Abstract
Abstract 3402
We discovered a novel BCR-ABL1 mutation (V304D) in pts with CML failing imatinib by DNA expansion of specific clones followed by DNA sequencing of ≥10 clones. BCR-ABL1V304D was detected in a median of 37% (range, 20% to 80%) resistant clones from 13 (18%) of 70 imatinib-resistant pts with CML in chronic phase (CP). Pts received imatinib for a median of 35 months (range 2 to 66) at doses ≥600 mg/d. No pt achieved a cytogenetic response. Four received nilotinib: 3 had hematologic resistance and 1 progressed to blast phase (BP). All pts died with negligible response to second-line TKIs: 8/12 pts on dasatinib had disease progression and 4 responded (2 hematologic and 2 transient minor cytogenetic responses). BCR-ABL1V304D failed to induce cytokine-independence or activate Stat5 in Ba/F3 cells. Phosphorylation of CrkL and specific BCR-ABL1 substrates were detectable but diminished compared to unmutated BCR-ABL1-transduced cells. BCR-ABL1V304D failed to catalyze autophosphorylation and the catalytic domain of ABL1V304D demonstrated deficient kinase activity. Enforced expression of BCR-ABL1V304Din CML cells induced quiescence and protection from imatinib-induced apoptosis. In vitro analyses of cells from a pt in CP expressing BCR-ABL1V304D in 50% of clones failed to detect CrKL phosphorylation in the presence of normal BCR-ABL1 protein levels, suggesting that BCR-ABL1V304D encodes a kinase-deficient protein and is associated with remarkable TKI resistance and extremely poor prognosis.
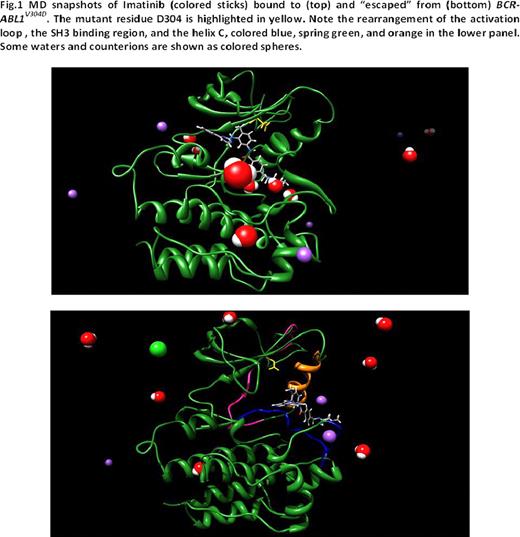
To determine the mechanism of resistance imposed by BCR-ABL1V304D, we modeled this mutation in water and counterions and compared it to unmutated and mutant BCR-ABL1 isoforms. We first correlated the free energy of binding (DGbind) to the corresponding IC50 (DGbind = -RT lnIC50) and calculated the difference in free energy of binding between wild-type and mutant kinases (DDGbind = DGbind(WT) – DGbind(MUT)). DGbind <0 indicates a tighter binding to a TKI of the unmutated kinase relative to the mutant kinase. A negative increase of 1.4 kcal/mol in DGbind corresponds to a decrease by a factor of 10 in the IC50 value. The DGbind (IC50) values of imatinib for wild-type, Y253H, and T315I kinases were -10.47kcal/mol (21nM), -7.45 kcal/mol (3.4mM), and -6.38 kcal/mol (21mM), similar to published experimental data (25nM, 1.8–3.9mM, and >10mM, respectively), thus validating our modeling. DGbind and IC50 values for imatinib and dasatinib against V304D are -9.86kcal/mol (59nM) and -12.27 kcal/mol (1.02nM), respectively. 3D images generated from an equilibrated frame of 10 ns molecular dynamics (MD) simulations demonstrated that the 304 position is not in direct contact with imatinib, nor does it directly alter imatinib binding. Rather, V304D disturbs the position of the regulatory αC helix (Figure1). Longer standard molecular dynamics simulations coupled with steered MD recipes indicate that V304D induces a rearrangement of the ATP/drug binding pocket and water-mediated disruption of some fundamental hydrogen bonds regulating the transition of the activation loop to a “semi-open” conformation and the apt overall conformation of the SH3-binding segment of the TK (residues K294-F311). Furthermore, a decrease in the number of total interactions causes unidirectional drug translation toward the binding site exit. Iterative simulations revealed significant ATP/inhibitor diversion with subsequent complete imatinib expulsion. Thus, the V304D-induced semi-opened conformation of the activation loop favors 1) the lateral escape of imatinib, thus increasing the rate of TKI dissociating from the kinase and 2) does not allow the passage of ATP to reach deep into the binding pocket, thus hampering tyrosine phosphorylation. A similar phenomenon is observed in the activation loop in the active conformation of the V304D kinase bound to dasatinib, which results in greater exposure to water solvent of a part of the binding site and almost complete loss of hydrophobic contacts in the opposite end of the binding site.
MD snapshots of Imatinib (colored sticks) bound to (top) and “escaped” from (bottom) SCTABLIV304D. The mutant residue D304 is highlighted in yellow. Note the rearrangement of the activation loop, the SH3 binding region, and the helix C, colored blue, spring green, and orange in the lower panel. Some waters and counterions are shown as colored spheres.
MD snapshots of Imatinib (colored sticks) bound to (top) and “escaped” from (bottom) SCTABLIV304D. The mutant residue D304 is highlighted in yellow. Note the rearrangement of the activation loop, the SH3 binding region, and the helix C, colored blue, spring green, and orange in the lower panel. Some waters and counterions are shown as colored spheres.
In summary, BCR-ABL1V304D results in kinase inactivation, pan-TKI resistance mediated by a novel mechanism of lateral escape at the kinase domain, less control of protein autoinhibition via perturbation of the SH3 binding domain and very poor prognosis. Complete modeling data against a panel of novel TKIs and potential modes of overcoming this novel mechanism of resistance will be presented.
Kantarjian:Bristol Myers Squibb: Research Funding; ARIAD: Research Funding; Nerviano: Research Funding. Cortes:Bristol Myers Squibb: Research Funding; ARIAD: Research Funding; Nerviano: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal