Abstract
Abstract 275
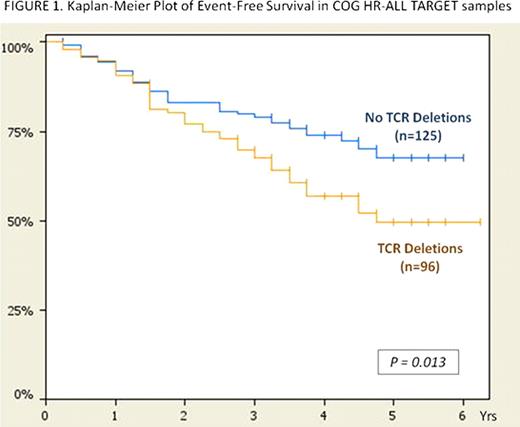
T-cell receptor (TCR) gene rearrangements occur in 50–80% of childhood precursor B-cell ALL and have been used for minimal residual disease (MRD) detection. However, TCR rearrangement has never been examined for its contribution to disease classification and outcome. We used Molecular Inversion Probe technology (Affymetrix) to analyze precursor B-cell ALL samples (n=48) from the University of Utah for genome-wide copy number changes, and evaluated the relationship between focal TCR deletions (indicating rearrangement) and clinical features. Copy number deletions were detected by Nexus Copy Number (BioDiscovery, Inc.) and were based on 5 consecutive probes having a value of <1.5 (diploid copy number=2). We then validated our findings in the diagnostic B-precursor ALL samples (n=221) from the Children's Oncology Group (COG) High Risk (HR)-ALL TARGET (Therapeutically Applicable Research to Generate Effective Targets) Project which includes 500K SNP array (Affymetrix) data from patients enrolled on COG P9906. In the Utah cohort, we observed the following TCR deletions: TCR-α (14q11.2)=23/48 (48%), TCR-β (7q34)=18/48 (38%), TCR-γ (7p14.1)=29/48 (60%), TCR-δ (14q11.2)=23/48 (48%), or any TCR deletion (TCR-‘any')=36/48 (75%). Utah samples contained 37.5% NCI-Rome designated “High Risk” patients, and 15/18 (83%) of these patients had TCR-‘any' deletions. Fisher's Exact Test (two-tailed) was used to look for associations in the Utah cohort between TCR-α,-β,-γ,-δ, or -‘any' deletions and high risk clinical features. TCR-α deletion was significantly associated with Day 28 MRD Positivity (p=0.0486); there was significant association between TCR-β deletion and Day 7 M3 Marrow (>25% blasts; p=0.019). TCR-δ deletion was also significantly associated with Day 28 MRD Positivity (p=0.0206). Due to the small sample size and insufficient follow-up, outcome analyses were not done in this cohort. The COG HR-ALL TARGET cohort had the following TCR deletions: TCR-α=53/221 (24%), TCR-β=24/221 (11%), TCR-γ=55/221 (25%), TCR-δ=53/221 (24%), or TCR-‘any'=96/221 (43%). Fisher's Exact test (two-tailed) was used to look for associations between TCR deletions and clinical features; outcome analyses were conducted using the Kaplan-Meier method and the log-rank test. In the TARGET cohort, we found TCR-α, TCR-γ, and TCR-δ deletions were significantly associated with occurrence of relapse and/or death (p=0.0209, p=0.0143, and p=0.0209, respectively). TCR-β deletion was associated with age at diagnosis ≥ 10 yo (p=0.0225), a high risk feature, and Day 29 MRD Positivity (p=0.0084). In the high-risk TARGET validation cohort, 43 of the 78 children (55%) who experienced a relapse or died contained TCR deletions vs. 53 of the 143 children (37%) who were event-free with TCR deletions (p=0.0109). There was a significant difference in event-free survival (EFS) between patients with TCR-‘any' deletions vs. those without the deletions (p=0.013), with the largest difference in survival occurring beyond two years of diagnosis (Figure 1). TCR deletions were uncommon in Hyperdiploid (0/4, 0%), E2A-PBX1 (n=1/25, 4%), and MLL (n=4/19, 21%) subtypes in the TARGET cohort, but were more frequent in TEL-AML1 (n=2/3, 67%) and Other (n=89/170, 52%) subtypes. In summary, both the Utah discovery cohort and the larger COG HR-ALL TARGET validation cohort showed focal TCR gene deletions that were linked to high risk clinical features, as well as associated with poor prognostic attributes such as end-of-induction MRD Positivity. Additionally, the larger TARGET cohort demonstrated that deletion of any TCR region was significantly associated with worse outcome. TCR gene deletions may indicate a more immature state of developmental arrest and could imply initial abortive T-cell lineage development prior to B-cell commitment and transformation. This represents the first study to reveal an association between TCR gene deletion status and clinical outcome in childhood B-precursor ALL, and these findings will need validation in additional pediatric and adult cohorts. Ultimately, focal TCR gene deletions may prove to be a valuable addition to the molecular risk classification schema currently applied to B-precursor ALL.
Disclosures:
No relevant conflicts of interest to declare.
Author notes
*
Asterisk with author names denotes non-ASH members.
© 2010 by The American Society of Hematology
2010

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal