Abstract
Abstract 784
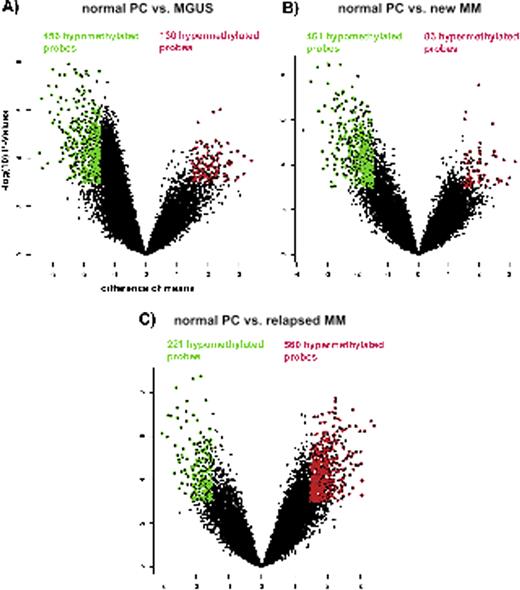
Gene expression is a tightly regulated process and is influenced by aberrant epigenetic changes that can lead to carcinogenesis. We used the HELP (HpaII tiny fragment Enrichment by Ligation-mediated PCR) assay to perform an unbiased genome-wide analysis of DNA methylation in 11 MGUS, 16 newly diagnosed myeloma, 17 relapsed myeloma, and 8 healthy control samples. The HELP assay uses differential methylation specific restriction digestion by HpaII and MspI followed by amplification, 2 color labeling, and co-hybridization to quantitatively determine individual promoter methylation of 25,626 loci. The methylome analysis was performed using CD 138+ sorted bone marrow plasma cells in all cases. We observed extensive DNA methylation changes in myeloma that were seen even in MGUS cases when compared to normal plasma cells. Unsupervised clustering of all samples showed that MGUS samples were distinct but epigenetically closer to normal plasma cells than to cases of newly diagnosed or relapsed Myeloma. MGUS cases were characterized predominantly by aberrant hypomethylation, with 456 hypomethylated probes versus 130 hypermethylated probes (cutoff criteria were defined as a difference of means > 1.5 and significance of this difference with p < 0.001, Figure 1) that affected pathways regulated by NF-kb transcription factor. Untreated newly diagnosed myeloma samples were also predominantly hypomethylated (461 hypomethylated probes, 83 hypermethylated probes) when compared to controls. In addition to NF-kB, the MAP kinase and PI3 kinase regulated pathways were affected by hypomethylated genes. In contrast, cases of relapsed myeloma showed predominantly hypermethylated loci (221 hypomethylated, 560 hypermethylated probes) when compared to controls and involved the TNF and retinoblastoma pathways. A large number of important genes including the tumor suppressors CDKN2A and CDKN2B were aberrantly hypermethylated in this cohort.
Volcano plot of differentially methylated genes, plotting difference of means against the −log(10) of the respective p-value. The cutoff for the definition for significant differentially methylated probes was set at a difference of means >1.5 or < −1.5 and a p-value < 0.001. Hypomethylated probes are depicted in green, hypermethylated probes in red. The respective numbers are documented in the same color. A) comparison of normal plasma cells with MGUS. B) comparison of normal plasma cells with newly diagnosed MM. C) comparison of normal plasma cell with relapsed myeloma
Volcano plot of differentially methylated genes, plotting difference of means against the −log(10) of the respective p-value. The cutoff for the definition for significant differentially methylated probes was set at a difference of means >1.5 or < −1.5 and a p-value < 0.001. Hypomethylated probes are depicted in green, hypermethylated probes in red. The respective numbers are documented in the same color. A) comparison of normal plasma cells with MGUS. B) comparison of normal plasma cells with newly diagnosed MM. C) comparison of normal plasma cell with relapsed myeloma
Analysis of our differentially methylated targets using the Molecular Signatures Database (MSigDB, Tamayo, et al. 2005, PNAS 102, 15545–15550), showed significant overlap for hypomethylated genes in MGUS and new multiple myeloma (MM). These gene sets contain genes with promoter regions around transcription start site containing motifs which match annotation for transcription factors SP1 and TCF3 as well as enrichment for genes with gene ontology annotations for plasma membrane proteins. Hypermethylated genes in new MM show significant overlap with genes in the neighborhood of RAD23A, CTBP1, G22P1 and SMC1, suggesting impairment of DNA repair, as well as enrichment for genes associated with apoptosis and programmed cell death.
In conclusion, genome-wide DNA methylation analysis is able to clearly differentiate between normal bone marrow plasma cells, MGUS as well as new MM and relapsed MM cells. Correlation of significantly differentially methylated genes with published gene sets reveals enrichment for genes involved in DNA repair, cell-cell signaling, cell death, apoptosis and cell cycle regulation.
Mehta:Celgene: Consultancy, Speakers Bureau; Takeda/Millennium: Speakers Bureau; Onyx: Research Funding. Singhal:Takeda/Millennium: Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Onyx: Research Funding; Celgene: Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal