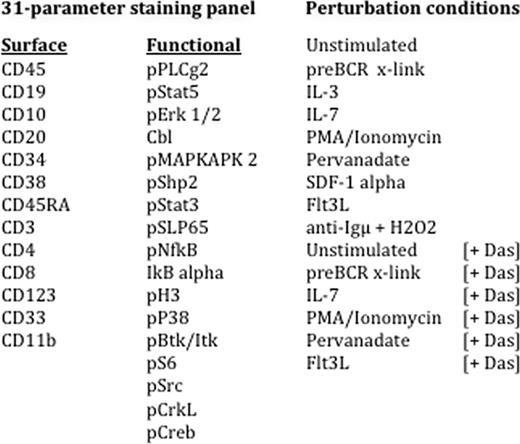
Abstract
Abstract 2761
High-risk pediatric B precursor lymphoblastic leukemia (preB ALL) remains a serious cause of morbidity and mortality despite therapeutic advances with consequent improvements in survival over the past 30 years. Intracellular protein networks are the functional mediators of differentiation decisions. Perturbing these networks with chemical and biological agents to unveil their signaling potential has shed insight into key networks in other hematologic malignancies such as B cell lymphoma, AML, JMML, and myelofibrosis. The current study was designed to profile evoked signaling responses in NCI high-risk pediatric preB ALL using a next-generation 'mass cytometry' platform (CyTOF.., DVS Sciences). This new technology platform utilizes antibodies conjugated to chelated metal ion tags, and thereby circumvents the limitations of overlapping spectral emission associated with more conventional fluorochrome-conjugated antibodies. Thus, in this preliminary study we measured 13 surface markers and 18 functional intracellular proteins simultaneously at the single-cell level in primary patient samples, with the goal of correlating distinct signaling responses with surface marker expression and response to the Src family tyrosine kinase inhibitor, dasatinib.
All procedures were approved by the Stanford University Human Subject Institutional Review Board. Signaling profiles were measured for cryopreserved diagnostic bone marrow samples from pediatric patients with NCI high-risk pediatric leukemia in the basal state and in response to a 15 minute treatment to 18 different extracellular modulators, with or without prior exposure to dasatinib for 30 minutes. An example is shown in Figure 1. Cell populations were gated based on surface marker expression. The activation states of 18 functional intracellular markers were measured within the leukemic blasts (Figure 1).
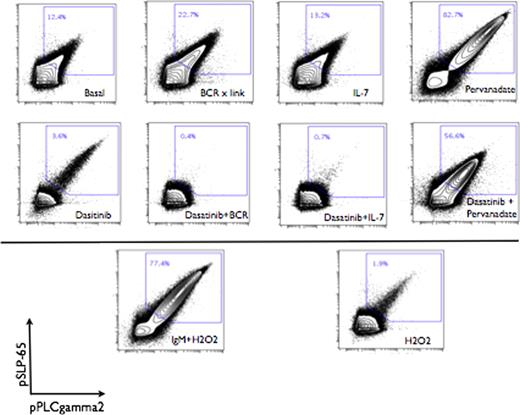
Within the bulk blast population, distinct signaling responses could be discerned correlating with surface marker heterogeneity. Blasts were manually gated into 5 distinct populations based on surface expression of CD45 and CD10. These populations demonstrated further heterogeneous expression of surface markers CD34, CD38 and CD19 as well as heterogeneous activation states of intracellular proteins such as pERK, NFkB, and pStat5. Robust activation of phospho-proteins proximal to the preB cell receptor was observed following treatment with preBCR cross-linking antibodies or with tyrosine phosphatase inhibitors. This robust signaling was abrogated by pre-treatment of leukemic blasts with dasatinib. Unexpectedly, concurrent treatment with IL-7 or preB cell receptor crosslinking antibodies and dasatinib sensitized cells to dasatinib inhibition, resulting in a synergistic suppression of phosphorylation below baseline values (Figure 2).
Together, these results suggest that intracellular signaling and response to therapeutic agents such as kinase inhibitors may differ based on the complex interaction of the leukemic cell and its microenvironment. This preliminary study shows the feasibility of using a novel mass cytometry platform to simultaneously monitor 18 intracellular signaling mediators on a per cell basis in primary samples from pediatric high risk ALL patients to a level of detail not previously possible. This approach allowed the identification of cells with a robust signaling phenotype, based on the activation of pERK, pStat5 and NFkB, members of pathways known to be involved proliferation and survival. These data identify potential targets for therapeutic agents such as the tyrosine kinase inhibitor dasatinib. inhibited preBCR signal transduction, and synergy with other biological agents. Further studies are ongoing to measure multidimensional signaling with CyTOF technology in additional high risk pediatric ALL samples with the goal of identifying and guiding the most beneficial therapeutic choices.
EFS and KLD contributed equally to this work.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal