Abstract
Abstract 4611
Chronic lymphocytic leukemia (B-CLL) is the most common adult leukemia in Western countries, and is characterized by a highly variable clinical course. Interphase fluorescent in situ hybridization (I-FISH) has been able to identify chromosomal abnormalities in ~80% of B-CLL, including deletions at 13q, 11q, 17p and trisomy 12, which has proven to be prognostic indicators for disease progression and survival. Although recent immunostimulatory methods have substantially improved analysis via conventional metaphase cytogenetics (CC), detection of chromosome changes is limited by the low mitotic activity of CLL cells in vitro. High-density single nucleotide polymorphism (SNP) arrays are commercially available technologies, which allow genome-wide detection of allelic copy number gains or losses, and loss-of-heterozygosity (LOH) regions.
To assess whether SNP-arrays are more sensitive than I-FISH and CC to detect specific chromosomal abnormalities associated with prognosis in B-CLL, i.e. del13q, del11q, del17p and tri12.
Blood samples from 24 patients with B-CLL at diagnosis were tested in parallel by I-FISH, CC and SNP-array. FISH: blood smear samples were hybridized with 4 probes, in order to detect deletions at 17p13.1, 11q22.3, 13q14.3, and trisomy 12 [respectively LSI p53, ATM, D13S319 and centromeric CEP12 probes, Abbott]. In normal lymphocytes, an average of 6.7% nuclei showed one signal (truncated nuclei), and we defined the cut-off level for detection of a deletion at 11% (mean+3SD). The cut-off for detection of tri12 was defined at 5% (mean+3SD). CC: blood lymphocytes are cultivated for 72 hours with immunostimulants (DSP30 and IL-2) and metaphases analyzed according to standard procedures. SNP-arrays: DNA samples (200 ng) were hybridized on the Illumina HumanCNV370-quad v3 BeadChips, which assess 373,397 markers with a median marker spacing of 4.9 kb (mean 7.8 kb). The I-Scan system was used to scan the BeadChips (primary data). GenomeStudio 1010 v1 and CNVPartition 2.4.4 package were used to process primary data and identify chromosomal deletions/amplifications and LOH regions.
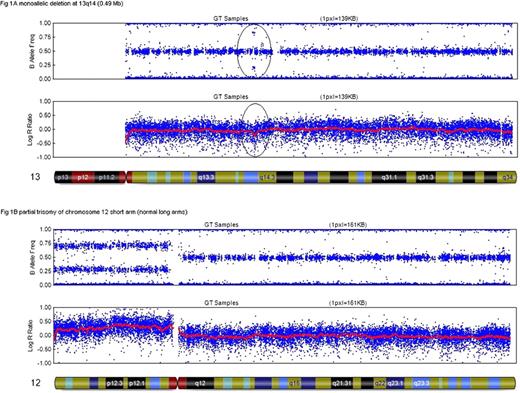
I-FISH identified deletions at 13q (15-95% of nuclei; mean=51%), 11q (35-54%; mean=43%) and trisomy 12 (32-49%; mean=37%) in 17 (71%) [including 2 cases of biallelic deletions], 3 (12.5%) and 4 (17%) cases, respectively. No del17p was detected. Five B-CLL cases presented associated FISH abnormalities. SNP-arrays identified all changes (100%) detected by I-FISH (del13q, median size: 13 Mb – range, 0.49–50 Mb; del11q, median size: 39 Mb – range, 34.8–42 Mb]. However, SNP-arrays showed 5 additional deletions at 13q14.3. Three patients had cryptic deletions (~52 to 82 kb), not detected by the FISH LSI D13S319 probe (~130 kb) or CC, and two others had large deletions (1.1 and 33.3 Mb) but with one signal below the cut-off of 11% by I-FISH (therefore considered as negative- both had normal CC). In these two cases, tumor cell enrichment before I-FISH allowed the detection of the deletion. In one case with del13q and tri12 (30% of nuclei by FISH), an additional del11q was also detected by SNP-array and CC (in 5/24 mitoses). The size of this deletion was 37.8 Mb and involved the cytogenetic bands 11q14.1 to 11q23.2 (including ATM). In addition, SNP-arrays enabled to define more precisely the size and location of the abnormalities. For instance, in one case with tri12 (as identified by the centromeric FISH probe), a partial trisomy of chromosome 12 short arm was indeed detected by SNP-array. No 17p abnormality was detected by SNP-array (either deletion or LOH). Only one of the 24 samples had no abnormality by SNP, I-FISH or CC for the loci studied.
Despites using a relatively low density SNP-array (~370,000 markers), a higher sensitivity of Illumina BeadChips was observed. “SNP+/FISH negative” cases can be explained by either cryptic deletions or low leukemic cell content (< cut-off level). “SNP+/CC negative” cases can be explained by either a higher resolution, or the presence of normal mitosis in excess following stimulation. Higher density SNP-arrays (> 5 million markers) may be used to improve the assay sensitivity, especially regarding del13q14 which seems to be present in a great majority of our cases. Our study confirms usefulness of SNP-arrays for prognostic evaluation of B-CLL patients at diagnosis, and suggests the use of this assay as a routine procedure.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal