Abstract
Although bone marrow (BM) is the main site of natural killer (NK)–cell development in adult mice, recent studies have identified a distinct thymic-dependent NK pathway, implicating a possible close link between NK- and T-cell development in adult hematopoiesis. To investigate whether a potential NK-/T-lineage restriction of multipotent progenitors might take place already in the BM, we tested the full lineage potentials of NK-cell progenitors in adult BM. Notably, although Lin−CD122+NK1.1−DX5− NK-cell progenitors failed to commit to the B and myeloid lineages, they sustained a combined NK- and T-cell potential in vivo and in vitro at the single-cell level. Whereas T-cell development from NK/T progenitors is Notch-dependent, their contribution to thymic and BM NK cells remains Notch-independent. These findings demonstrate the existence of bipotent NK-/T-cell progenitors in adult BM.
Introduction
All blood cell lineages are derived from hematopoietic stem cells (HSCs) that reside in the adult bone marrow (BM).1 Lineage commitment, or restriction, is the process by which a multipotent stem or progenitor cell becomes increasingly restricted in its lineage potentials, to ultimately develop into a fully committed progenitor of a single-cell lineage. The characterization of lineage-restricted progenitors represents an important step toward a better understanding of the cellular pathways and molecular regulation of lineage commitment of the different hematopoietic lineages. Decades of extensive research have provided solid evidence that the lineage restriction process from multipotent HSCs to lineage-committed progenitors occurs as a stepwise process; however, the cellular pathways by which lymphoid lineage restriction occurs remain the subject of debate and continued investigation.1-4 This is of particular interest for the T-cell lineages, in which it remains unclear which stages of lineage restriction occur in the BM and subsequently in the thymus.5-7
The recently identified lymphoid primed multipotent progenitor (LMPP), which lacks erythroid/megakaryocyte potentials but maintains combined lymphoid and granulocyte-monocyte potentials,8 and the common lymphoid progenitor9 represent lymphomyeloid and lymphoid-restricted candidate thymus seeding progenitors in adult BM. Both of these, however, are shared progenitors between the T- and B-cell lineages, of which the B-cell commitment is finalized in the BM, whereas the final stages of T-cell commitment are thought to take place predominantly or exclusively in the thymus.5,7,10,11 Thus, an important question is whether in adult BM also occurs lineage restriction toward bipotent progenitors with combined lineage potentials for T cells and other lineages thought to be derived in part in the thymus.
Natural killer (NK) cells, large granular lymphocytes, a lymphoid lineage distinct from B and T cells, are major components of innate immunity and play an important role in eliminating tumor cells as well as in the defense against viral infections.12,13 Despite significant progress in understanding NK-cell biology, the early stages of NK-lineage commitment from multipotent progenitors remain unclear. In adult mice and humans, BM is the main site of NK-cell development14-16 ; however, recent studies have demonstrated the existence of another distinct thymic-dependent adult NK-cell developmental pathway.17 The finding that in adult mice T cells as well as a fraction of NK cells are generated in the thymus implicates a possible close link between T- and NK-cell development; however, the source of thymic NK cells remains unknown.17
Studies supporting the existence of bipotent NK/T progenitors during early development support the prethymic restriction of thymus seeding progenitors during development, but the existence and location of NK/T lineage-restricted progenitors as a lineage commitment step in adult hematopoiesis have yet to be identified.18-20
A candidate NK lineage-restricted progenitor (NKP) has been identified in adult mouse BM as having a Lin−CD122+NK1.1−DX5− phenotype,21 although the lineage potentials of this NKP progenitor have not been extensively studied and its in vivo lineage-reconstituting potential has yet to be evaluated. Thus, we further investigated whether the Lin−CD122+NK1.1−DX5− NKPs in adult mouse BM are truly NK cell–restricted or whether they maintain other blood cell lineage potentials. Herein, we show that, although Lin−CD122+NK1.1−DX5− cells lack B and myeloid cell potential, they sustain a combined NK- and T-cell potential in vivo and in vitro at the single-cell level. These BM bipotent NK-/T-cell progenitors can generate both thymic and BM-dependent NK cells, through Notch-independent mechanisms, whereas their T-cell generation is strictly Notch-dependent. Taken together, our data demonstrate the existence of bipotent NK/T progenitors in adult BM providing direct evidence for NK/T lineage restriction taking place prethymically in adult BM.
Methods
Mouse strains
C57BL/6 mice were used at 6 to 10 weeks of age for isolation of thymic progenitors, and 10- to 12-week-old mice were used for purification of BM and thymic progenitors. Mice deficient in Il2rg expression22,23 purchased from The Jackson Laboratory were used as recipients in transplantation experiments. Genotyping was performed by polymerase chain reaction (PCR) on genomic DNA derived from the tails,22 and C57BL/6 mice were used as wild-type (WT) controls and sorting in all experiments. All mice were maintained under specific pathogen–free conditions at the Lund University Animal Facility. The Ethical Committee at Lund University approved all performed experiments. Tissues were collected as previously described.24,25
Monoclonal antibodies, flow cytometry, and cell sorting
Before staining single-cell suspensions with antibodies against specific surface markers, Fc receptors were blocked by incubation with 2.4G2 (anti-FcRIII). All antibodies were from BD Biosciences unless otherwise indicated. Monoclonal antibodies (conjugated with different fluorochromes) used to stain cell-surface antigens were: B220 (RA3-6B2), CD19 (1D3), KIT (2B8), SCA-1 (E13-161.7), FLT3 (AZF10.1), T-cell receptor-β (TCR-β; H57-597), CD11c (N418), CD3 (17A2), CD122 (TM-β1), CD49b (DX5), NK1.1 (PK136), CD11b/Mac-1 (M1/70), CD127 (A7R34), Ly49D (4E5), CD25 (3C7, 7D4), Ter119 (LY-76), Gr-1 (RB6-8C5), CD45.1 (A20), CD45.2 (104), CD4 (H129.19), CD8-α (53-6.7), Thy1.2 (53-2.1), TCR-δγ (GL3), CD27 (LG.7F9), CD94 (18d3), NKp46 (29A1.4), and Ly49mix included Ly49G2 (AT8) plus Ly49C/I/F/H (14B11) plus Ly49A (A1). Isotype-matched controls labeled with appropriate fluorochromes were used. Biotinylated antibodies were visualized by streptavidin-phycoerythrin or streptavidin-phycoerythrin Cy7 and purified antibodies by polyclonal goat antirat-Tricolor (both from Caltag). 7-Amino-actinomycin D (Sigma-Aldrich), TO-PRO-1 iodide, 1mM (Invitrogen), and propidium iodide (1.0 mg/mL; Invitrogen) were used to exclude dead cells from the analysis. For the evaluation of NKP and NK-cell compartments in BM, the mixture of the following antibodies was used to define the lineage-negative (Lin−) population: anti-CD19, Gr-1, Mac-1, CD3, CD4, CD8, and Ter119. Samples were analyzed on LSR II (BD Biosciences), between 50 000 and 500 000 events were collected, and analysis was performed using FlowJo software (Version 8.8.6; TreeStar).
For sorting the Lin−CD122+NK1.1−DX5− NK progenitors, BM cells were first enriched for CD122+ cells by incubation with anti-CD122 biotinylated antibody, antibiotin Micro beads (Miltenyi Biotec), and then positive selection was done on 25 LS MACS column (Miltenyi Biotec). The enriched CD122+ cells were next stained with directly conjugated antibodies against lineage markers: anti-CD19, Gr-1, Mac-1, CD3, CD4, CD8, and Ter119 as well as anti-NK1.1 and DX5.
The sorting of the Lin−KIThighCD25− thymic progenitors was performed as follows: thymocyte single-cell suspension was first incubated with low concentration of purified anti-CD4 and CD8-α antibodies, and double-negative (DN) cells were isolated on a magnetic particle concentrator (MPC-6; Dynal) after incubation with antibodies and subsequent incubation with sheep anti–rat IgG (Fc)–conjugated immunomagnetic beads (Dynal). The DN cells were then stained with goat F(ab)2 antirat Ig(H+L) tricolor conjugate (Caltag) to visualize remaining positive cells and directly conjugated antibodies against lineage markers: CD19, Gr-1, Mac-1, TCR-β, CD8-α, Ter119, NK1.1, CD11c, as well as CD25 and KIT.
Transplantation assay
Sublethally irradiated (350 cGy) 10- to 12-week-old CD45.2 Il2rg−/− recipient mice were transplanted with 1300 NKPs sorted from 10- to 12-week-old WT CD45.1 donors.
At 14, 21, and 28 days after transplantation, recipient mice were analyzed for mutilineage donor-derived reconstitution by fluorescence-activated cell sorter (FACS). The peripheral blood (PB), spleen, and BM cells were stained with antibodies against CD45.1, CD45.2 (to distinguish donor and recipient); Mac-1, Gr-1 (myeloid cells, also defined as negative for NK1.1); CD19 (B cells); CD3, CD4, and CD8-α (T cells); and NK1.1 and DX5 (NK cells).
OP9 and OP9-DL1 cultures
The OP9 and OP9-DL1 cells were kindly provided by J. C. Zúñiga-Pflücker (Department of Immunology, University of Toronto, Sunnybrook Research Institute, Toronto, ON) and cultures were carried out as previously described.8,27 The 20, 10, 5, or single-sorted NKP and DN1 cells were plated on previously established, approximately 80% confluent stroma cell monolayers in 48-well plates in 1 mL of Opti MEM plus Gluta Max 1 time medium (Invitrogen) containing 10% fetal calf serum (FCS; Sigma-Aldrich), 1% penicillin/streptomycin (Sigma-Aldrich), 1% 10−2M 2-mercaptoethanol (Sigma-Aldrich) supplemented with cytokines with final concentrations: FLT3 ligand (FL, 25 ng/mL), interleukin-7 (IL-7, 20 ng/mL), KIT ligand (KL, 25 ng/mL), IL-15 (25 ng/mL), and IL-2 (50 ng/mL) (all cytokines purchased from PeproTech). Cells were cultured at 37°C for 14 to 21 days, and half of the coculture medium was replaced weekly. At indicated days, cells were harvested and analyzed by FACS for B (CD19+), T (CD3+CD4+/CD8+NK1.1−, NK1.1−CD25+TCR-β+Thy1.2+, NK1.1−CD25+Thy1.2+, TCR-β+CD4+/CD8+NK1.1−, or TCR-β+CD25−NK1.1−CD122−), NK (CD3−NK1.1+DX5+, TCR-β−NK1.1+DX5+, Lin−CD122+CD25−NK1.1+IL-7Rα−, or CD3−NK1.1+Mac-1+), NK.1.1+TCR+ (TCR-β+NK1.1+ DX5+, CD3+NK1.1+, or TCR-β+NK1.1+), or thymic NK (Lin−CD122+CD25−NK1.1+Ly49D−IL-7Rα+) committed progeny.
Terasaki culture assay to detect myeloid potential
Purified single Lin−CD122+NK1.1−DX5− NK progenitors and Lin−SCA-1+KIT+FLT3high LMPPs (used as positive controls) were sorted directly into Terasaki plates containing serum-free medium (X-vivo 15; Lonza Walkersville) with 0.5% bovine serum albumin (StemCell Technologies), 10% FCS (Sigma-Aldrich), 1% penicillin/streptomycin (Sigma-Aldrich), 1% l-glutamine (Sigma-Aldrich), and 1% 10−2M 2-mercaptoethanol (Sigma-Aldrich), supplemented with cytokines with final concentrations: FL (50 ng/mL), KL (50 ng/mL), thrombopoietin (50 ng/mL), IL-3 (10 ng/mL), granulocyte colony-stimulating factor (50 ng/mL), and granulocyte-macrophage colony-stimulating factor (20 ng/mL) and cultured for 12 days at 37°C. After 8 and 12 days, cultures were evaluated under the inverted microscope, and the size of proliferating clones was scored.
Gene expression analysis by RT-PCR
Clones generated from single Lin−CD122+NK1.1−DX5− NKPs in OP9DL1 stroma cocultures that were positive for the NK- and T-cell lineage potentials by FACS were analyzed for the T lineage specific gene expression. Thymocytes were used as positive control, and OP9DL1 stroma cells cultured without hematopoietic cells were used as negative control. RNA extraction and reverse-transcribed (RT)–PCR were performed as previously described.26 The primers used for nested muliplex RT-PCR were as follows:
Hprt.
Hprt-1: 5′GGGGGCTATAAGTTCTTTGC; Hprt-2: 5′GTTCTTTGCTGACCTGCTGG; Hprt-3: 5′TGGGGCTGTACTGCTTAACC; Hprt-4: 5′TCCAACACTTCGAGAGGTCC.
Ptcra.
Ptcra-1: 5′CTCTACCATCAGGCATCG; Ptcra-2: 5′GGCAGTGCCCTAGACGCC; Ptcra-3: CTCCTGGCTGTCGAAGATTCC; Ptcra-4: 5′GAAGCAGTTTGAAGAGGAGC.
Cd3e.
Cd3e-1: 5′CCTCAGAAGCATGATAAGC; Cd3e-2: 5′GTTGACATCTGTATCACTCTGG. Cd3e-3: 5′ACTGCTCTCTGATTCAGGC; Cd3e-4: 5′AGCAAGGTTCTAGGACACG. Primers 1 plus 4 and 2 plus 3 are outer and inner primer pairs, respectively.
Statistics
All results are expressed as mean (SD). The statistical significances between groups were determined using the Student t test.
Results
BM Lin−CD122+NK1.1−DX5− NKPs efficiently generate NK and T cells in vitro
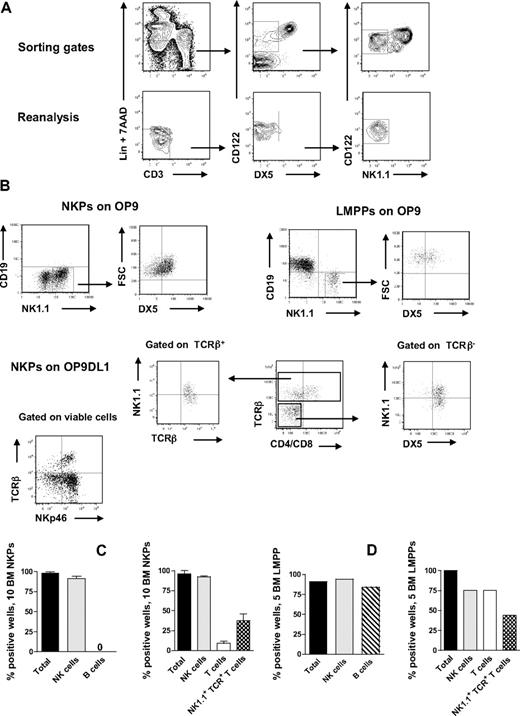
NKPs have been identified in the BM of adult mice as having a Lin−CD122+NK1.1−DX5− phenotype.21 However, previous in vitro characterization of the lineage potentials of NKPs was pursued using large cell numbers, and the T-cell potential was evaluated using a fetal thymic organ culture,21 whereas more efficient T-cell assays have since been developed and applied at the single-cell level with high efficiency.8,27,28 To further investigate the lineage potentials of Lin−CD122+NK1.1−DX5− cells, we first optimized the culture conditions by testing different cytokine combinations known to support in vitro NK-cell generation.14,29 In agreement with previous studies,21,30 approximately 40% of BM NKPs expressed IL-7Rα, 60% of NKPs were Thy1.2+, and there was no CD25 expression detectable within NKPs (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). When as few as 10 NKPs were cocultured on OP9 or OP9DL1 stroma cell lines for 14 days in the presence of KL, FL, IL-7 (first week only), IL-2, and IL-15, essentially all wells, contained large numbers of TCR-β−NK1.1+DX5+ NK cells (Figure 1A-C). To further confirm the NK-cell phenotype, the generated clones were evaluated for the expression of NKp46 (the most specific NK-cell marker)31,32 and essentially all wells in which NKPs were cocultured on O9DL1 stroma with cytokines contained TCR-β−NKp46+ NK cells (Figure 1B). In agreement with previous studies,21 NKPs did not produce any detectable levels of CD19+ B cells (Figure 1A-C). Notably, however, approximately 10% of the wells seeded with 10 Lin−CD122+NK1.1−DX5− cells on OP9DL1 stroma contained a distinct population of TCR-β+NK1.1−CD4+/CD8+ T cells, and 37% of the wells contained TCR-β+NK1.1+DX5+ cells (Figure 1B-D). Previous studies have identified a subset of T cells that coexpress TCR together with NK1.1 and DX5 molecules.33 However, NK T cells and NK-like T cells represent heterogeneous populations, and the exact characteristics of TCR+NK1.1+ T cells produced from NKPs here remain to be established.
BM NKPs can efficiently generate NK and T cells in vitro. Ten Lin−CD122+NK1.1−DX5− NKPs and 5 Lin−KIT+SCA-1+FLT3high LMPPs sorted from BM of 8- to 12-week-old mice were cultured on OP9 and OP9DL1 stroma cell lines with cytokines: KL, IL-7 (first week only), FL, IL-2, and IL-15. After 14 and 21 days, cells were harvested and evaluated for NK (TCR-β−NK1.1+DX5+ or CD3−NK1.1+DX5+), B (CD19+), T (TCR-β+NK1.1−CD4+/CD8+ or NK1.1−CD3+CD4+/CD8+), and NK1.1+TCR+ T cells (TCR-β+NK1.1+DX5+) by FACS. ToPro was used to exclude dead cells. (A) Representative FACS profiles of gating and the reanalysis of sorting the Lin−CD122+DX5−NK1.1− NKPs. After gating on Lin−CD3− cells, the CD122+DX5− gate was set and finally the CD122+NK1.1− gate was selected. (Bottom panels) Reanalysis after sorting; the purity was reproducibly more than 98%. (B) Representative FACS profiles of cells generated from NKPs and LMPPs cultured on OP9 and OP9DL1 stroma. The specific gates are indicated in the text above the plot and by the arrow. (C) Mean (SD) proportion of total positive wells and wells containing NK, B, T, and NK1.1+TCR+ T cells generated from 10 NKPs cultured on OP9 (left panel) and OP9DL1 (right panel). Data represent mean (SD) values from 6 or 7 independent experiments with at least 16 to 24 wells analyzed in each experiment. (D) Proportion of total positive wells and wells containing NK, B, T, and NK1.1+TCR+ T cells, generated from 5 LMPPs cultured on OP9 (left panel) and OP9DL1 (right panel). Data are from representative experiment with 16 to 36 wells analyzed.
BM NKPs can efficiently generate NK and T cells in vitro. Ten Lin−CD122+NK1.1−DX5− NKPs and 5 Lin−KIT+SCA-1+FLT3high LMPPs sorted from BM of 8- to 12-week-old mice were cultured on OP9 and OP9DL1 stroma cell lines with cytokines: KL, IL-7 (first week only), FL, IL-2, and IL-15. After 14 and 21 days, cells were harvested and evaluated for NK (TCR-β−NK1.1+DX5+ or CD3−NK1.1+DX5+), B (CD19+), T (TCR-β+NK1.1−CD4+/CD8+ or NK1.1−CD3+CD4+/CD8+), and NK1.1+TCR+ T cells (TCR-β+NK1.1+DX5+) by FACS. ToPro was used to exclude dead cells. (A) Representative FACS profiles of gating and the reanalysis of sorting the Lin−CD122+DX5−NK1.1− NKPs. After gating on Lin−CD3− cells, the CD122+DX5− gate was set and finally the CD122+NK1.1− gate was selected. (Bottom panels) Reanalysis after sorting; the purity was reproducibly more than 98%. (B) Representative FACS profiles of cells generated from NKPs and LMPPs cultured on OP9 and OP9DL1 stroma. The specific gates are indicated in the text above the plot and by the arrow. (C) Mean (SD) proportion of total positive wells and wells containing NK, B, T, and NK1.1+TCR+ T cells generated from 10 NKPs cultured on OP9 (left panel) and OP9DL1 (right panel). Data represent mean (SD) values from 6 or 7 independent experiments with at least 16 to 24 wells analyzed in each experiment. (D) Proportion of total positive wells and wells containing NK, B, T, and NK1.1+TCR+ T cells, generated from 5 LMPPs cultured on OP9 (left panel) and OP9DL1 (right panel). Data are from representative experiment with 16 to 36 wells analyzed.
Importantly, NKPs cocultured on OP9DL1 stroma produced both TCR-αβ and TCR-γδ T cells (supplemental Figure 2).
Taken together, these results suggest that a fraction of Lin−CD122+NK1.1−DX5− NKPs, although having lost B-cell potential (in contrast to control multipotent LMPPs8 ) sustain the ability to produce T cells (Figure 1B,D).
To test the myeloid potential of NKPs, we performed a single-cell myeloid culture assay (“Teraski culture assay to detect myeloid potential”), again using multipotent LMPPs as a positive control. In agreement with previous studies,21 no myeloid cells were generated from Lin−CD122+NK1.1−DX5− cells, regardless of the different time points evaluated, confirming their loss of myeloid potential (Table 1). Taken together, these data demonstrate that Lin−CD122+NK1.1−DX5− NKPs, in addition to having an extensive NK-cell potential, also sustain an ability to produce T cells.
Lin−CD122+NK1.1−DX5− NKPs lack myeloid lineage potential
| Experiment no. . | Cell population . | No. of cells plated per well . | No. of wells with small size clones (3-50 cells) . | No. of wells with medium size clones (50 cells; 10%) . | No. of wells with big clones > 10% . | Total no. of wells with clones . |
|---|---|---|---|---|---|---|
| 1 | LMPP | 1 | 4 | 23 | 71 | 98/120 |
| 1 | NKP | 1 | 0 | 0 | 0 | 0/120 |
| 2 | LMPP | 1 | 3 | 17 | 73 | 93/120 |
| 2 | NKP | 1 | 0 | 0 | 0 | 0/120 |
| Experiment no. . | Cell population . | No. of cells plated per well . | No. of wells with small size clones (3-50 cells) . | No. of wells with medium size clones (50 cells; 10%) . | No. of wells with big clones > 10% . | Total no. of wells with clones . |
|---|---|---|---|---|---|---|
| 1 | LMPP | 1 | 4 | 23 | 71 | 98/120 |
| 1 | NKP | 1 | 0 | 0 | 0 | 0/120 |
| 2 | LMPP | 1 | 3 | 17 | 73 | 93/120 |
| 2 | NKP | 1 | 0 | 0 | 0 | 0/120 |
Indicated cell populations were sorted as single cells directly into X-vivo 15 medium containing 10% of FCS and cytokines: KL + granulocyte-macrophage colony-stimulating factor + granulocyte colony-stimulating factor + IL-3 + FL + thrombopoietin. The plates were scored at day 8 (not shown) and day 12 (data in table) of the culture, with similar results.
BM Lin−CD122+NK1.1−DX5− NKPs generate NK and T cells in vivo after transplantation
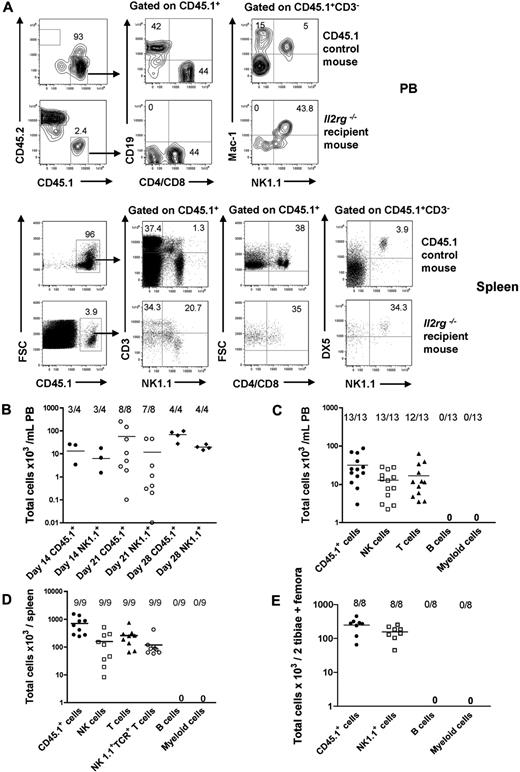
As the lineage potentials of Lin−CD122+NK1.1−DX5− NKPs have not previously been investigated in vivo, we next examined the ability of NKPs to reconstitute different lineages, with a particular emphasis on whether Lin−CD122+NK1.1−DX5− NKPs could reconstitute physiologically relevant numbers of T cells. Specifically, NKPs sorted from CD45.1 BM were injected intravenously into sublethally irradiated CD45.2 Il2rg−/− recipients. Evaluation of CD45.1 donor reconstitution in PB showed a maximum reconstitution at 21 days after transplantation (Figure 2A-B). At 21 days, all the recipient mice showed donor-derived NK-cell reconstitution (CD45.1+CD3−NK1.1+DX5+) in the PB and in the spleen, and as many as 12 of 13 and 9 of 9 recipients also demonstrated T-cell reconstitution (CD45.1+CD4+/CD8+NK1.1−) in the PB and in the spleen, respectively (Figure 2A,C-D). In addition, in all the recipient mice, donor-derived NK1.1+TCR+ T cells (CD45.1+CD3+NK1.1+) were present in the spleen at 21 days after transplantation (Figure 2B,D).
BM NKPs reconstitute NK and T cells in vivo. Sublethally irradiated 10- to 12-week-old Il2rg−/− CD45.2 recipient mice were transplanted with 1300 BM Lin−CD122+NK1.1−DX5− NKPs sorted from 9- to 12-week-old CD45.1 WT mice. Donor-derived lineage reconstitution was evaluated in the PB, spleen, and BM at 14, 21, and 28 days after transplantation by FACS. (A) Representative FACS plots of donor-derived CD45.1+ NK, T, B, and myeloid cell reconstitution in the PB and in the spleen from representative mice at 21 days after transplantation. (Top panels) Nontransplanted CD45.1 control mouse. (Bottom panels) Il2rg−/− transplanted recipient mouse. The specific gates are indicated in the text above the plot and by the arrow. The numbers show the proportion of cells within the gate, of total (CD45.1 plus CD45.2) cells and represent mean values from 9 to 13 mice from 2 independent experiments. (B) Donor-derived total reconstitution (CD45.1+) and donor-derived NK cells (CD45.1+CD3−NK1.1+DX5+) at 14, 21, and 28 days after transplantation in the PB. Data represent mean (SD) of total cells per 1 mL of PB; 4 to 8 mice were analyzed per group in 1 or 2 experiments. (C) Donor-derived total reconstitution (CD45.1+), donor-derived NK cells (CD45.1+CD3−NK1.1+DX5+), donor-derived T cells (CD45.1+CD4+/CD8+NK1.1−), donor-derived B cells (CD45.1+CD19+), and donor-derived myeloid cells (CD45.1+NK1.1−Mac-1+) at 21 days after transplantation in the PB. Data represent mean (SD) values of total cells per 1 mL of PB; 13 mice were analyzed per group in 2 independent experiments. (D) Donor-derived total (CD45.1+), NK (CD45.1+CD3−NK1.1+DX5+), T (CD45.1+CD4+/CD8+NK1.1−), and NK1.1+TCR+ T cells (CD45.1+CD3+NK1.1+) at 21 days after transplantation in the spleen. Data represent mean (SD) values of total cells per spleen; 9 mice were analyzed per group in 2 independent experiments. (E) Donor-derived total (CD45.1+), B cell (CD45.1+CD19+), myeloid cell (CD45.1+NK1.1−Mac-1+), and NK1.1+ cell (CD45.1+NK1.1+Mac-1−) reconstitution at 21 days after transplantation in the BM. Data represent mean (SD) values of total cells per 2 tibiae and 2 femora; 8 mice were analyzed per group. Representative FACS profiles are shown in supplemental Figure 4.
BM NKPs reconstitute NK and T cells in vivo. Sublethally irradiated 10- to 12-week-old Il2rg−/− CD45.2 recipient mice were transplanted with 1300 BM Lin−CD122+NK1.1−DX5− NKPs sorted from 9- to 12-week-old CD45.1 WT mice. Donor-derived lineage reconstitution was evaluated in the PB, spleen, and BM at 14, 21, and 28 days after transplantation by FACS. (A) Representative FACS plots of donor-derived CD45.1+ NK, T, B, and myeloid cell reconstitution in the PB and in the spleen from representative mice at 21 days after transplantation. (Top panels) Nontransplanted CD45.1 control mouse. (Bottom panels) Il2rg−/− transplanted recipient mouse. The specific gates are indicated in the text above the plot and by the arrow. The numbers show the proportion of cells within the gate, of total (CD45.1 plus CD45.2) cells and represent mean values from 9 to 13 mice from 2 independent experiments. (B) Donor-derived total reconstitution (CD45.1+) and donor-derived NK cells (CD45.1+CD3−NK1.1+DX5+) at 14, 21, and 28 days after transplantation in the PB. Data represent mean (SD) of total cells per 1 mL of PB; 4 to 8 mice were analyzed per group in 1 or 2 experiments. (C) Donor-derived total reconstitution (CD45.1+), donor-derived NK cells (CD45.1+CD3−NK1.1+DX5+), donor-derived T cells (CD45.1+CD4+/CD8+NK1.1−), donor-derived B cells (CD45.1+CD19+), and donor-derived myeloid cells (CD45.1+NK1.1−Mac-1+) at 21 days after transplantation in the PB. Data represent mean (SD) values of total cells per 1 mL of PB; 13 mice were analyzed per group in 2 independent experiments. (D) Donor-derived total (CD45.1+), NK (CD45.1+CD3−NK1.1+DX5+), T (CD45.1+CD4+/CD8+NK1.1−), and NK1.1+TCR+ T cells (CD45.1+CD3+NK1.1+) at 21 days after transplantation in the spleen. Data represent mean (SD) values of total cells per spleen; 9 mice were analyzed per group in 2 independent experiments. (E) Donor-derived total (CD45.1+), B cell (CD45.1+CD19+), myeloid cell (CD45.1+NK1.1−Mac-1+), and NK1.1+ cell (CD45.1+NK1.1+Mac-1−) reconstitution at 21 days after transplantation in the BM. Data represent mean (SD) values of total cells per 2 tibiae and 2 femora; 8 mice were analyzed per group. Representative FACS profiles are shown in supplemental Figure 4.
Furthermore, and in line with the findings from the in vitro studies, transplanted NKPs did not contribute to detectable B-cell (CD45.1+CD19+) and myeloid (CD45.1+NK1.1−Mac-1+) reconstitution, not in the PB (Figure 2A,C), the spleen, or the BM (Figure 2E; supplemental Figure 3). These data demonstrate that Lin−CD122+NK1.1−DX5− NKPs engraft and generate NK cells in vivo after transplantation; and consistent with the data from in vitro studies, NKPs also have a considerable potential to reconstitute T cells but lack the ability to produce cells of the B and myeloid lineages.
Single Lin−CD122+NK1.1−DX5− BM cells have a combined NK- and T-cell potential
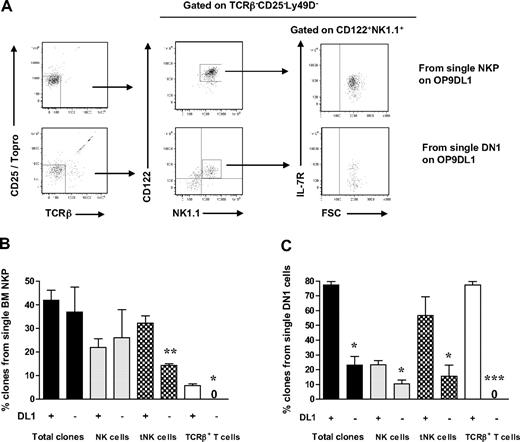
To establish whether the combined NK- and T-cell potentials of Lin−CD122+NK1.1−DX5− BM cells reflect the existence of separate NK and T cell-restricted progenitors within the Lin−CD122+NK1.1−DX5− compartment, or the existence in adult BM of a bipotent NK-/T-cell progenitor, we further optimized the cell culture conditions to efficiently promote generation of NK cells and T cells from single Lin−CD122+NK1.1−DX5− cells.
Previous studies have shown that the generation of T cells in OP9DL1 cultures is IL-7 dose dependent34 and that high levels of IL-7 block T-cell generation.35,36 Therefore, IL-7 was excluded from the medium, and the time of the culture was extended to 21 days based on previous studies.27 In addition, T-cell potential was in agreement with previous studies,37 defined as generation of cells with coexpression of CD25 and Thy1.2, and/or CD4+/CD8+TCR-β+ T cells. Under these conditions, as much as 42% of single Lin−CD122+NK1.1−DX5− cells generated sizable clones, which could be analyzed by multicolor FACS. As much as 25% of Lin−CD122+NK1.1−DX5− cells (60% of the clones) produced NK1.1+DX5+TCR-β− NK cells and 14% NK1.1−CD25+Thy1.2+ T cells (34% of the clones; most of them also coexpressing TCR-β) (Figure 3A-B; supplemental Figure 4).
BM single Lin−CD122+NK1.1−DX5− NKPs have combined NK- and T-cell potential. Single BM Lin−CD122+NK1.1−DX5− NKPs were directly sorted into OP9DL1 stroma layers and cultured with KL, IL-7 (first week), FL, IL-2, and IL-15. After 14 and 21 days, clones were harvested and analyzed by FACS. ToPro was used to eliminate dead cells. (A-B) FACS profiles of representative clones generated from a single Lin−CD122+NK1.1−DX5− NKP. Text above profiles indicates prior gating. (C) Mean (SD) proportion of total proliferating clones and clones containing NK (TCR-β−NK1.1+DX5+) and T (NK1.1−CD25+Thy1.2+ or NK1.1−CD25+ Thy1.2+ TCR-β+) cells generated from single NKPs. Data expressed in relation to plated single cells, from 3 independent experiments, with total 167 plated cells and 105 clones analyzed. (D) Mean (SD) proportion of total clones and clones containing NK (TCR-β−NK1.1+ DX5+), T (TCR-β+NK1.1−CD4+/CD8+, NK1.1− CD25+Thy1.2+, or NK1.1−CD25+Thy1.2+ TCR-β+), and TCR+NK1.1+ T cells, or combinations of these, generated from single NKPs. Data are expressed in relation to number of plated single cells, from 6 independent experiments, with a total 382 single cells plated and 201 clones analyzed. (E) Clones generated from single NKPs (shown in panels A and C), shown to have combined NK- and T-cell potential by FACS, were analyzed by RT-PCR for Cd3e, Ptcra, and Hprt gene expression. Thymocytes were used as positive control and OP9DL1 stroma cells cultured without hematopoietic cells as a negative control. Ethidium bromide–stained agarose gels with resulting PCR products from RT-PCR analysis of Cd3e, Ptcra, and Hprt. The photo has been taken using Gel logic 100 (Kodak). Data from 1 of 2 experiments with similar results. Note that clone 2 is the same clone derived from a single Lin−CD122+NK1.1−DX5− NKP and analyzed by FACS in panel A.
BM single Lin−CD122+NK1.1−DX5− NKPs have combined NK- and T-cell potential. Single BM Lin−CD122+NK1.1−DX5− NKPs were directly sorted into OP9DL1 stroma layers and cultured with KL, IL-7 (first week), FL, IL-2, and IL-15. After 14 and 21 days, clones were harvested and analyzed by FACS. ToPro was used to eliminate dead cells. (A-B) FACS profiles of representative clones generated from a single Lin−CD122+NK1.1−DX5− NKP. Text above profiles indicates prior gating. (C) Mean (SD) proportion of total proliferating clones and clones containing NK (TCR-β−NK1.1+DX5+) and T (NK1.1−CD25+Thy1.2+ or NK1.1−CD25+ Thy1.2+ TCR-β+) cells generated from single NKPs. Data expressed in relation to plated single cells, from 3 independent experiments, with total 167 plated cells and 105 clones analyzed. (D) Mean (SD) proportion of total clones and clones containing NK (TCR-β−NK1.1+ DX5+), T (TCR-β+NK1.1−CD4+/CD8+, NK1.1− CD25+Thy1.2+, or NK1.1−CD25+Thy1.2+ TCR-β+), and TCR+NK1.1+ T cells, or combinations of these, generated from single NKPs. Data are expressed in relation to number of plated single cells, from 6 independent experiments, with a total 382 single cells plated and 201 clones analyzed. (E) Clones generated from single NKPs (shown in panels A and C), shown to have combined NK- and T-cell potential by FACS, were analyzed by RT-PCR for Cd3e, Ptcra, and Hprt gene expression. Thymocytes were used as positive control and OP9DL1 stroma cells cultured without hematopoietic cells as a negative control. Ethidium bromide–stained agarose gels with resulting PCR products from RT-PCR analysis of Cd3e, Ptcra, and Hprt. The photo has been taken using Gel logic 100 (Kodak). Data from 1 of 2 experiments with similar results. Note that clone 2 is the same clone derived from a single Lin−CD122+NK1.1−DX5− NKP and analyzed by FACS in panel A.
In addition, essentially all the NK1.1+DX5+TCR-β− NK cells generated from single NKPs coexpressed NKp46 and CD94, further supporting their NK-cell identity (Figure 3B). The T-cell identity of clones containing NK1.1−CD25+Thy1.2+ cells was also verified through expression of Cd3ϵ and Ptcra by RT-PCR (Figure 3A,D; supplemental Figure 4). The fact that not all clones expressed both Cd3e and Ptcra but expressed TCR-β most probably reflects the different maturation stages of cells generated from individual clones, as it has been shown that Ptcra expression is down-regulated at the DN4-ISP transition.38 Thus, the cells that show expression of Cd3e but lack Ptcra probably had already reached the later stage.
Importantly, almost all single Lin−CD122+NK1.1−DX5− cells generating T cells also produced NK cells (Figure 3C), demonstrating the existence of Lin−CD122+NK1.1−DX5− bipotent NK-/T-cell progenitors in adult BM.
Thymic NK cells can be generated from BM Lin−CD122+NK1.1−DX5− progenitors and early thymic progenitors
Although the BM is the main site of NK-cell development, a distinct thymic-dependent NK pathway has recently been identified.17 Thymic and BM-dependent NK cells have different phenotypes and properties, but the developmental relationship between these 2 NK-cell populations remains to be established.
Recent studies have shown that early thymic progenitors (ETPs; Lin−CD25−Kithigh) are multipotent10,11 and both double-negative 1 (DN1) and double-negative 2 (DN2) thymocytes can generate NK cells.39 Thus, we used the ETP population to establish the optimal conditions to generate thymic NK (tNK) cells in vitro. When 10 ETPs were plated on OP9DL1 stroma with KL, FL, and IL15, essentially all the wells were positive and contained BM-dependent NK and tNK cells as well as T cells and NK1.1+TCR+ T cells (Figure 4A).
Thymic NK cells can be generated from early thymic progenitors as well as Lin−CD122+NK1.1−DX5− NK-/T-cell progenitors in the BM. (A) Ten DN1 cells (Lin−CD25−Kithigh) sorted from the thymus of 6- to 9-week-old mice and (B) 5 Lin−CD122+NK1.1−DX5− NKPs sorted from BM of 9- to 12-week-old mice were cultured on OP9DL1 stroma for 21 days in the presence of KL, FL, and IL-15. Generated clones were evaluated by FACS for the presence of BM-dependent NK (TCR-β−NK1.1+CD122+IL-7Rα−), thymic NK (tNK; TCR-β−CD25−Ly49D− NK1.1+CD122+IL-7Rα+), T (TCR-β+NK1.1−CD4+/CD8+), and NK1.1+TCR+ (NK1.1+TCR-β+) T cells. Data represent mean (SD) proportion of positive wells from 2 independent experiments, with 24 to 36 wells being analyzed in each experiment.
Thymic NK cells can be generated from early thymic progenitors as well as Lin−CD122+NK1.1−DX5− NK-/T-cell progenitors in the BM. (A) Ten DN1 cells (Lin−CD25−Kithigh) sorted from the thymus of 6- to 9-week-old mice and (B) 5 Lin−CD122+NK1.1−DX5− NKPs sorted from BM of 9- to 12-week-old mice were cultured on OP9DL1 stroma for 21 days in the presence of KL, FL, and IL-15. Generated clones were evaluated by FACS for the presence of BM-dependent NK (TCR-β−NK1.1+CD122+IL-7Rα−), thymic NK (tNK; TCR-β−CD25−Ly49D− NK1.1+CD122+IL-7Rα+), T (TCR-β+NK1.1−CD4+/CD8+), and NK1.1+TCR+ (NK1.1+TCR-β+) T cells. Data represent mean (SD) proportion of positive wells from 2 independent experiments, with 24 to 36 wells being analyzed in each experiment.
Next, we applied the same culture conditions to test whether tNK cells can be generated from Lin−CD122+NK1.1−DX5− BM NKPs. When 5 sorted BM NKPs were cocultured on OP9DL1 stroma with KL, FL, and IL-15, 83% of wells were positive and contained both BM NK cells and tNK cells (80% and 69%, respectively; Figure 4B).
Importantly, tNK cells generated in vitro from NKPs and ETPs maintained the same phenotype as NK cells in the thymus, in that they expressed IL-7Rα, lacked the expression of Ly49 receptors, and coexpressed CD94, NKp46, and high levels of CD2717 (supplemental Figure 5).
Role of Notch in generation of thymic and BM-dependent NK cells
Whereas Notch signaling is critical for T-cell development,40,41 it is not clear whether Notch might be involved in regulation of distinct stages of NK-cell development. To investigate whether Notch signaling is required for generation of BM and thymic-dependent NK cells, single BM Lin−CD122+NK1.1−DX5− NKPs, or ETPs, were directly sorted onto OP9 stroma cells expressing Notch ligand Delta1 (DL1) or not, in the presence of KL, FL, and IL-15. Approximately 40% of single NKPs generated clones irrespective of the expression of DL1, and also the contribution toward the generation of BM-dependent NK cells was Notch-independent (Figure 5A-B). Although reduced, BM Lin−CD122+NK1.1−DX5− cells also generated tNK cells in the absence of DL1, whereas T-cell generation from BM Lin−CD122+NK1.1−DX5− cells was strictly Notch-dependent (Figure 5A-B). In contrast to BM NKPs, single ETPs showed reduced overall clonal growth in the absence of DL1 (Figure 5B-C). Notably, although not strictly dependent on DL1 activation, tNK-cell production from ETPs was enhanced in the presence of DL1, whereas T-cell generation from ETPs was entirely DL1-dependent. Taken together, both Lin−CD122+NK1.1− DX5− NKPs and ETPs contribute to generation of thymic and BM-dependent NK cells in vitro; and unlike the T-cell generation from these progenitors, the generation of both of these NK developmental pathways appears to be largely Notch-independent.
Notch-independent generation of thymic and BM NK cells from Lin−CD122+NK1.1−DX5− NK/T progenitors. Single Lin−CD122+NK1.1−DX5− NKPs from BM from 10- to 12-week-old mice and single DN1 (Lin−CD25−Kithigh) cells from the thymus from 6- to 8-week-old mice were directly sorted onto OP9DL1 and OP9 stroma layers and cultured for 21 days with KL, FL, and IL-15. Clones were harvested and analyzed by FACS for the presence of BM-dependent NK (TCR-β−CD25−NK1.1+CD122+IL-7Rα−) and thymic NK (tNK) cells (TCR-β−CD25−Ly49D−NK1.1+CD122+IL-7Rα+), as well as TCR-β+CD25−NK1.1−CD122− T cells. (A) Representative FACS profiles of clones generated from single NKPs and DN1 thymocytes cultured on OP9DL1 stroma. Text above panels indicates prior gating. (B) Mean (SD) proportion of total generated clones and clones containing BM and thymic NK cells and T cells from single NKPs cultured on OP9DL1 and OP9. (C) Mean (SD) proportion of total generated clones and clones containing BM and thymic NK cells and T cells from single DN1 cells cultured on OP9DL1 and OP9. Data expressed in relation to plated single cells, from 2 to 4 independent experiments, were 24 to 40 clones analyzed in each experiment. *P < .05; **P < .01; ***P < .001.
Notch-independent generation of thymic and BM NK cells from Lin−CD122+NK1.1−DX5− NK/T progenitors. Single Lin−CD122+NK1.1−DX5− NKPs from BM from 10- to 12-week-old mice and single DN1 (Lin−CD25−Kithigh) cells from the thymus from 6- to 8-week-old mice were directly sorted onto OP9DL1 and OP9 stroma layers and cultured for 21 days with KL, FL, and IL-15. Clones were harvested and analyzed by FACS for the presence of BM-dependent NK (TCR-β−CD25−NK1.1+CD122+IL-7Rα−) and thymic NK (tNK) cells (TCR-β−CD25−Ly49D−NK1.1+CD122+IL-7Rα+), as well as TCR-β+CD25−NK1.1−CD122− T cells. (A) Representative FACS profiles of clones generated from single NKPs and DN1 thymocytes cultured on OP9DL1 stroma. Text above panels indicates prior gating. (B) Mean (SD) proportion of total generated clones and clones containing BM and thymic NK cells and T cells from single NKPs cultured on OP9DL1 and OP9. (C) Mean (SD) proportion of total generated clones and clones containing BM and thymic NK cells and T cells from single DN1 cells cultured on OP9DL1 and OP9. Data expressed in relation to plated single cells, from 2 to 4 independent experiments, were 24 to 40 clones analyzed in each experiment. *P < .05; **P < .01; ***P < .001.
Discussion
The present studies investigated and established several important and novel aspects of NK-cell lineage restriction and development. Although there is strong evidence that the lineage commitment of HSCs occurs as a stepwise process, the cellular pathways and restriction sites to generate bipotent and unipotent progenitors of lymphoid lineages remain the subject of debate and investigation.1-4 Of relevance for our studies, it has remained an unresolved question whether adult multipotent hematopoietic progenitors become NK/T-cell lineage-restricted already in the BM or in the thymus.5,6 In the present studies, using highly sensitive and specific assays, down to the single-cell level, we demonstrate that, although adult BM Lin−CD122+NK1.1−DX5− progenitors have lost B and myeloid cell potentials, they sustain a combined NK- and T-cell potential in vivo and in vitro at the single-cell level. Interestingly, these NK/T progenitors contribute to both thymic and BM-dependent NK-cell pathways, and this process is neither suppressed, as the B-cell fate of progenitors with combined B- and T-cell potential, nor dependent on Notch signaling, as the T-cell potential of these bipotent NK-/T-cell progenitors.
Although it has been previously suggested that the BM Lin−CD122+NK1.1−DX5− NKP population might be heterogeneous,21,30 previous in vitro evaluation of the lineage potentials did not indicate that the candidate NKP committed Lin−CD122+NK1.1−DX5− progenitors sustained any ability to generate blood cell lineages other than NK cells.21 Our finding that a large fraction of BM Lin−CD122+NK1.1−DX5− progenitors also sustain T-cell potential might be explained by the fact that we have used the OP9DL1 stroma culture assay, which allows detection of the T-cell potential with very high efficiency at the single-cell level,8,27 compared with the fetal thymic organ culture used in previous studies.21 In addition, the use of more optimal cytokine combinations, promoting both NK- and T-cell development from Lin−CD122+NK1.1−DX5− progenitors, might have been important. This might also explain why BM Lin−CD122+NK1.1−DX5− NKPs cultured in vitro at the single-cell level had a very high cloning efficiency and extensive NK-cell potential in the present studies, whereas in previous studies only one in 12 sorted BM Lin−CD122+NK1.1−DX5− NKPs formed clones.21 The potential differences between cells generated from NKPs and LMPPs (used as a positive control in our studies) could reflect the developmental hierarchical relationship between LMPPs and NKPs, but most probably differences in in vitro kinetics of NK-cell generation and differentiation between the 2 progenitor types, and potentially also the impact of the other lymphoid cells generated in cultures of LMPPs. Importantly, NK cells generated in vitro from NKPs expressed multiple NK-cell specific markers, including NKp46, CD94, and a variety of Ly49 receptors, further supporting their NK-cell identity. To demonstrate that the combined NK- and T-cell potential did not reflect the existence of Lin−CD122+NK1.1−DX5− NK cell as well as T-cell committed progenitors, single cell experiments were critical. The analysis of the lineage potentials of Lin−CD122+NK1.1−DX5− cells cultured as single cells demonstrated that a majority of the clones that generated T cells also produced NK cells, indicating that the Lin−CD122+NK1.1−DX5− NKP population contains a large fraction of bipotent NK-/T-cell progenitors.
Importantly, phenotypic and gene expression analysis was used to confirm that clones generated from single NKPs contain cells committed to the T-cell lineage (CD25+Thy1.2+ cells that coexpress CD4 and CD8 as well as TCR-β, CD3ϵ, and Ptα). The potential to generate T cells from NKPs is further supported by transplantation studies, clearly showing in vivo donor-derived reconstitution of CD3+CD4+CD8+NK1.1− and CD4+CD8+NK1.1− T cells from NKPs.
Previous studies did not evaluate the lineage potentials of candidate NKPs in vivo.21 Herein, by evaluating the progeny derived from transplanted BM Lin−CD122+NK1.1−DX5− progenitors, we confirmed in vivo the combined NK- and T-cell potential (and lack of myeloid and B-cell potential). In addition to donor-derived NK and T cells, reconstituted recipient mice had a donor-derived CD3−NK1.1− population in the spleen. Although the identity of these cells is unknown, they may represent transplanted undifferentiated NKPs or transitional NKPs.
NKT cells represent a sublineage of T cells with unique properties that play an important role against microbial, viral, and parasitic infections and are defined as T cells expressing the NK lineage receptors, such as NK1.1 as well as αβT cell receptors (TCRs).42,43 The “true” mouse NK T cells are characterized by CD1d reactivity that can be identify either by the positive staining of the TCR with the aGC-loaded CD1d tetramer or the detection of invariant Vα14-Jα18: Vβ8.2, 7 2 TCR.33,44 In the adult mouse, only 1% of the T cells in the spleen is aGC-CD1d-tetramer positive. The experimental setup used in our studies, with analysis of the progeny of cells generated from very low numbers of precursors, would not allow us to conclusively determine whether CD1d-restricted NK T cells are generated from NKPs. Therefore, the exact identity and functional properties of the NK1.1+TCR+ T cells generated from NKPs remain to be established. It is possible that NK1.1+ TCR+ T cells might be a fraction of T cells expressing NK1.1 or NK T cells. Regardless of whether the NK1.1+TCR+ cells represent NK1.1+ T cells or CD1d-restricted NK T cells, their generation from BM NKPs further substantiates that Lin−CD122+NK1.1−DX5− NKPs are not NK lineage-restricted in their potential and can generate not only NK cells but also cells of non-NK lineage, such as different subset of T cells (conventional NK1.1− TCR+ T cells and NK1.1+ TCR+ T cells). Separate studies using the CD1d-deficient mice will help to conclusively establish whether CD1d-restricted NK T cells can be generated from NKPs and to address the developmental relationship between NK and NK T cells.
The identification of bipotent NK-/T-cell progenitors in adult BM demonstrate that this prethymic lineage restriction is not unique during development but remains an important lineage restriction point in adult BM hematopoiesis.18-20
The recent finding that in adult mice a distinct population of NK cells is produced by a thymic-dependent pathway is also compatible with the prethymic lineage restriction of bipotent NK-/T-cell progenitors, although the identity of the progenitors capable of producing thymic NK cells has not been established.17
The origin of the thymic NK cells remains unclear and controversial. Whereas the data from Vosshenrich et al17 support that the IL-7Rα+ NK cells are thymus-derived, studies by Stewart et al45 demonstrated that these NK cells are also present in athymic mice, suggesting that the thymus might not be essential for the development of IL-7Rα+ NK cells, and might also be generated in secondary lymphoid tissues, for example, in the lymph nodes, before they reenter the thymus. In addition, a fraction of γδ T cells coexpress NK1.1 and IL-7Rα, and these “NK- like γδ T cells” highly resemble the thymic NK-cell phenotype.45
In agreement with previous studies, approximately 40% of Lin−CD122+NK1.1−DX5− NKPs expressed IL-7Rα. Studies using mice deficient in IL-7R signaling have shown that, whereas BM NK-cell development is IL-7–independent, thymic NK cells are IL-7–dependent.24,30
Here we show through in vitro studies that the same BM Lin−CD122+NK1.1−DX5− NKPs might contribute to both BM- and thymic-dependent NK pathways. However, it remains unclear whether thymic NK cells can be generated from NKPs after in vivo transfer and whether their development depends on the thymus. In studies in which we transplanted NKPs intravenously into IL-2rg–deficient recipient mice (data not shown), the thymus remained rudimental, not allowing conclusive analysis of IL-7Rα+ NK cells in the thymus, and there were also no detectable donor-derived IL-7Rα+ NK cells present in the spleen. However, even in steady state, IL-7Rα+ NK cells represent only 1% to 5% of total NK cells in the spleen17 (and data not shown). We also performed intrathymic transfers of NKPs as well as whole BM cells but failed to detect any NK-cell reconstitution in the thymus or in the periphery (data not shown). The inability to reconstitute thymic NK cells upon transplantation appears to be a general problem, probably reflecting technical limitations. Therefore, further studies are required to address whether Lin−CD122+NK1.1−DX5− cells may represent thymic immigrants or whether the IL-7Rα+ NK cells reenter the thymus from the lymph nodes.
The critical role of Notch signaling in T-cell development is well established,40,41,46 whereas the role of Notch in regulation of NK lymphopoiesis is unclear, although in vitro studies have suggested that Notch might be important for the NK lineage.47 Herein, we demonstrate that, in contrast to B-cell development,40,41 in vitro generation of BM as well as thymic NK cells from bipotent NK-/T-cell progenitors is neither restricted nor dependent on Notch signaling, whereas the generation of T cells from BM Lin−CD122+NK1.1−DX5− NK-/T-cell progenitors is entirely Notch-dependent.
Our studies identify Lin−CD122+NK1.1−DX5− cells in adult BM as bipotent NK-/T-cell progenitors, contributing to thymic as well as BM-dependent NK developmental pathways. The identification of a distinct NK-/T-cell progenitor will facilitate the identification of the molecular determinants of the lineage restriction of these progenitors from multipotent progenitors in the BM to NK and T cell-restricted progenitors in the BM and thymus.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Susanna Cardell, David Bryder, and William Agace for helpful discussions and Lilian Wittmann, Zhi Ma, and Shabnam Kharazi for expert advice and technical assistance.
This work was supported by the Swedish Medical Research Council, the Swedish Cancer Society, the Swedish Pediatric Cancer Society, and the Hemato-Linne grant. E.S. has an Assistant Professor position from the Swedish Cancer Society. H.N.C. is supported by the Iranian Ministry of Health. The Lund Stem Cell Center is supported by a Center of Excellence grant from the Swedish Foundation for Strategic Research.
Authorship
Contribution: H.N.C. and E.S. designed and conceptualized the research, analyzed the data, and wrote the manuscript; H.N.C., Y.T., M.C., and C.M.C. purified cell populations, performed in vitro cultures and transplantation studies, and analyzed the data; and S.E.W.J. participated in the study design, analysis, and discussions of data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ewa Sitnicka, Hematopoietic Stem Cell Laboratory, Lund Research Center for Stem Cell Biology and Cell Therapy, Lund University, 221-84 Lund, Sweden; e-mail: ewa.sitnicka@med.lu.se.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal