = .027), 54% (P = .0004), and 55% (P = .0006) respectively. The effect of knockdown on TTA was estimated using Cox regression, and stratifying by experimental day and the effect on TA was estimated using a linear mixed-effects model as described in “Methods.”
Proteomic analysis of the interactome of LRRFIP1
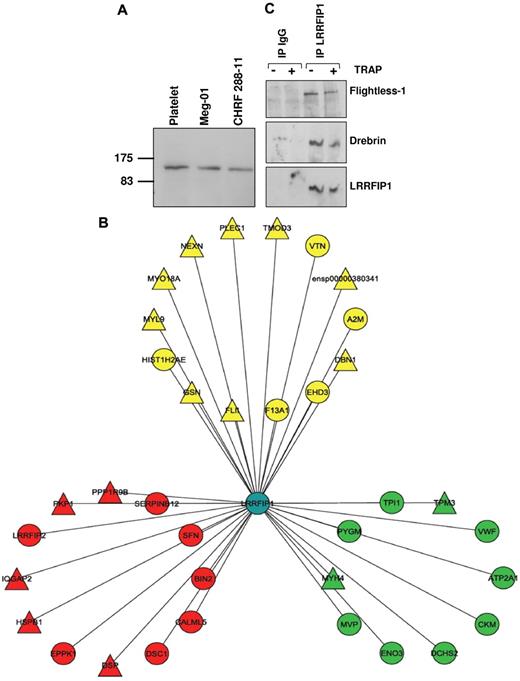
In nucleated cells, LRRFIP1 acts in the nucleus as a repressor of transcription27,28 ; therefore, its function in the anucleate platelet cannot obviously be inferred. To determine a putative mechanism underlying the effect of variation in the LRRFIP1 transcript level on the functional response of human platelets and the profound changes in thrombus formation in D rerio injected with the lrrfip1 MO, we used a coimmunoprecipitation/mass spectrometry approach to identify components of the protein-protein interaction network (or “interactome”) for LRRFIP1. First, we confirmed its presence at the protein level in platelets and the cytoplasm of the megakaryocytic cell lines, Meg-01 and CHRF 288-11, by Western blotting (Figure 6A). We then performed an analysis of the LRRFIP1 pull-downs from resting and TRAP-activated platelets by tandem mass spectrometry, which identified 25 and 27 interactors (Figure 6B; and Table 2) in resting and activated platelets, respectively. The interaction of 2 proteins with LRRFIP1, FLI1 (Flightless-1), and DBN1 (Drebrin 1), both identified by a large number of peptide hits, were verified in the resting and activated LRRFIP1 interactomes by Western blotting (Figure 6C). FLI1, an actin severing and capping protein shown previously to interact with LRRFIP1,29,30 and DBN1, a previously unattributed platelet protein with a role in the formation of dendritic spines and stabilization of tight junctions,31 both form part of a functional cassette of cytoskeletal proteins involved in actin binding and remodeling (Figure 6B; and Table 2).
LRRFIP1 interactions in resting and activated human platelets imply a role in the regulation of the platelet cytoskeleton. (A) Western blotting with anti-LRRFIP1 identifies a 160-kDa protein in platelets and the megakaryocytic cell lines, Meg-01 and CHRF-288-11. (B) Tandem mass spectrometry analysis LRRFIP1 coimmunoprecipitation (co-IP) from resting and activated platelets identified novel and known protein-protein interactions (Table 2). Shown is a Cytoscape visualization of the LRRFIP1 protein-protein interaction network in platelets (Cytoscape Version 2.5). Node colors indicate interactions detected in resting only (green), activated only (red), or both resting and activated (yellow) platelets. LRRFIP1, which was detected in both resting and activated co-IPs; LRRFIP1 is shown centrally in blue. Node diameters indicate the number of peptide hits; a semiquantitative measure of protein abundance. (C) Western blotting of LRRFIP1 co-IPs with specific anti-Flightless 1 and anti-Drebrin antibodies confirmed these protein-protein interactions.
LRRFIP1 interactions in resting and activated human platelets imply a role in the regulation of the platelet cytoskeleton. (A) Western blotting with anti-LRRFIP1 identifies a 160-kDa protein in platelets and the megakaryocytic cell lines, Meg-01 and CHRF-288-11. (B) Tandem mass spectrometry analysis LRRFIP1 coimmunoprecipitation (co-IP) from resting and activated platelets identified novel and known protein-protein interactions (Table 2). Shown is a Cytoscape visualization of the LRRFIP1 protein-protein interaction network in platelets (Cytoscape Version 2.5). Node colors indicate interactions detected in resting only (green), activated only (red), or both resting and activated (yellow) platelets. LRRFIP1, which was detected in both resting and activated co-IPs; LRRFIP1 is shown centrally in blue. Node diameters indicate the number of peptide hits; a semiquantitative measure of protein abundance. (C) Western blotting of LRRFIP1 co-IPs with specific anti-Flightless 1 and anti-Drebrin antibodies confirmed these protein-protein interactions.
Proteins identified by mass spectrometry in the anti-LRRFIP1 pull-down from resting and activated platelets
| IPI ID . | HGNC gene ID* . | Description . | No. of peptides present . | |||
|---|---|---|---|---|---|---|
| Xtandem . | Sequest . | |||||
| Resting . | Activated . | Resting . | Activated . | |||
| IPI00478003.1 | A2M | Alpha-2-macroglobulin | 2 | 1 | 1 | 1 |
| IPI00297550.8 | F13A1 | Coagulation factor xiiia | 2 | 6 | 3 | 4 |
| IPI00021458.1 | EHD3 | eh domain-containing protein 3 | 1 | 2 | 0 | 2 |
| IPI00513782.3 | GSN | Gelsolin | 4 | 13 | 0 | 0 |
| IPI00026272.2 | HIST1H2AB/HIST1H2AE | Histone h2a type 1-b | 2 | 1 | 2 | 3 |
| IPI00003406.3 | DBN1 | Isoform 1 of drebrin | 14 | 33 | 11 | 35 |
| IPI00017030.10 | MYO18A | Isoform 1 of myosin-xviiia | 15 | 6 | 16 | 4 |
| IPI00180404.7 | NEXN | Isoform 1 of nexilin | 4 | 16 | 3 | 12 |
| IPI00014898.2 | PLEC1 | Isoform 1 of plectin-1 | 1 | 15 | 1 | 12 |
| IPI00006207.2 | LRRFIP1 | Leucine-rich repeat flightless-interacting protein 1 | 1 | 4 | 0 | 5 |
| IPI00220278.5 | MYL9 | Myosin regulatory light chain2; smooth muscle | 3 | 5 | 4 | 7 |
| IPI00031023.1 | FLII | Protein flightless-1 homolog | 16 | 26 | 17 | 21 |
| IPI00005087.1 | TMOD3 | Tropomodulin-3 | 5 | 13 | 6 | 8 |
| IPI00176593.3 | No symbol | Uncharacterized protein ensp00000380341 | 4 | 10 | 8 | 11 |
| IPI00298971.1 | VTN | Vitronectin precursor | 3 | 2 | 1 | 1 |
| IPI00218474.5 | ENO3 | Beta-enolase | 5 | 0 | 3 | 0 |
| IPI00399257.2 | DCHS2 | CDNA flj45804 fis | 2 | 0 | 0 | 0 |
| IPI00027487.3 | CKM | Creatine kinase m-type | 2 | 0 | 3 | 0 |
| IPI00218130.3 | PYGM | Glycogen phosphorylase; muscle form | 3 | 0 | 4 | 0 |
| IPI00024804.3 | ATP2A1 | Serca1b of sarcoplasmic | 2 | 0 | 2 | 0 |
| IPI00000105.4 | MVP | Major vault protein | 4 | 0 | 1 | 0 |
| IPI00001753.1 | MYH4 | Myosin-4 | 8 | 0 | 6 | 0 |
| IPI00383071.1 | TPI1 | Triosephosphate isomerase (fragment) | 2 | 0 | 2 | 0 |
| IPI00183968.4 | TPM3 | Tropomyosin 3 isoform 1 | 3 | 0 | 3 | 0 |
| IPI00023014.1 | VWF | Von willebrand factor | 2 | 0 | 2 | 0 |
| IPI00021536.1 | CALML5 | Calmodulin-like protein 5 | 0 | 1 | 0 | 3 |
| IPI00007425.1 | DSC1 | Desmocollin 1 | 0 | 2 | 0 | 2 |
| IPI00010951.2 | EPPK1 | Epiplakin | 0 | 5 | 0 | 7 |
| IPI00025512.2 | HSPB1 | Heat shock protein beta-1 | 0 | 1 | 0 | 2 |
| IPI00013890.2 | SFN | 14-3-3 protein sigma | 0 | 1 | 0 | 3 |
| IPI00550792.4 | BIN2 | Bridging integrator 2 | 0 | 2 | 0 | 3 |
| IPI00007277.2 | LRRFIP2 | Leucine-rich repeat flightless-interacting protein 2 | 0 | 2 | 0 | 3 |
| IPI00299048.6 | IQGAP2 | ras GTPase-activating-like protein iqgap2 | 0 | 2 | 0 | 3 |
| IPI00071509.1 | PKP1 | Plakophilin-1 | 0 | 3 | 0 | 3 |
| IPI00013933.2 | DSP | DPI of desmoplakin | 0 | 28 | 0 | 24 |
| IPI00045550.4 | PPP1R9B | Protein phosphatise-1; regulatory subunit 9b | 0 | 3 | 0 | 0 |
| IPI00033583.3 | SERPINB12 | Serpin b12 | 0 | 2 | 0 | 3 |
| IPI ID . | HGNC gene ID* . | Description . | No. of peptides present . | |||
|---|---|---|---|---|---|---|
| Xtandem . | Sequest . | |||||
| Resting . | Activated . | Resting . | Activated . | |||
| IPI00478003.1 | A2M | Alpha-2-macroglobulin | 2 | 1 | 1 | 1 |
| IPI00297550.8 | F13A1 | Coagulation factor xiiia | 2 | 6 | 3 | 4 |
| IPI00021458.1 | EHD3 | eh domain-containing protein 3 | 1 | 2 | 0 | 2 |
| IPI00513782.3 | GSN | Gelsolin | 4 | 13 | 0 | 0 |
| IPI00026272.2 | HIST1H2AB/HIST1H2AE | Histone h2a type 1-b | 2 | 1 | 2 | 3 |
| IPI00003406.3 | DBN1 | Isoform 1 of drebrin | 14 | 33 | 11 | 35 |
| IPI00017030.10 | MYO18A | Isoform 1 of myosin-xviiia | 15 | 6 | 16 | 4 |
| IPI00180404.7 | NEXN | Isoform 1 of nexilin | 4 | 16 | 3 | 12 |
| IPI00014898.2 | PLEC1 | Isoform 1 of plectin-1 | 1 | 15 | 1 | 12 |
| IPI00006207.2 | LRRFIP1 | Leucine-rich repeat flightless-interacting protein 1 | 1 | 4 | 0 | 5 |
| IPI00220278.5 | MYL9 | Myosin regulatory light chain2; smooth muscle | 3 | 5 | 4 | 7 |
| IPI00031023.1 | FLII | Protein flightless-1 homolog | 16 | 26 | 17 | 21 |
| IPI00005087.1 | TMOD3 | Tropomodulin-3 | 5 | 13 | 6 | 8 |
| IPI00176593.3 | No symbol | Uncharacterized protein ensp00000380341 | 4 | 10 | 8 | 11 |
| IPI00298971.1 | VTN | Vitronectin precursor | 3 | 2 | 1 | 1 |
| IPI00218474.5 | ENO3 | Beta-enolase | 5 | 0 | 3 | 0 |
| IPI00399257.2 | DCHS2 | CDNA flj45804 fis | 2 | 0 | 0 | 0 |
| IPI00027487.3 | CKM | Creatine kinase m-type | 2 | 0 | 3 | 0 |
| IPI00218130.3 | PYGM | Glycogen phosphorylase; muscle form | 3 | 0 | 4 | 0 |
| IPI00024804.3 | ATP2A1 | Serca1b of sarcoplasmic | 2 | 0 | 2 | 0 |
| IPI00000105.4 | MVP | Major vault protein | 4 | 0 | 1 | 0 |
| IPI00001753.1 | MYH4 | Myosin-4 | 8 | 0 | 6 | 0 |
| IPI00383071.1 | TPI1 | Triosephosphate isomerase (fragment) | 2 | 0 | 2 | 0 |
| IPI00183968.4 | TPM3 | Tropomyosin 3 isoform 1 | 3 | 0 | 3 | 0 |
| IPI00023014.1 | VWF | Von willebrand factor | 2 | 0 | 2 | 0 |
| IPI00021536.1 | CALML5 | Calmodulin-like protein 5 | 0 | 1 | 0 | 3 |
| IPI00007425.1 | DSC1 | Desmocollin 1 | 0 | 2 | 0 | 2 |
| IPI00010951.2 | EPPK1 | Epiplakin | 0 | 5 | 0 | 7 |
| IPI00025512.2 | HSPB1 | Heat shock protein beta-1 | 0 | 1 | 0 | 2 |
| IPI00013890.2 | SFN | 14-3-3 protein sigma | 0 | 1 | 0 | 3 |
| IPI00550792.4 | BIN2 | Bridging integrator 2 | 0 | 2 | 0 | 3 |
| IPI00007277.2 | LRRFIP2 | Leucine-rich repeat flightless-interacting protein 2 | 0 | 2 | 0 | 3 |
| IPI00299048.6 | IQGAP2 | ras GTPase-activating-like protein iqgap2 | 0 | 2 | 0 | 3 |
| IPI00071509.1 | PKP1 | Plakophilin-1 | 0 | 3 | 0 | 3 |
| IPI00013933.2 | DSP | DPI of desmoplakin | 0 | 28 | 0 | 24 |
| IPI00045550.4 | PPP1R9B | Protein phosphatise-1; regulatory subunit 9b | 0 | 3 | 0 | 0 |
| IPI00033583.3 | SERPINB12 | Serpin b12 | 0 | 2 | 0 | 3 |
The number of peptides is a semiquantitative measure of protein abundance. This experimental information has been used to create the LRRFIP1 interaction network, as shown in Figure 6B. List of the International Protein Index identifiers (IPI ID) based on the peptides that have been detected by mass spectrometry in the anti-LRRFIP1 pull-downs obtained from platelets.
Gene identifier (ID) is by the HUGO Gene Nomenclature Committee (HGNC). Proteins shown in bold were validated by Western blotting.
Discussion
Platelets are key cells in the processes of hemostasis, atherothrombosis, and wound healing. Platelet response, volume, and count are independent risk factors for MI, particularly in the immediate postevent period, supported by compelling evidence of increased morbidity and mortality in patients undergoing percutaneous intervention with hyperactive platelets, despite antiplatelet therapy.32 The response of platelets to activatory signals from agonists, such as collagen and ADP, ranges widely,1,–3,33 and this variation is, to a large extent, inherited.3 Until recently, the number of robustly validated genetic variants affecting the platelet functional phenotype has remained small, but in the era of large-scale “-omics” strategies, novel genes and variants are emerging.34 A candidate gene study in the PFC identified 16 novel SNPs associated with platelet function,10 and a GWAS in healthy subjects identified 2 additional QTLs for platelet function.35 A separate GWAS in ∼ 5000 healthy volunteers identified 3 QTLs for platelet count and 12 for platelet volume.7,–9 Interestingly, 2 volume QTLs at chromosomes 7q22.3 and 10q21.2-q21.3 also exerted an effect on function, indicating pleiotropic effects of sequence variation on several distinct phenotypic platelet traits.7,8,35 The identification of platelet QTLs leads to new insights in MK and platelet biology and also provides an alternative approach to discover risk genes for MI and CAD.8 However, the platelet QTLs for function, volume, and count discovered so far only explain a fraction of the heritability of these traits.
Although many regulatory proteins of platelet function are known, transcription profiling of platelets,11,–13,36 and their precursor cell, the MK,4,37 identifies more than 5500 transcripts, many of which encode proteins with no known function in platelets.38 We reasoned that profiling of the transcripts of platelets from a substantial number of subjects selected from across the PFC, who represented the full spectrum of the platelet response, could be used to discover novel proteins with functional roles. We applied a stepwise approach to identify, and functionally verify, possible novel regulators of platelet function.
The arraying of the RNA samples of 37 subjects showed that 63 transcripts were correlated with 1 or more of the 4 functional parameters (Figure 3A). Only 2 of the transcripts, encoded by the genes DNM3 and MPL, were uniquely present in MKs and platelets and absent from the other 7 blood cell types. Interestingly, the former gene, which encodes Dynamin 3, a protein that possesses mechanochemical properties involved in actin-membrane processes,39 has also been identified as a QTL for platelet volume.8 MPL encodes the receptor for thrombopoietin (TPO), and activation of platelets with TPO has been shown to modify their function.40,41
To explore the function of some of the genes hitherto unknown to be implicated in platelet function, we initially genotyped 173 SNPs in the 9 most highly correlated of the 63 genes, to identify possible associations with platelet function in the PFC. This identified robust associations for COMMD7 and PFKL (Table 1; Figure 4). Integration of the association results with data from the WTCCC MI/CAD GWAS identified 6 genes for a case-control genotyping study in MI cases and controls (Table 1; supplemental Table 4). This genotyping identified the SNP, rs6141803, in COMMD7 and the nsSNP, rs3739038, in LRRFIP1 as putative novel association signals for MI, with P values 5.3 × 10−6 and 7.9 × 10−4, respectively. The remaining 4 candidate genes (ARPC4, GRB2, GTF2A2, and PFKL) showed no association with MI (supplemental Table 4).
Both COMMD7 and LRRFIP1 are genes that have previously been associated with the regulation of gene transcription, so their novel role in regulating platelet function would not be immediately apparent. COMMD7, like other members of the 10-member family of COMMD genes,42 functions as a repressor of transcription of the NFKB1 gene,42,43 a key transcription factor controlling many proinflammatory genes through binding to RelA (nuclear factor-κB p65).42 Interestingly, the nuclear factor-κB protein has recently been identified as playing a nonclassical role in platelet shape change and thrombus stability.44 Here, we show, in gene-silencing studies in D rerio, that the lack of commd7 results in less robust thrombus formation, but it does not alter early events of cell attachment after vessel wall injury. Whether this effect on platelet function is imprinted during megakaryopoiesis, when the COMMD7 protein could operate at the level of the nucleus by altering the levels of the NFKB1 transcript, or whether the level of cytoplasmic COMMD7 protein alters the function of platelets by an alternative mechanism, requires further study.
LRRFIP1 is also demonstrated to play a role in the regulation of gene transcription. It has been identified as a repressor of the transcription of genes, such as EGFR, PDGFA and TNFA, by binding to their promoter regions.27,28 Silencing of lrrfip1 led to a dramatic reduction in thrombus formation in D rerio, affecting both TTA and TA to a level similar to, or greater than, that observed when silencing the itga2b gene, which encodes the α-chain of the α-IIb-β3 integrin (Figure 5). Although the TA is indicative of the role of each gene played in thrombus formation over time, the TTA is more representative of the initial stages. Our data in the D rerio thrombosis model and the proteomic analysis of the interactome suggest Lrrfip1 could be involved in thrombus formation through its role in cytoskeletal rearrangement. The significantly reduced ability of thrombocytes to support thrombus formation in the morphants is wholly consistent with the observation from the WGE study, in which subjects' LRRFIP1 expression in platelets correlated positively with the level of fibrinogen binding to αIIb-β3 in response to ADP (Figure 3B). This was further corroborated by a significant effect of a relatively rare nsSNP, rs3213869 (MAF 0.0439, encoding a Q251 > R polymorphism in LRRFIP1), on the level of fibrinogen binding to platelets after activation with ADP (Figure 4). The observation that differences in both the level of the LRRFIP1 transcript and a rare amino acid variant in the protein can alter platelet function is compatible with the notion that different types of genetic mechanisms may underlie the observed relationships between sequence variation in genes encoding regulatory nodes in pathways and cellular phenotypes.
The parallel observations in both humans and D rerio strongly suggest that LRRFIP1 may form part of an activatory, or feed-forward, signaling network in platelets, and that the protein may have additional cytoplasmic roles, unrelated to its nuclear role as a repressor of transcription.29,45 Cytoplasmic LRRFIP1 has been shown, in both Drosophila and in yeast-2 hybrid pull-downs, to be associated with FLI1,29,45 which plays a key role in actin severing and capping and cytoskeletal rearrangement. Its cytoplasmic role in both nucleated cells and platelets, however, remains elusive. Here, we provide several lines of evidence strongly supporting cytoplasmic LRRFIP1 as a key component in the cytoskeletal regulation of platelet activation and thrombus formation. First, the protein is present abundantly in the cytoplasm of both megakaryocytic cells and platelets (Figure 6A). The LRRFIP1 interactome, as determined by mass spectrometry analysis of the LRRFIP1 pull-down in resting and TRAP-activated platelets (a strong agonist to maximize cytoskeletal rearrangement), shows a host of cytoskeletal remodeling proteins (Figure 6B; and Table 2). By immunoblotting with specific antibodies, we confirmed the interaction with FLI1 that has already been observed in other species and identified DBN1 as one of its new interactors (Figure 6C). The protein, DBN1, is known to be involved in cytoskeletal rearrangement of actin filaments and is implicated in the stabilization of tight junction formation in neuronal cells,31 although its presence in platelets has not been previously demonstrated. Interestingly, the level of the DBN1 transcript in cultured MKs is rather low (log2 SI, 8.37), compared with platelets (log2 SI, 9.41), suggesting it may be one of a limited set of transcripts and proteins that are enriched in the final stages of MK development and proplatelet formation. Many other LRRFIP1 interactors detected have well-recognized roles in cytoskeletal function, including gelsolin, nexilin, plectin-1, and desmoplakin (DSP).21,22 The composition of the LRRFIP1 interactome changes upon platelet activation with the addition of DSP, LRRFIP2, RAP and 9 other new binding partners, and the loss of 10. The association of LRRFIP1 with LRRFIP2 after platelet activation is worth noting; we carried out extensive analysis of the protein interaction database, IntAct (http://www.ebi.ac.uk/intact/), that established a link in activated platelets with the pivotal WNT effector,46 Dishevelled. As such, LRRFIP1 and LRRFIP2 may modulate canonical WNT signaling, which has recently been identified by us as a novel negative regulator of platelet function.47 The question of whether the same changes would occur upon activation with ADP has not been answered by this study, but taken together, the proteomic analysis of the interactome complements the observation of the reduced thrombus formation in D rerio, suggesting that the loss of the Lrrfip1 protein prevents the cytoskeletal reorganization required for rapid, effective shape change and spreading.
In conclusion, we have examined the effect of the abundance of individual transcripts within the platelet transcriptome on platelet function in a cohort of 500 healthy subjects. Two transcripts, COMMD7 and LRRFIP1, in addition to the correlation between their level and platelet responsiveness, harbored SNPs that showed an association with the platelet response and a modest association with MI. The silencing of both genes significantly modified thrombus formation in D rerio. Taken together, this provides compelling evidence that these 2 proteins are novel regulators of thrombus formation in both fish and man. We postulate that the cytoplasmatically localized forms of both proteins are responsible for the observed effects on platelet function. This assumption is supported by the results of the interactome studies, which are fully compatible with the assumption that in platelets, LRRFIP1 mainly plays a cytoskeletal role. We reason that both proteins may also modify the function of other cells because the genes are ubiquitously transcribed, which may explain the possible association with MI that was observed in the case-control genotyping study. This notional association needs to be confirmed in larger association studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We gratefully acknowledge the generosity of the healthy blood donors and patients with CAD who made their DNA samples available to this project as a “free gift.” We thank the Conway Mass Spectrometry Core Facility, particularly Dr Giuliano Elia and Mr Kieran Wynne, for technical assistance. P.B. was a PhD candidate at Cambridge University, and this work was submitted in partial fulfillment of the requirement for the PhD.
The Bloodomics project was funded by the 6th Framework Program of the European Union (LSHM-CT-2004-503485). The project made use of volunteer nonremunerated NHSBT donors from the Cambridge BioResource. This local resource for genotype-phenotype association studies was supported by a grant from the National Institute for Health Research to the Cambridge Biomedical Research Center. N.W., S.F.G., and W.H.O. and A.G. and N.S. received National Institute for Health Research Program and Unit grant funding, respectively.
Authorship
Contribution: A.H.G. designed the experiment and wrote the manuscript; P.B. performed and analyzed WGE studies; I.S. performed D rerio studies; I.C.M. performed proteomics studies; C.I.J. performed the function studies in the PFC; D.A. contributed DNA samples from MI cases and related clinical information; B.d.B. contributed to bioinformatical analysis; S.L.B. performed the statistical analysis of the case-control genotyping and D rerio studies; H.D. contributed to the design of the experiment; F.D. was responsible for the design of analysis plans and oversight of statistical analysis; D.J.F. was responsible for oversight of proteomics studies; S.F.G. performed the function studies in the PFC; A.G. performed statistical analysis of WGE studies; K.K. coordinated the Bloodomics project; C.L. had oversight of WGE studies; M.N.O. performed D rerio studies; C.M.R. was responsible for bioinformatics; D.S. had oversight of D rerio studies; J.S. was responsible for the Bloodomics DNA repository; M.D.T. contributed DNA samples from MI cases and related clinical information, and integrated clinical information from all clinical MI collections; J.-J.Z. contributed DNA samples from controls and related clinical information; N.J.S. contributed DNA samples from MI cases and related clinical information; N.A.W. had oversight of WGE studies; P.B.M. had oversight of proteomics and proteomics data analysis; and W.H.O. designed the experiment and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alison H. Goodall, Department of Cardiovascular Sciences, University of Leicester, Clinical Sciences Wing, Glenfield Hospital, Leicester, LE3 9QP, United Kingdom; e-mail: ahg5@le.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal