Abstract
Abstract 2269
Point mutations in the BCR-ABL tyrosine kinase domain (TKD) are an important mechanism of resistance to treatment with tyrosine kinase inhibitors (TKIs). However, detection of mutant subclones may be difficult to interpret with regard to the risk of imminent onset of resistant disease. The biological relevance of several mutations is not well understood, and some may indeed be biologically neutral (bystander mutations). Moreover, even subclones carrying mutations known to confer resistant disease (driver mutations) may disappear below the detection limit without any alteration of treatment. Surveillance of the size of mutant subclones during treatment could therefore provide better understanding of their biological behavior in vivo, and facilitate timely assessment of impending resistant disease.
To address this issue, we have established a ligase-dependent (LD) PCR technique permitting quantitative monitoring of point-mutated subclones (Preuner et al. Leukemia 2008). The LD-PCR approach displays a detection limit ≥1%, and permits reproducible assessment of changes in the size of mutant subclones exceeding ±5%. We have investigated a series of prospectively collected PB specimens derived from CML patients displaying a variety of mutations including M244V, L248V, G250E, Q252H, E255K, T315I, F317L-A/G, M351T, and F359V with the aim to monitor the proliferation kinetics of mutant leukemic cells during the course of treatment.
Our observations revealed that a) the appearance of new mutant subclones at a level detectable by LD-PCR occurred as early as a few weeks after initiation of treatment with a TKI, b) the time span until mutant leukemic cells became the dominant BCR-ABL positive clone was variable, ranging between 1–18 months following first detection, c) different mutant subclones appeared sequentially in individual patients upon changes of treatment (Fig.), d) disappearance of a dominant subclone carrying the highly resistant mutation T315I could be documented, e) the response of mutant subclones to treatment and the rise of new mutant subclones could be readily documented by the quantitative LD-PCR approach, f) changes in total BCR-ABL transcripts assessed by RQ-PCR mostly occurred in parallel to corresponding kinetics of mutant subclones, but expanding mutant clones could also be observed at a time when BCR-ABL transcripts were still decreasing, and g) documentation of expanding mutant subclones occasionally preceded the rise in BCR-ABL transcripts by several weeks. Quantitative mutational screening revealed the appearance and expansion of expected mutations, such as G250E, M244V during treatment with imatinib, the F317L on dasatinib or E255K on nilotinib, but also unexpected findings such as selection and outgrowth of a subclone carrying the L248V mutation upon onset of nilotinib.
Our observations indicate that quantitative monitoring of mutant subclones during treatment with TKIs provides information on their responsiveness to therapy and the imminent onset of resistant disease. Judicious implementation of quantitative diagnostic approaches such as the LD-PCR technique in the surveillance of CML patients can improve our current options for timely treatment decisions, and could help optimizing disease management in patients displaying point mutations in the BCR-ABL TKD or other sites of potential relevance.
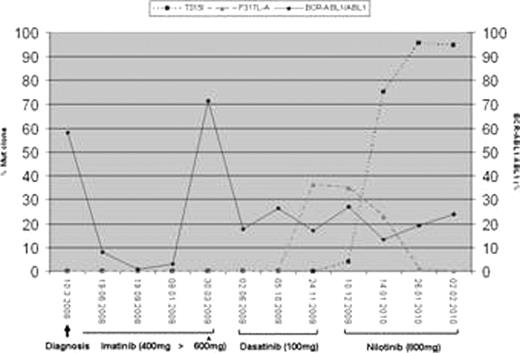
After inadequate molecular response to different doses of imatinib (solid line=BCR-ABL1IS transcript levels),a subclone carrying a F317L-A mutation (dashed line) appeared after onset of dasatinib. This subclone disappeared upon switch to nilotinib, but concomitantly, a subclone carrying the T315I mutation (dotted line) evolved rapidly and became the dominant BCR-ABL positive clone.
Example of consecutive occurrence of mutant subclones during treatment with TK inhibitors.
Example of consecutive occurrence of mutant subclones during treatment with TK inhibitors.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal