Abstract
Treatment of acute promyelocytic leukemia (APL) with all-trans-retinoic acid (ATRA) results in terminal differentiation of leukemic cells toward neutrophil granulocytes. Administration of ATRA leads to massive changes in gene expression, including down-regulation of cell proliferation–related genes and induction of genes involved in immune function. One of the most induced genes in APL NB4 cells is transglutaminase 2 (TG2). RNA interference–mediated stable silencing of TG2 in NB4 cells (TG2-KD NB4) coupled with whole genome microarray analysis revealed that TG2 is involved in the expression of a large number of ATRA-regulated genes. The affected genes participate in granulocyte functions, and their silencing lead to reduced adhesive, migratory, and phagocytic capacity of neutrophils and less superoxide production. The expression of genes related to cell-cycle control also changed, suggesting that TG2 regulates myeloid cell differentiation. CC chemokines CCL2, CCL3, CCL22, CCL24, and cytokines IL1B and IL8 involved in the development of differentiation syndrome are expressed at significantly lower level in TG2-KD NB4 than in wild-type NB4 cells upon ATRA treatment. Based on our results, we propose that reduced expression of TG2 in differentiating APL cells may suppress effector functions of neutrophil granulocytes and attenuate the ATRA-induced inflammatory phenotype of differentiation syndrome.
Introduction
Acute promyelocytic leukemia (APL), a subtype of acute myeloid leukemia, is characterized by a specific chromosome translocation t(15;17), which results in the rearrangement of the promyelocytic leukemia (PML) and the retinoic acid receptor-α (RARα) genes, causing the expression of a PML-RARα chimeric protein.1-3 PML-RARα acts as a transcriptional repressor, which disrupts the retinoic acid signaling pathway, precludes the differentiation program, and arrests cells at the promyelocytic stage of myeloid differentiation. Treatment of APL patients with high dosage of all-trans-retinoic acid (ATRA) is the prototype of differentiation therapy resulting in terminal differentiation of APL cells.4-6
Despite the high cure rate achieved by ATRA therapy, induction of hemorrhage, infection, and differentiation syndrome (DS; formerly retinoic acid syndrome) may cause death during the early treatment phase of APL. The excessive systemic inflammatory response during DS is associated with tissue infiltration of differentiating APL cells and induces pulmonary, renal, or cardiac failure.7,8 Recently published results showed that infiltrating differentiating APL cells enhance migration of leukemic cells into the lung by the massive chemokine induction (eg, CCL2/MCP-1, CCL3/MIP-1a, CCL22/MDC, CCL24/MPIEF-2) provoked by ATRA treatment and pulmonary infiltration of APL cells induce an uncontrollable hyperinflammatory reaction in the lung.9
NB4 is a well-characterized human promyelocytic leukemia cell line isolated and established from an APL patient.10 ATRA induces terminal differentiation of these immature leukemic promyelocytes into granulocytes. Differentiated NB4 cells bear the most common features of natural granulocytes (ie, they express differentiation markers CD11b, CD11c, CD44, become adherent and capable of cell migration, and acquire the ability to phagocytose and to kill pathogens).11,12 Therefore, ATRA-induced differentiation of NB4 cells provides an excellent in vitro model to investigate molecular events occurring during terminal differentiation of myeloid cells.
Type-2 transglutaminase (TG2), a member of the transglutaminase family, is a multifunctional, Ca2+-dependent acyltransferase that catalyzes the covalent cross-linking of proteins by the formation of γ-glutamil-ϵ-lysyl bond or the incorporation of monoamines into proteins.13 TG2 acts as a G protein in signal transduction processes and interacts with integrins and fibronectin after its secretion. Contrary to other transglutaminases, TG2 is characterized by broad tissue distribution and subcellular localization (cytoplasmic, mitochondrial, and nuclear).14-16 TG2 is abundantly expressed in immune cells including bone marrow–derived white blood cell. In macrophages, it may constitute 0.1%-1% of total cell protein and has been suspected to mediate inflammatory reactions, adhesion and spreading of these cells, phagocytosis, and clearance of apoptotic cells.17-21 However, the exact role of TG2 in various functions of the immune system and in the differentiation process of immune cells is not completely understood.
In our previous study, we demonstrated that TG2 plays an important role in neutrophil granulocyte differentiation and gene expression. We found that inhibition of the cross-linking activity of TG2 decreased expression of GP91phox NADPH oxidase and impeded several neutrophil cellular functions such as migration and superoxide anion production.16 Lack of TG2 protein in TG2−/− mouse neutrophils was also accompanied by a significant decrease in GP91phox mRNA and protein expression with diminished nitroblue tetrazolium test (NBT) positivity and superoxide anion production as well as impaired neutrophil extravasation.16
To further investigate the role of TG2 in differentiation of promyelocytic cells toward neutrophil granulocytes, we stably silenced TG2 in NB4 cells by RNA interference and then analyzed the consequent gene expression changes by microarray during the differentiation. We demonstrated that TG2 plays an important role in modulating transcriptional regulation in NB4 cells upon ATRA treatment. Silencing its expression leads to a significant delay in the differentiation process and down-regulation of genes related to the innate immune system. These cells failed to develop the features of totally matured neutrophils, as they remained proliferative for a longer time, were less adherent, and had lower phagocytic ability than those expressing TG2 at normal levels. In the context of DS, these cells showed low-level expression of inflammatory cytokines and decreased adhesion and migration ability.
Methods
Cell culture
NB4 (purchased from DSMZ GmbH) cell line was cultured in RPMI 1640 Medium (Sigma-Aldrich) supplemented with (10% vol/vol) fetal bovine serum (FBS; Sigma-Aldrich), 2mM glutamine (Sigma-Aldrich), 100 U/mL penicillin, and 100 μg/mL streptomycin solution (Sigma-Aldrich). Differentiation of NB4 was induced at 1 × 105 cells/mL by administration of 1μM ATRA (Sigma-Aldrich). The 293FT packaging cell line was purchased from Invitrogen and maintained in Dulbecco modified Eagle medium (Sigma-Aldrich) supplemented with 10% FBS, 4mM glutamine, 100 U of penicillin, 100 μg/mL streptomycin solution, and 1mM sodium pyruvate (Sigma-Aldrich).
Virus production and generation of stable TG2 knockdown NB4 cell line
A set of 5 antihuman TG2 short hairpin RNA (shRNA) expressing lentiviral plasmids (pLKO.1-puro) was purchased from Sigma-Aldrich. As a negative control, a nontargeting shRNA vector was used (Sigma-Aldrich), which expresses a shRNA sequence containing 4-bp mismatches to any known human gene. Lentiviral vectors were produced in 293FT cells according to the manufacturer's protocol. For NB4 transduction 2 × 104 cells were transduced separately with the 5 different anti-TG2 shRNA lentiviral vectors or with the nontargeting shRNA lentiviral vector at a final multiplicity of infection of 1.0 in 100 μL of medium. Parallel infections (3-5) were performed with each viral vector. After 24-36 hours stably transduced cells were selected in the presence of puromycin (5 μg/mL; Sigma-Aldrich). The efficiency of TG2 gene silencing was determined at different time points by real-time quantitative polymerase chain reaction (Q-PCR) and by Western blot analysis after induction of TG2 by ATRA.
Microarray experiment: sample preparation, labeling, hybridization, and data analysis
Differentiation of NB4 was induced at 1 × 105 cells/mL by the administration of 1μM ATRA, and cells were harvested at 0, 48, or 72 hours thereafter. Total RNA from 1010 cells was isolated using the RNeasy kit (QIAGEN). Experiments were performed in biological triplicates representing samples from different differentiations. Further processing and labeling, hybridization to GeneChip Human Gene 1.0 ST Arrays (Affymetrix), and scanning were conducted at the Microarray Core Facility of the European Molecular Biology Laboratory (Heidelberg, Germany). Image files were imported to GeneSpring 10.1 (Agilent). All microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE23702 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE23702). Raw signal intensities were normalized per chip (to the 50th percentile) and per gene (to the median). To identify significantly regulated genes between 2 compared samples, we identified probe sets that showed at least 2-fold up- or down-regulation by eliminating probe sets with a ratio of signal intensity between 0.5 and 2. Finally, we performed a t test for each pair of probe sets and filtered for values of P ≤ .05. The PANTHER classification system (www.pantherdb.org/tools/genexAnalysis.jsp) was used for functional classification of genes. Expression levels of ATRA-regulated genes are presented in log-transformed (base 2) form as supporting information in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Real-time Q-PCR
Total RNA was isolated from cells using Trizol Reagent (Invitrogen) and reverse transcribed to cDNA by High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturers' instructions. Transcript levels were determined by real-time Q-PCR using TaqMan probes. Real-time monitoring was carried out using an ABI Prism 7900 (Applied Biosystems). Transcript levels were normalized to the level of cyclophilin D, and gene expression was determined by the comparative cycle threshold (CT) method.
Western blot analysis, cross-linking activity assay of TG2, NBT reducing assay, and determination of superoxide anion production
These methods were published previously.16
Flow cytometry
Flow cytometric analysis was performed on a FACSCalibur (BD Biosciences) using Cell Quest Pro software. To detect the surface expression of CD11c 8 × 105 cells were incubated in dark with fluorescein isothiocyanate (FITC)-conjugated antihuman CD11c monoclonal antibody (mAb) or with anti-immunoglobulin G1 (IgG1) mAb as isotype negative control for 2 hours at 4°C in 1% bovine serum albumin phosphate-buffered saline. Cells were washed, fixed by 1% paraformaldehyde, and then analyzed by flow cytometry. The geometric mean fluorescence of the FITC-positive cells was used to calculate CD11c surface expression labeled by antiIgG1 mAb.
For detection of phagocytic capacity, 105 3-days differentiated NB4 cells were incubated in dark with FITC-labeled Listeria monocytogenes or Staphylococcus aureus at a 1:50 ratio (cells/bacteria) for 2 hours at 37°C or at 4°C (to assess the aspecific sticking of bacteria to the phagocytes). After 2 hours, cells were put on ice, washed 2× with 5-fold volume of ice-cold phosphate-buffered saline, fixed by 1% paraformaldehyde, and then analyzed by flow cytometry. Percent of FITC-positive cells were used as a measure of phagocytosis, with uptake at 4°C as a control for bacterial adhesion.
Cell-cycle analysis by flow cytometry was carried out by standard method (propidium iodide staining after alcoholic fixation).
Cell adhesion and migration
For determination of cell adhesion, cells were differentiated for 48 and 72 hours, than plated into 1% bovine serum albumin-coated (Sigma-Aldrich) plastic tissue culture dishes for 1 hour. Nonadherent cells were removed by washing, then the remaining cells were fixed with ice-cold methanol:acetic acid (4:1), stained with hematoxylin solution for 5 minutes, and counted microscopically. Migration assays were performed in BD Matrigel Invasion Chamber (BD Biosciences) according to the manufacturer's protocol using 200nM fMLP, 10 ng/mL interleukin-8 (IL-8), and 10% FBS as chemoattractants. Differentiated cells (2.5 × 104) were allowed to migrate for 14 hours. Chemoattractive effect of ATRA-treated wild-type, virus control, and TG2-KD NB4 cell supernatants on white blood cells was evaluated using transwell polycarbonate inserts (6.5-mm diameter) with 5-μm pores (Costar Corning) and carried out as described previously.9
CC chemokine level measurements
Supernatant of undifferentiated, 2- and 3-day ATRA-treated NB4 cells and its sublines were used for quantification of secreted CC chemokines. Concentration of CCL2 and CCL24 were measured using the Quantikine human enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems), CCL3 and CCL22 were quantified by the RayBio human ELISA kit (RayBiotech). Assays were performed according to the manufacturer's protocols.
Results
Generation of stable TG2 knockdown NB4 cell line
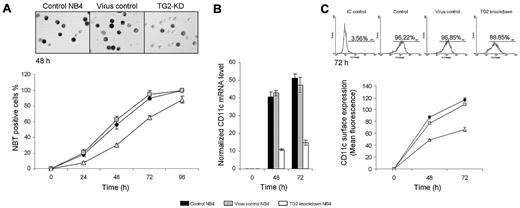
To evaluate the role of TG2 in neutrophil granulocyte differentiation, the TG2 gene was silenced in NB4 acute promyelocyte cell line through RNA interference. NB4 cells were transduced by anti-TG2 shRNA expressing lentiviral vector (TG2-KD NB4). Nontargeting shRNA control vector was also used to create virus control NB4. The expression of TG2 mRNA was dramatically induced by ATRA treatment and continuously increased during the 4 days of differentiation (Figure 1A). In virus control, NB4 TG2 mRNA levels were not affected, whereas expression of specific shRNA sequence against TG2 (TG2-KD NB4) significantly kept TG2 mRNA and protein levels down even at the fourth day of differentiation (Figure 1A-B).
Generation of stabile TG2 knockdown NB4 cell line. (A) NB4 cells (control NB4), NB4 cells transduced by random, nontarget shRNA expressing lentivirus (virus control NB4) and NB4 cells transduced by specific shRNA against TG2 expressing lentivirus (TG2 knockdown NB4) were differentiated in the presence of 1μM ATRA for 0, 24, 48, 72, and 96 hours. mRNA expression of TG2 was determined at the indicated time points by real-time Q-PCR, measurements were conducted in triplicates, values are expressed as mean % ± SD of the mean. (B) Control, virus control, and TG2 knockdown NB4 cells were differentiated in the presence of 1μM ATRA for 96 hours. Top panel shows cytosolic activity of TG2 measured at the indicated time point by detecting incorporation of [3H]putrescine into casein. The amount of incorporated [3H]putrescine was determined in a β-counter. Bars depict the means of 3 separate experiments each performed in duplicates. Inset in top panel shows the cytosolic protein levels of TG2 at the indicated time points detected by Western blot. Total protein (25 μg) was loaded in each line, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, blotted and developed with CUB7402 monoclonal antibody against TG2. Bottom panel shows nuclear activity of TG2 measured by detecting incorporation of [3H]putrescine into casein. Inset in bottom panel shows the Western blot analysis of nuclear TG2. TG2 enzyme activity assays and Western blot analyses were performed as described above.
Generation of stabile TG2 knockdown NB4 cell line. (A) NB4 cells (control NB4), NB4 cells transduced by random, nontarget shRNA expressing lentivirus (virus control NB4) and NB4 cells transduced by specific shRNA against TG2 expressing lentivirus (TG2 knockdown NB4) were differentiated in the presence of 1μM ATRA for 0, 24, 48, 72, and 96 hours. mRNA expression of TG2 was determined at the indicated time points by real-time Q-PCR, measurements were conducted in triplicates, values are expressed as mean % ± SD of the mean. (B) Control, virus control, and TG2 knockdown NB4 cells were differentiated in the presence of 1μM ATRA for 96 hours. Top panel shows cytosolic activity of TG2 measured at the indicated time point by detecting incorporation of [3H]putrescine into casein. The amount of incorporated [3H]putrescine was determined in a β-counter. Bars depict the means of 3 separate experiments each performed in duplicates. Inset in top panel shows the cytosolic protein levels of TG2 at the indicated time points detected by Western blot. Total protein (25 μg) was loaded in each line, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, blotted and developed with CUB7402 monoclonal antibody against TG2. Bottom panel shows nuclear activity of TG2 measured by detecting incorporation of [3H]putrescine into casein. Inset in bottom panel shows the Western blot analysis of nuclear TG2. TG2 enzyme activity assays and Western blot analyses were performed as described above.
We have previously demonstrated that during ATRA-induced differentiation of NB4 cells, TG2 translocates into the nucleus. To examine the effects of shRNA on both endogenous expression and translocation of TG2 into the nucleus, we analyzed the cytosolic and the nuclear fraction of control, virus control, and TG2 knockdown NB4 cells by Western blot (Figure 1B). Expression of TG2 was detectable at the first day and increased up to the fourth day in cytosolic fraction during the differentiation process. Virus control NB4 cells showed similar expression pattern. In TG2-KD NB4 cells treated in the same way, there were no detectable signs of TG2 in the cytosol at the first and the second day, and it remained at a low level at the third and fourth day as compared with the control or the virus control samples. Determination of TG2 crosslinking activity in cytosolic fractions also confirmed that TG2 activity was not affected in virus control cells, but markedly decreased in TG2-KD NB4 cells (Figure 1B). In nuclear fractions of control and virus control cells, TG2 appeared at the second day of the differentiation process. However, in TG2-KD NB4 cells, TG2 was hardly detectable, even on the third day. Nuclear cross-linking activity and protein level of TG2 changed in a parallel manner.
Taken together, these results indicated that ATRA-treated TG2-KD NB4 cells prove an appropriate model for studying the role of TG2 in neutrophil granulocyte differentiation; furthermore, the virus control NB4 cells served as the proper control to interpret results of the silencing experiments.
shRNA-induced knocking down of TG2 delays the differentiation process of NB4 cells
NBT reducing ability and the expression of CD11c are considered as reliable markers of differentiation of myeloid leukemia cells. Untreated cells were not able to reduce NBT, although in the case of control and virus control NB4 cells, the reducing activity of NBT were observed 24 hours after ATRA treatment and reached maximum levels at 72-96 hours. Differentiated TG2-KD NB4 cells were also able to reduce NBT at 24 hours, but the number of NBT-positive cells was 3 times less than those in control and the virus control NB4 cells and did not reach the maximum level even into 96 hours of differentiation (Figure 2A).
shRNA-induced knocking down of TG2 delays the differentiation process of NB4 cells. NB4 cells were cultured in standard medium in presence of 1μM ATRA for 0, 24, 48, 72, and 96 hours. (A) NBT reduction assays were performed (5 × 104 cells) at the indicated time points in triplicates in a 96-well plate by the addition of 1 mg/mL NBT and 2 μL (100 μg/mL) phorbol myristate acetate for 30 minutes. At least 400 cells were counted in each sample and according to their intracellular blue formazan deposit contents percentage of NBT positivity was determined. Percentage of NBT positivity is expressed as mean % ± SD for n = 3 parallel experiments. Light-microscopic image inserts show typical NBT positivity of the ATRA-treated control, virus control, and TG2 knockdown NB4 at the second-day stage of the differentiation (Zeiss Axiovert 135 inverted microscope, equipped with Achroplan 10×/0.25 Ph1; Carl Zeiss). (B) Induction of CD11c differentiation marker in ATRA-treated control, virus control, and TG2 knockdown NB4 cells were determined at the given time points of differentiation by real-time Q-PCR. Measurements were conducted in triplicates, values are expressed as mean % ± SD of the mean. (C) Differences in surface expression of CD11c between control, virus control, and TG2 knockdown NB4 cells were analyzed by flow cytometry at the indicated time points. Cells (8 × 105) were labeled with mouse antihuman CD11c IgG. Graph shows the average of the mean values of fluorescence measured in FL2 channel with ± SD. Typical flow cytometric profiles of CD11c surface expression in each cell types at the third day are shown by the inserted histograms.
shRNA-induced knocking down of TG2 delays the differentiation process of NB4 cells. NB4 cells were cultured in standard medium in presence of 1μM ATRA for 0, 24, 48, 72, and 96 hours. (A) NBT reduction assays were performed (5 × 104 cells) at the indicated time points in triplicates in a 96-well plate by the addition of 1 mg/mL NBT and 2 μL (100 μg/mL) phorbol myristate acetate for 30 minutes. At least 400 cells were counted in each sample and according to their intracellular blue formazan deposit contents percentage of NBT positivity was determined. Percentage of NBT positivity is expressed as mean % ± SD for n = 3 parallel experiments. Light-microscopic image inserts show typical NBT positivity of the ATRA-treated control, virus control, and TG2 knockdown NB4 at the second-day stage of the differentiation (Zeiss Axiovert 135 inverted microscope, equipped with Achroplan 10×/0.25 Ph1; Carl Zeiss). (B) Induction of CD11c differentiation marker in ATRA-treated control, virus control, and TG2 knockdown NB4 cells were determined at the given time points of differentiation by real-time Q-PCR. Measurements were conducted in triplicates, values are expressed as mean % ± SD of the mean. (C) Differences in surface expression of CD11c between control, virus control, and TG2 knockdown NB4 cells were analyzed by flow cytometry at the indicated time points. Cells (8 × 105) were labeled with mouse antihuman CD11c IgG. Graph shows the average of the mean values of fluorescence measured in FL2 channel with ± SD. Typical flow cytometric profiles of CD11c surface expression in each cell types at the third day are shown by the inserted histograms.
The expression of the CD11c differentiation marker was measured at 48 and 72 hours of differentiation, and its expression in TG2-KD NB4 cells was 3-fold lower than in the control or in the virus control NB4 cells at each day of differentiation (Figure 2B). CD11c surface expression was also evaluated at the second and the third day of the differentiation by flow cytometric analysis, and it was further confirmed that whereas control virus infection did not influence CD11c surface expression, knockdown of TG2 lead to decreased expression of CD11c on the cell surface (Figure 2C).
Comparison of gene expression profiling of differentiating control and TG2 knockdown NB4 cells
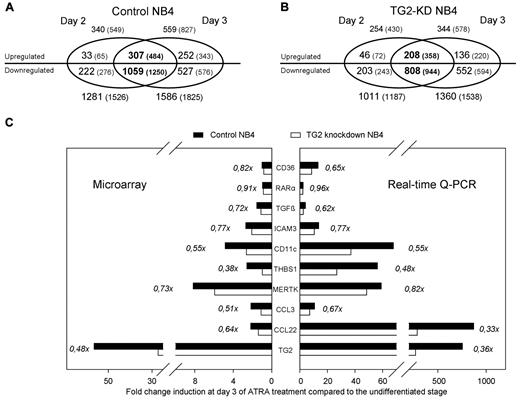
To determine the influence of TG2 on NB4 differentiation, gene expression changes induced by ATRA in control NB4 cells were determined during the differentiation process (on days 0, 2, and 3) and compared with those in TG2-KD NB4 cells (see supplemental Table 2). During the differentiation process, we detected 340 up-regulated genes on day 2 and 559 on day 3, and among these up-regulated genes, 307 showed at least 2-fold increase on both days of the sampling in NB4 control cells (Figure 3A). Interestingly, we identified approximately 3-4× more suppressed than induced genes by ATRA at both time points: 1281 genes showed decreased expression on day 2 and 1586 on day 3. Genes (1059) were found to be repressed on both days of differentiation in control NB4 cells (Figure 3A).
Number of genes (and entities) regulated by ATRA in differentiating control and TG2 knockdown NB4. (A-B) Number of annotated genes (and all entities in parentheses) regulated by ATRA in control NB4 (A) and in TG2-KD NB4 cells (B) are visualized as a Venn diagram. Two-fold change difference was set, and significantly changed genes were selected (P ≤ .05 and Bonferoni Multiple Correction was used). The numbers of up- and down-regulated genes are separated. Genes significantly regulated at the both stages of the differentiation (day 2 and day 3) are presented in the overlapping region of Venn diagram (in bold). Note that among the genes up-regulated in both stages of the differentiation in TG2-KD NB4 cells, almost 100 (307-209) genes were not up-regulated, whereas approximately 250 (1059-808) genes were not down-regulated compared with the control cells. (C) Real-time Q-PCR verification of microarray data. Fold change induction of 10 selected genes upon ATRA-treatment in control (black bars) and TG2-KD cells (white bars) are shown among the X-axis on the third day of differentiation. Left panel shows the results obtained from the microarray analysis, and right panel demonstrates a representative expression pattern of the same genes determined by real-time Q-PCR. The genes analyzed were the cell surface antigen CD36, the RARα, the TGFβ, the intercellular adhesion molecule 3 (ICAM3), the cell surface antigen CD11c/integrin αX (CD11c/ITGAX), the thrombospondin-1 (THBS1), the MER receptor tyrosine kinase (MERTK), the CC chemokines CCL3 and CCL22, and the TG2. Ratios of fold change differences of analyzed genes in the TG2-KD NB4 cells and the control cells are presented.
Number of genes (and entities) regulated by ATRA in differentiating control and TG2 knockdown NB4. (A-B) Number of annotated genes (and all entities in parentheses) regulated by ATRA in control NB4 (A) and in TG2-KD NB4 cells (B) are visualized as a Venn diagram. Two-fold change difference was set, and significantly changed genes were selected (P ≤ .05 and Bonferoni Multiple Correction was used). The numbers of up- and down-regulated genes are separated. Genes significantly regulated at the both stages of the differentiation (day 2 and day 3) are presented in the overlapping region of Venn diagram (in bold). Note that among the genes up-regulated in both stages of the differentiation in TG2-KD NB4 cells, almost 100 (307-209) genes were not up-regulated, whereas approximately 250 (1059-808) genes were not down-regulated compared with the control cells. (C) Real-time Q-PCR verification of microarray data. Fold change induction of 10 selected genes upon ATRA-treatment in control (black bars) and TG2-KD cells (white bars) are shown among the X-axis on the third day of differentiation. Left panel shows the results obtained from the microarray analysis, and right panel demonstrates a representative expression pattern of the same genes determined by real-time Q-PCR. The genes analyzed were the cell surface antigen CD36, the RARα, the TGFβ, the intercellular adhesion molecule 3 (ICAM3), the cell surface antigen CD11c/integrin αX (CD11c/ITGAX), the thrombospondin-1 (THBS1), the MER receptor tyrosine kinase (MERTK), the CC chemokines CCL3 and CCL22, and the TG2. Ratios of fold change differences of analyzed genes in the TG2-KD NB4 cells and the control cells are presented.
Remarkably, in differentiating TG2-KD NB4 cells there were less up-regulated genes, 254 on day 2, 344 on day 3, and 208 overlapped, than in control ATRA-treated NB4 cells (Figure 3B). On the other hand, there were 1011 down-regulated genes on day 2, 1360 on day 3, and 808 overlapped in differentiating TG2-KD NB4 cells, as compared with control. These data suggest that the presence of TG2 has significant impact on ATRA-induced changes in gene expression in NB4 cells. To validate the microarray results, we confirmed the expression patterns for 10 selected genes by using real-time Q-PCR (Figure 3C).
Functional classification of genes altered by ATRA in differentiating control and TG2-KD NB4 cells
Inasmuch as the gene expression changes are more pronounced on day 3, we focused on the third day differentiated stage for functional classification of the genes with altered expression. Gene sets representing the regulated genes in control and TG2-KD NB4 cells were classified into biological categories using the PANTHER Protein Classification System. As shown in Table 1, several functional categories were found to be significantly enriched (P ≤ .05) among the 2144 genes regulated by ATRA in control NB4 cells at day 3. The identified categories are in accordance with the current knowledge about NB4 cells differentiation, ATRA-induced cell cycle arrest is accompanied by the repression of genes serving the metabolite requirements of cell division or necessary for the cell cycle and protein assembly. Differentiating NB4 cells develop the features of neutrophil granulocytes associated with induction of several immune function-related genes.
Classification of biological processes significantly enriched in the set of genes regulated by ATRA in control and TG2 knockdown NB4 cells at day 3
| Biological process . | No. of genes in reference list . | Control NB4 . | TG2-KD NB4 . | ||||
|---|---|---|---|---|---|---|---|
| No. of genes observed . | No. of genes expected . | P . | No. of genes observed . | No. of genes expected . | P . | ||
| Immunity and defense | 1318 | 199 | 98.16 | 5.00E−19 | 140 | 78.15 | 1.55E−09 |
| Interferon-mediated immunity | 63 | 20 | 4.69 | 1.77E−05 | 6 | 3.74 | NS |
| MHCI-mediated immunity | 22 | 12 | 1.64 | 3.35E−05 | 9 | 1.3 | .0018 |
| Granulocyte-mediated immunity | 64 | 16 | 4.77 | .0056 | 10 | 3.8 | NS |
| T cell–mediated immunity | 194 | 30 | 14.45 | .0311 | 22 | 11.5 | NS |
| Cytokine/chemokine–mediated immunity | 125 | 22 | 9.31 | .0384 | 12 | 7.41 | NS |
| Nucleoside, nucleotide, and nucleic acid metabolism | 3343 | 360 | 248.97 | 1.59E−11 | 272 | 198.23 | 1.44E−06 |
| DNA metabolism | 360 | 57 | 26.81 | 3.04E−05 | 44 | 21.35 | .0014 |
| DNA repair | 169 | 34 | 12.59 | 7.85E−05 | 27 | 10.02 | .0012 |
| tRNA metabolism | 42 | 13 | 3.13 | .0035 | 12 | 2.49 | .0017 |
| Cell cycle | 1009 | 132 | 75.15 | 2.47E−08 | 102 | 59.83 | 7.42E−06 |
| Cell-cycle control | 418 | 67 | 31.13 | 1.72E−06 | 48 | 24.79 | .0028 |
| Protein metabolism and modification | 3040 | 315 | 226.41 | 4.22E−08 | 249 | 180.27 | 3.61E−06 |
| Protein modification | 1157 | 123 | 86.17 | .0108 | 97 | 68.61 | NS |
| Protein complex assembly | 68 | 16 | 5.06 | .0112 | 12 | 4.03 | NS |
| Intracellular protein traffic | 1008 | 103 | 75.07 | .0318 | 86 | 59.77 | .0202 |
| Biological process . | No. of genes in reference list . | Control NB4 . | TG2-KD NB4 . | ||||
|---|---|---|---|---|---|---|---|
| No. of genes observed . | No. of genes expected . | P . | No. of genes observed . | No. of genes expected . | P . | ||
| Immunity and defense | 1318 | 199 | 98.16 | 5.00E−19 | 140 | 78.15 | 1.55E−09 |
| Interferon-mediated immunity | 63 | 20 | 4.69 | 1.77E−05 | 6 | 3.74 | NS |
| MHCI-mediated immunity | 22 | 12 | 1.64 | 3.35E−05 | 9 | 1.3 | .0018 |
| Granulocyte-mediated immunity | 64 | 16 | 4.77 | .0056 | 10 | 3.8 | NS |
| T cell–mediated immunity | 194 | 30 | 14.45 | .0311 | 22 | 11.5 | NS |
| Cytokine/chemokine–mediated immunity | 125 | 22 | 9.31 | .0384 | 12 | 7.41 | NS |
| Nucleoside, nucleotide, and nucleic acid metabolism | 3343 | 360 | 248.97 | 1.59E−11 | 272 | 198.23 | 1.44E−06 |
| DNA metabolism | 360 | 57 | 26.81 | 3.04E−05 | 44 | 21.35 | .0014 |
| DNA repair | 169 | 34 | 12.59 | 7.85E−05 | 27 | 10.02 | .0012 |
| tRNA metabolism | 42 | 13 | 3.13 | .0035 | 12 | 2.49 | .0017 |
| Cell cycle | 1009 | 132 | 75.15 | 2.47E−08 | 102 | 59.83 | 7.42E−06 |
| Cell-cycle control | 418 | 67 | 31.13 | 1.72E−06 | 48 | 24.79 | .0028 |
| Protein metabolism and modification | 3040 | 315 | 226.41 | 4.22E−08 | 249 | 180.27 | 3.61E−06 |
| Protein modification | 1157 | 123 | 86.17 | .0108 | 97 | 68.61 | NS |
| Protein complex assembly | 68 | 16 | 5.06 | .0112 | 12 | 4.03 | NS |
| Intracellular protein traffic | 1008 | 103 | 75.07 | .0318 | 86 | 59.77 | .0202 |
NS indicates not significant.
In TG2-KD NB4 cells, the categories of biological process characteristic for differentiation of control NB4 cells, identified above, contained less number of genes and several subcategories were not significantly overrepresented at reduced TG2 level. These data clearly indicate that induction of TG2 in NB4 cells contributes to the transcriptional remodeling, which leads to the terminal differentiation program of these leukemic cells.
TG2 facilitates the transcriptional alterations induced by ATRA
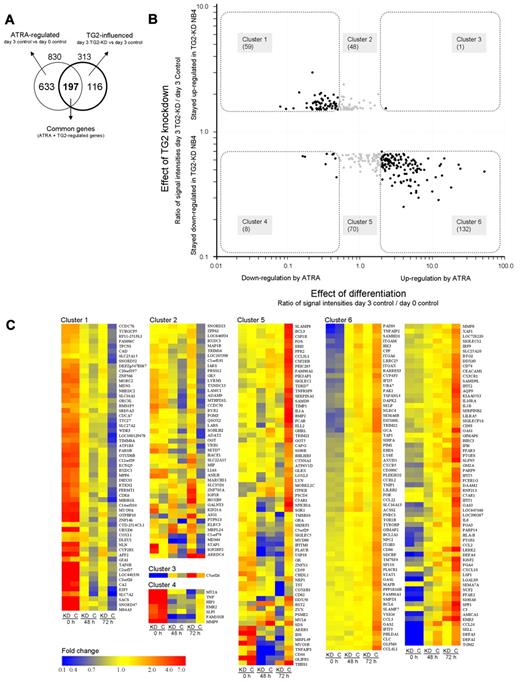
Because the effect of the knockdown of TG2 was found to be the most pronounced on the third day upon the phenotypic features of differentiating NB4, we identified the genes whose expression were affected by TG2 at that stage. We also considered that the regulatory effect of TG2 on gene expression might not be reflected in our results to its full extent because of incomplete (less than 100% efficient) silencing of the gene and the temporary nature of the knock-down effect. To avoid losing potentially TG2-regulated genes, threshold for inclusion in the study was set to 1.5-fold change, which is a relatively low value but a strict significance criterion (P ≤ .05) was retained to select relevant genes. Figure 4A shows in a Venn diagram the overlapping genes regulated by ATRA and modulated by TG2 at the third day of differentiation. We identified 313 genes that are affected by TG2. An important observation is that 197 annotated genes from the 313 (63%) influenced by TG2, are also regulated by ATRA, showing that TG2 has a significant impact mainly on the expression of differentiation related genes.
TG2 influences the expression of ATRA-regulated genes in differentiation of NB4 cell. (A) Genes (830) regulated by ATRA on day 3 belonging in the 4 most affected biological process categories (Table 1) were further investigated in the respect of TG2 effect. The Venn diagram illustrates the number of genes regulated by ATRA (830), the genes influenced by TG2 (313), and the number of the common part of these 2 gene sets (197). (B). The ratios of normalized transcript levels of 3-day ATRA-treated control NB4 cells versus untreated control NB4 cells were calculated in case of selected genes (effect of differentiation). Similarly, the ratios of normalized transcript levels of 3-day ATRA-treated TG2-KD NB4 cells versus 3-day ATRA-treated control NB4 cells were determined (effect of TG2 knockdown). The 2 ratios of transcript levels were plotted against each other resulting in a scatter plot that shows the relationship of transcriptional changes caused by the differentiation process and the TG2 knockdown effect. Down-regulation of 59 genes is behind (cluster 1) and the expression of 8 genes (cluster 4) is suppressed at higher degree than it occurs in ATRA-treated NB4 cells. Whereas 1 gene is more up-regulated by ATRA in TG2-KD cells than normally expected (cluster 3), 132 genes are less induced in TG2-KD NB4 cells (cluster 6) as compared with the control. TG2-influenced genes regulated by ATRA are highlighted, and genes that belong to a certain cluster are indicated. (C) Gene sets regulated by TG2 during ATRA-induced differentiation of NB4 cells. The heat map represents the differentially expressed genes in control and TG2-KD NB4 cells upon ATRA-treatment at undifferentiated, 2-day, and 3-day differentiated stages. Cluster 1 contains the genes (59 genes) that are down-regulated by ATRA in normal case, but remained significantly at higher level in TG2-KD NB4 cells. Cluster 2 (48 genes) and cluster 5 (69 genes) contain those genes whose expression change did not reach the 2-fold threshold upon ATRA-treatment, but were up-regulated or down-regulated in TG2-KD cells, respectively. In cluster 3 (1 gene) and cluster 4 (8 genes) the suppressed TG2 expression further increased the up- or down-regulating effect of ATRA, respectively. Cluster 6 represents the genes (132 genes) significantly induced by ATRA, but showing lower expression in TG2-KD NB4 cells. Heat maps were generated by GeneSpring 10.1 (Agilent) using hierarchical clustering algorithm in all cases of gene lists. Fold change differences are indicated by colors.
TG2 influences the expression of ATRA-regulated genes in differentiation of NB4 cell. (A) Genes (830) regulated by ATRA on day 3 belonging in the 4 most affected biological process categories (Table 1) were further investigated in the respect of TG2 effect. The Venn diagram illustrates the number of genes regulated by ATRA (830), the genes influenced by TG2 (313), and the number of the common part of these 2 gene sets (197). (B). The ratios of normalized transcript levels of 3-day ATRA-treated control NB4 cells versus untreated control NB4 cells were calculated in case of selected genes (effect of differentiation). Similarly, the ratios of normalized transcript levels of 3-day ATRA-treated TG2-KD NB4 cells versus 3-day ATRA-treated control NB4 cells were determined (effect of TG2 knockdown). The 2 ratios of transcript levels were plotted against each other resulting in a scatter plot that shows the relationship of transcriptional changes caused by the differentiation process and the TG2 knockdown effect. Down-regulation of 59 genes is behind (cluster 1) and the expression of 8 genes (cluster 4) is suppressed at higher degree than it occurs in ATRA-treated NB4 cells. Whereas 1 gene is more up-regulated by ATRA in TG2-KD cells than normally expected (cluster 3), 132 genes are less induced in TG2-KD NB4 cells (cluster 6) as compared with the control. TG2-influenced genes regulated by ATRA are highlighted, and genes that belong to a certain cluster are indicated. (C) Gene sets regulated by TG2 during ATRA-induced differentiation of NB4 cells. The heat map represents the differentially expressed genes in control and TG2-KD NB4 cells upon ATRA-treatment at undifferentiated, 2-day, and 3-day differentiated stages. Cluster 1 contains the genes (59 genes) that are down-regulated by ATRA in normal case, but remained significantly at higher level in TG2-KD NB4 cells. Cluster 2 (48 genes) and cluster 5 (69 genes) contain those genes whose expression change did not reach the 2-fold threshold upon ATRA-treatment, but were up-regulated or down-regulated in TG2-KD cells, respectively. In cluster 3 (1 gene) and cluster 4 (8 genes) the suppressed TG2 expression further increased the up- or down-regulating effect of ATRA, respectively. Cluster 6 represents the genes (132 genes) significantly induced by ATRA, but showing lower expression in TG2-KD NB4 cells. Heat maps were generated by GeneSpring 10.1 (Agilent) using hierarchical clustering algorithm in all cases of gene lists. Fold change differences are indicated by colors.
To visualize the relationship between the TG2 modulatory effect and gene expression changes related to the differentiation process, the transcript level ratios of TG2-KD NB4 cells versus control cells at day 3 (effect of TG2 knockdown), as well as differentiated control NB4 versus undifferentiated NB4 (effect of differentiation), were determined and plotted against each other in case of the 313 TG2-dependent genes (Figure 4B). The modulatory effect of TG2-KD is represented along the y-axis, whereas the effect of the ATRA-induced differentiation is visualized along the x-axis, and genes regulated by ATRA are highlighted by black spots.
As shown by the scatter plot, a large proportion of the ATRA-inhibited genes fell into cluster 1, meaning that their expression remained high in TG2-KD NB4 cells. Knocking down of TG2 augments the ATRA-induced repression only in a few cases (cluster 4). These results indicate that the inhibitory effect of ATRA, in case of these genes, is mediated by TG2. In the ATRA-induced genes, there was only one gene that remained up-regulated in TG2-KD cells (cluster 3), whereas 131 showed reduced expression in cells lacking TG2 (cluster 6). This means that TG2 has an important facilitating role in the induction of certain genes characteristic to the differentiation process. Taken together, these results suggest that TG2 and ATRA have mainly synergistic effects during differentiation. Relative expression levels during the differentiation process in control and TG2-KD NB4 cells are presented as a heat map in Figure 4C. Significantly overrepresented gene ontology categories derived from clusters 1 and 6 are presented in supplemental Table 2. Important genes from these 2 clusters, which are relevant to cell-cycle regulation and immune function, are also listed.
TG2 knockdown NB4 cells retain cell proliferation potential
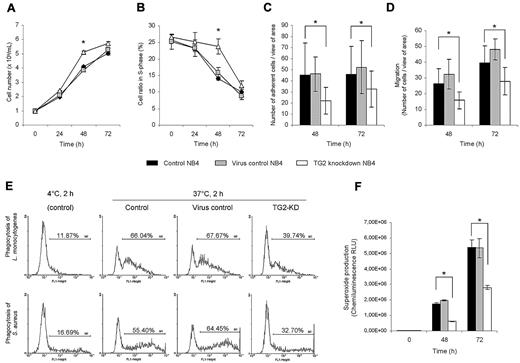
ATRA induces cell-cycle arrest and terminal differentiation of NB4 cells by down-regulating genes, which are responsible for the extensive proliferation of these leukemic cells. Based on the gene expression data, there are several cell proliferation-related genes (such as E2F5, CDK6) whose expression remained at a markedly higher level in TG2-KD NB4 cells. Studying the changes of cell proliferation and cell-cycle distribution profiles, a significant effect of antiTG2 shRNAs expression was observed on both. At 48 hours, there were approximately 27% more cells, and even at 72 hours approximately 10% more proliferating cells, in culture of TG2-KD NB4 than in controls (Figure 5A). Cell-cycle distribution in NB4 cell lines revealed that when TG2 is silenced, significantly more cells were present in S phase on the third day of differentiation (Figure 5B).
Knocking down of TG2 in NB4 cells enhances the cell proliferation potency and reduces the immunological functions during ATRA-induced differentiation. NB4 cells (105 cells/mL) and its sublines (virus control and TG2-KD NB4 cells) were cultured in standard medium in presence of 1μM ATRA for 48 and 72 hours. (A) Cell numbers were determined by the trypan blue dye exclusion method at the 24, 48, and 72 hours states of the differentiation. Figure shows growth curves of control, virus control, and TG2-KD NB4 cells conducted in 3 parallel experiments. The increase in TG2-KD NB4 cell number was statistically significant (P ≤ .05) at 48 and 72 hours. (B) Cell-cycle of NB4 cells and its sublines (minimum 20.000 events collected from each) were analyzed by flow cytometry. Figure shows mean of S-phase ratios of cell cycles from 3 independent experiments at 0, 24, 48, and 72 hours of differentiating cells. The error bars represent SD of means. Increase number of TG2-KD NB4 cells in S-phase at 48 hours was calculated to be statistically significant (P ≤ .05). (C) After 48 and 72 hours of initiation of differentiation, 105 cells in 1 mL were allowed to adhere to plastic surface of 24-well plate for 30 minutes. After removal of nonadherent cells, the number of adherent cells were determined by randomly selected 10 fields of view seen through the eyepieces of the microscope performed in 3 independent experiments. Decrease in adherence in case of TG2-KD NB4 cells was statistically significant (P ≤ .05). (D) Cells (2.5 × 105) from each cell line were placed into the upper chamber of Matrigel Invasion Chamber after 48 hours and 72 hours of initiation of differentiation. Migration was elicited by 200nM fMLP or 10 ng/mL IL-8 containing medium in lower chamber. Numbers of cells that migrated through the chambers were determined as described previously in panel A. The migration time was 14 hours. The decrease in chemotactic activity was calculated to be significant at 48 and 72 hours (P ≤ .05). (E) ATRA differentiated control, virus control, and TG2-KD NB4 cells were fed with 2 types of FITC-labeled bacteria (L monocytogenes and S aureus) with the ratio of 1:50 cell/bacterium for 2 hours in the presence of serum at 72-hour stage of differentiation. Phagocytic capacity was evaluated by flow cytometric analysis. The bars represent mean and SEM of 3 independent experiments (P ≤ .05). Typical flow cytometric profiles of L monocytogenes (top panel) and S aureus (bottom panel) phagocyting 72 hours differentiated cells are shown. (F) Intracellular NADPH-oxidase activity of 48 and 72 hours ATRA differentiated control, virus control, and TG2-KD NB4 cells (106 cells/mL) was induced by 50nM phorbol myristate acetate in 1-mL reaction volume containing 5 μL of L-012 (100μM). Results are the mean ± SD of 3 experiments. Chemiluminescence was detected by MOONLIGHT 2010 luminometer at intervals of 10 seconds. The decrease in ROS production in TG2 knockdown NB4 was calculated to be statistically significant (P ≤ .05).
Knocking down of TG2 in NB4 cells enhances the cell proliferation potency and reduces the immunological functions during ATRA-induced differentiation. NB4 cells (105 cells/mL) and its sublines (virus control and TG2-KD NB4 cells) were cultured in standard medium in presence of 1μM ATRA for 48 and 72 hours. (A) Cell numbers were determined by the trypan blue dye exclusion method at the 24, 48, and 72 hours states of the differentiation. Figure shows growth curves of control, virus control, and TG2-KD NB4 cells conducted in 3 parallel experiments. The increase in TG2-KD NB4 cell number was statistically significant (P ≤ .05) at 48 and 72 hours. (B) Cell-cycle of NB4 cells and its sublines (minimum 20.000 events collected from each) were analyzed by flow cytometry. Figure shows mean of S-phase ratios of cell cycles from 3 independent experiments at 0, 24, 48, and 72 hours of differentiating cells. The error bars represent SD of means. Increase number of TG2-KD NB4 cells in S-phase at 48 hours was calculated to be statistically significant (P ≤ .05). (C) After 48 and 72 hours of initiation of differentiation, 105 cells in 1 mL were allowed to adhere to plastic surface of 24-well plate for 30 minutes. After removal of nonadherent cells, the number of adherent cells were determined by randomly selected 10 fields of view seen through the eyepieces of the microscope performed in 3 independent experiments. Decrease in adherence in case of TG2-KD NB4 cells was statistically significant (P ≤ .05). (D) Cells (2.5 × 105) from each cell line were placed into the upper chamber of Matrigel Invasion Chamber after 48 hours and 72 hours of initiation of differentiation. Migration was elicited by 200nM fMLP or 10 ng/mL IL-8 containing medium in lower chamber. Numbers of cells that migrated through the chambers were determined as described previously in panel A. The migration time was 14 hours. The decrease in chemotactic activity was calculated to be significant at 48 and 72 hours (P ≤ .05). (E) ATRA differentiated control, virus control, and TG2-KD NB4 cells were fed with 2 types of FITC-labeled bacteria (L monocytogenes and S aureus) with the ratio of 1:50 cell/bacterium for 2 hours in the presence of serum at 72-hour stage of differentiation. Phagocytic capacity was evaluated by flow cytometric analysis. The bars represent mean and SEM of 3 independent experiments (P ≤ .05). Typical flow cytometric profiles of L monocytogenes (top panel) and S aureus (bottom panel) phagocyting 72 hours differentiated cells are shown. (F) Intracellular NADPH-oxidase activity of 48 and 72 hours ATRA differentiated control, virus control, and TG2-KD NB4 cells (106 cells/mL) was induced by 50nM phorbol myristate acetate in 1-mL reaction volume containing 5 μL of L-012 (100μM). Results are the mean ± SD of 3 experiments. Chemiluminescence was detected by MOONLIGHT 2010 luminometer at intervals of 10 seconds. The decrease in ROS production in TG2 knockdown NB4 was calculated to be statistically significant (P ≤ .05).
Several immune functions are compromised in differentiating TG-KD NB4
Based on the microarray data, several functional deficiencies in differentiating TG2-KD NB4 cells could be predicted. As many genes with reduced expression levels in TG2-KD NB4 cells belonged to the cell motility or cell adhesion categories (ie, integrins and selectins), we tested whether these expression changes appear at the functional level. Adhesion of neutrophils is crucial for physiological processes of cell migration, chemotaxis, and phagocytosis. Undifferentiated NB4 cells are floating in culture, but after the administration of ATRA, they start to adhere to plastic surfaces. Numbers of adherent cells were determined in the second and third day of the differentiation and a significant reduction in the adherence of TG2-KD NB4 cells was observed (Figure 5C). Microscopic analysis also revealed that differentiating NB4 cells without TG2 are less spread, remain spherical, and are only weekly attached to surface. We also tested whether down-regulation of TG2 expression would affect the migration and chemotaxis of differentiating NB4 cells in response to the chemoattractant IL-8 and/or fMLP, and we observed a reduction in migration of TG2-KD NB4 cells (Figure 5D).
After chemotaxis, phagocytosis of microorganisms by neutrophils is an important prompt response to inflammation. As differentiated TG2-KD NB4 shows marked reduction in the expression of several phagocytosis-related genes, such as CD14, CD36, MERTK, and others, we were interested to see whether phagocytic capacity was influenced by the reduced TG2 level. Approximately 1.5-fold and 2-fold decreases in phagocytosis of L monocytogenes and S aureus were detected, respectively (Figure 5E).
Measurement of highly reactive oxygen species (ROS) generation by neutrophils during activation of respiratory burst is of great importance to evaluate the bactericidal activity of neutrophils. Neutrophils increase their consumption of O2 to generate ROS, as superoxide anion (O2−) and H2O2, by NADPH-oxidase. As neutrophil cytosolic factor 2 (NCF2; P67PHOX/NOXA2), a major component of neutrophil NADPH-oxidase system, was found to be less induced in TG2-KD NB4 cells, ROS production was determined during the differentiation process at the second and the third day. TG2-KD NB4 cells generated approximately 2.5-fold less superoxide anion than controls in accordance with the lower expression of NCF2 (Figure 5F).
Production of CC chemokines involved in the differentiation syndrome is restricted in TG2-KD NB4 cells upon ATRA treatment
In the induction of the members of CC chemokine family microarray profiling showed that CCL2, CCL3, CCL22, and CCL24 were expressed significantly lower in TG2-KD cells at 48 and 72 hours after ATRA-stimulation (Figure 6A). We confirmed their reduced expressions both by real-time Q-PCR and measuring their concentration in the culture fluid (Figure 6B-C). These results reveal that the suppressed TG2 expression can restrict the ATRA-induced CC chemokine productions of APL cells. We could also verify that the lower production of chemokines in TG2-KD NB4 cells leads to reduced chemotaxis of white blood cells in a transwell system (Figure 6D).
Production of CC chemokines induced by ATRA are restricted in TG2 knockdown NB4 cells. NB4 cells (105 cells/mL) and its sublines (virus control and TG2-KD NB4 cells) were cultured in presence of 1μM ATRA for 48 and 72 hours. (A) Relative expressions of CCL2, CCL3, CCL20, and CCL24 chemokines during the ATRA-induced differentiation of control and TG2-KD NB4 cells are presented by a heat map. Results are derived from the microarray experiment. (B) Relative mRNA expressions of CC chemokines were determined at the indicated time points by real-time Q-PCR. Expression levels are normalized to the level of the cyclophilin. (C) Protein levels of CC chemokines were quantified in supernatants of NB4 cells at the indicated time points by ELISA. Figures show typical protein level patterns from 2 or 3 independent experiments measured in duplicates. (D) Chemoattractive effect of supernatant of differentiating control, virus control, and TG2-KD NB4 cells on peripheral white blood cells was evaluated by migration assay. Migration experiments were performed 2 times with 3 parallels in each experiment. Statistical significance between wild-type and TG2-KD cells was determined by using the Student t test P < .05.
Production of CC chemokines induced by ATRA are restricted in TG2 knockdown NB4 cells. NB4 cells (105 cells/mL) and its sublines (virus control and TG2-KD NB4 cells) were cultured in presence of 1μM ATRA for 48 and 72 hours. (A) Relative expressions of CCL2, CCL3, CCL20, and CCL24 chemokines during the ATRA-induced differentiation of control and TG2-KD NB4 cells are presented by a heat map. Results are derived from the microarray experiment. (B) Relative mRNA expressions of CC chemokines were determined at the indicated time points by real-time Q-PCR. Expression levels are normalized to the level of the cyclophilin. (C) Protein levels of CC chemokines were quantified in supernatants of NB4 cells at the indicated time points by ELISA. Figures show typical protein level patterns from 2 or 3 independent experiments measured in duplicates. (D) Chemoattractive effect of supernatant of differentiating control, virus control, and TG2-KD NB4 cells on peripheral white blood cells was evaluated by migration assay. Migration experiments were performed 2 times with 3 parallels in each experiment. Statistical significance between wild-type and TG2-KD cells was determined by using the Student t test P < .05.
Discussion
In our experiments, we have clarified the role of TG2 in the ATRA-induced differentiation of the NB4 promyelocytic leukemia cell line. Undifferentiated NB4 cells do not express TG2, but after the administration of ATRA TG2, a direct target gene of RARα, is strongly induced. Using lentivirus-mediated gene silencing, we generated stable TG2 knockdown NB4 cells that allowed us to study the differentiation process at reduced TG2 expression. Taking into account, that besides the numerous advantages of shRNA-mediated gene silencing, complete and sustained elimination of the expression of a particular gene cannot be achieved, which may attenuate the consequences of knocking down of TG2. Nevertheless, we managed to reduce the level of TG2 during differentiation, as it was confirmed by the determination of protein amount of TG2 by Western blot and by the measurement of its activity (Figure 1).
ATRA-treated TG2-KD NB4 cells lag behind significantly in differentiation as indicated by the decreased level of CD11c mRNA, its surface expression and the diminished NBT reducing capacity of these cells (Figure 2A-C). The delayed differentiation of the knockdown cells is also revealed in several functional consequences such as decreased chemotaxis, adherence, phagocytic capacity, superoxide production, and their sustained proliferative ability. TG2 has been implicated in various physiological phenomena, which can individually bear importance to the function of neutrophils.13 For instance, TG2 was identified as a cell surface receptor for fibronectin and therefore implicated in the adhesion and migration of monocytes or fibroblasts.22,23 Ex vivo–maturated macrophages showed significantly decreased phagocytosis when they were differentiated in the presence of TG2 inhibitor.24 TG2 has been shown to influence the phagocytosis of apoptotic corps by macrophages, most probably through its interaction with integrins and MFG-E8.25
Impairment of neutrophil function by the reduction of TG2 could, in principle, result from the compromise of one or more of the aforementioned processes. The unexpected comprehensiveness of phenotypic changes in TG2-KD NB4, however, suggest these do not represent isolated instances of interference with the molecular machinery of specific granulocyte tasks (phagocytosis, adherence, migration), rather a break-up of the blueprint for the granulocyte-specific molecular apparatus as a whole. This must entail a principally unique regulatory effect of TG2, executed higher upstream during the course of the signaling events that induce NB4 cell differentiation.
The immediate effect of ATRA on gene transcription results in comprehensive reprogramming of the transcriptome. In literature, there is ample evidence that TG2 may influence effectors of gene expression (histones, Sp1) or components of signaling pathways, which in turn impact on transcription. Nuclear TG2 is able to directly cross-link and inactivate the Sp1 transcription factor participating in the development of alcoholic liver disease.26 It was also demonstrated that TG2 modulates the transcriptional activity of the retinoblastoma protein by its cross-linking or kinase activity.27-29 In the cytoplasm, TG2 can catalyze polymer formation of I-κBα monomers, thereby it contributes to the activation of nuclear factor-κB in microglia and breast cancer cells.30,31 To ascertain whether TG2 may be implicated in ATRA-induced gene expression regulation in NB4 cells, total gene expression profiling was carried out.
So far, a whole genome gene expression analysis of ATRA-treated NB4 cells has not been performed. Previous similar analyses involved approximately 13 thousand genes, while our study concerned more than 28 thousand genes.32,33 Besides identifying several new ATRA-regulated genes (supplemental Table 1), we managed to confirm the changes in the expression of several markers of differentiation such as the members of the CCAAT/enhancer-binding protein family (C/EBPβ and ϵ), the helix-loop-helix (HLH) family (BHLHB2, ID-2), the interferon regulatory factors (IRFs), signal transducer and activator of transcription (STATs), and SWI/SNF family of proteins (SMARCD).
The complex modulatory role of TG2 in the differentiation process affects the remodeling of gene expression profile in ATRA-induced differentiated NB4 cells. We identified at least 300 genes whose expression was dependent on the presence of TG2 at some level, and it turned out that they were mostly ATRA-dependent as well. Genes affected by both differentiation (ATRA-dependent) and TG2 seem to be regulated in an opposite direction (Figure 4B). In details, 88.1% of the genes down-regulated by ATRA remained at a higher level in TG2-KD cells, and most genes induced by ATRA in control NB4 cells showed lower expression level in the TG2-KD one. These results raise a possible synergistic/additive effect of TG2 with regulatory role of ATRA.
Detailed analysis of the genes whose expressions were modulated by TG2 revealed 3 important findings. First, among the down-regulated genes by ATRA but held at higher level in TG2-KD cells, the cell-cycle and cell proliferation–related genes were significantly overrepresented (eg, E2F5, CDK6). This result led us to assume that TG2-KD cells remain more proliferative for a longer time; therefore TG2 is necessary for the attenuation of cell proliferation. Indeed, TG2-KD cells have higher dividing capacity with more cells distributed in S-phase of the cell cycle in the early stage of the differentiation program (Figure 5A-B). However, it has to be noticed that we did not observe difference in the surviving capability of differentiated TG2-KD cells as compared with the differentiated control NB4 cells during long-term culturing (see supplemental Figure 1), or in the rate of apoptosis induced by arsenic-trioxide treatment (date not shown).
Second is that the genes, which are ATRA-induced but less expressed in TG2-KD cells during differentiation, mostly belong to the immunity and defense (granulocyte-, interferon-, and cytokine/chemokine-mediated immunity), the cell motility and the cell adhesion categories. In TG2-KD cells, expression of key molecules involved in chemotaxis, phagocytic capacity, and superoxide production are missing from the normally up-regulated genes in control ATRA-treated cell. One of these is the paxillin that is necessary for adhesion and motility of leukocytes. PAK1/p21protein (Cdc42/Rac)–activated kinase 1 is known for taking part in the regulation of chemotaxis, chemokin-indused cytoskeletal actin polymerization, and oxidative burst.34,35 We found that the expression of S100A8, a recently described proinflammatory protein expressed by phagocytes and implicated both in NADPH oxidase activation by interaction with NCF2/p67PHOX or transepithelial migration of neutrophils also remained at lower level in TG2-KD NB4 cells.36,37 These examples suggest that TG2 is required for the development of the full innate immune function of differentiating NB4 cells including their full inflammatory responsiveness (Figure 5C-F).
Third is related to the role of TG2 in the process of inflammation. TG2 is implicated in the enhancement of inflammation, inasmuch as TG2 inhibitory peptide significantly decreased the production of inflammatory cytokines and neutrophil infiltration into lung in lipopolysaccharide-treated mice.38 Furthermore, TG2 knockout mice are partially resistant to lipopolysaccharide-elicited experimental septic shock with increased survival, a diminished inflammatory response, and attenuated organ damage.9,39 The expression level of the CCL2 inflammatory mediator was consistently up-regulated in ATRA-treated APL patients.9 The simultaneous expression of cytokines such as TNF-α, IL-1β, IL8, and CCL2 by ATRA-treated leukemia cells, such as NB4 cells, may result in both increased binding to epithelial cells and chemotactic transmigration and thereby further accelerate tissue infiltration.40-42 Despite the corticosteroid treatment, a massive induction of CC chemokines (eg, CCL2) appears in ATRA-treated APL patients, which might lead to the differentiation syndrome with excessive inflammatory response.8,9,41 Because knocking down of TG2 in NB4 cells is accompanied by reduced chemokine production (CCL2, CCL3, CCL22, and CCL24) upon ATRA-treatment and therefore leads to decreased development of chemoattractive capacity, we propose that there is an essential role of TG2 in the regulation of inflammatory responsiveness and development of differentiation syndrome (Figure 6).
Based on these findings presented here, it may be a viable option to interfere with such a pathological condition through modulating TG2 level in the differentiating leukemic cells. Specific down-regulation of TG2 expression by gene silencing or using appropriate compounds may lead to gene-specific suppression of cytokine secretion in DS patients. Such a therapeutic approach may also work in other TG2-related in gain-of-function diseases like neurodegeneration, fibrosis, inflammation, cardiac failure, and celiac disease.13
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Iván Uray and Dr Máté Demény for their helpful comments and providing critical reading of the manuscript.
This work was supported by the following grants: Hungarian Scientific Research Funds (OTKA NI 67 877, K 61 868, K 77 587) and EU grants MRTN-CT-2006-036032, MRTN-CT-2006-035624, and LSHB-CT-2007-037730.
Authorship
Contribution: K.C. designed, performed experiments, analyzed microarray data, quantified gene expression, and wrote the manuscript; I.N. performed the cell adhesion and migration experiments; L.F. provided guidance for the study design, financial support, interpretation, took part in writing of the manuscript, and final approval of the version to be submitted; and Z.B. developed the concept and interpretation, designed the study, performed experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zoltán Balajthy, Department of Biochemistry and Molecular Biology, University of Debrecen, Medical and Health Science Center, H-4012 Debrecen, Nagyerdei krt 98, Hungary; e-mail:balajthy@dote.hu.

![Figure 1. Generation of stabile TG2 knockdown NB4 cell line. (A) NB4 cells (control NB4), NB4 cells transduced by random, nontarget shRNA expressing lentivirus (virus control NB4) and NB4 cells transduced by specific shRNA against TG2 expressing lentivirus (TG2 knockdown NB4) were differentiated in the presence of 1μM ATRA for 0, 24, 48, 72, and 96 hours. mRNA expression of TG2 was determined at the indicated time points by real-time Q-PCR, measurements were conducted in triplicates, values are expressed as mean % ± SD of the mean. (B) Control, virus control, and TG2 knockdown NB4 cells were differentiated in the presence of 1μM ATRA for 96 hours. Top panel shows cytosolic activity of TG2 measured at the indicated time point by detecting incorporation of [3H]putrescine into casein. The amount of incorporated [3H]putrescine was determined in a β-counter. Bars depict the means of 3 separate experiments each performed in duplicates. Inset in top panel shows the cytosolic protein levels of TG2 at the indicated time points detected by Western blot. Total protein (25 μg) was loaded in each line, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, blotted and developed with CUB7402 monoclonal antibody against TG2. Bottom panel shows nuclear activity of TG2 measured by detecting incorporation of [3H]putrescine into casein. Inset in bottom panel shows the Western blot analysis of nuclear TG2. TG2 enzyme activity assays and Western blot analyses were performed as described above.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/19/10.1182_blood-2010-01-266064/5/m_zh89991061230001.jpeg?Expires=1769180210&Signature=D86AciJOqTIjbqu98oW7sx9KNhWJKeqU4BazNXuaJGhD2luNGnPlMIBrhsURSBORUi8BXPzdLhfSkxxOsx2M5NZeaPJ6-4ITMJO9muxg3EUi38a3SNkjstbguvYh46bDPbEJhBM4YrRZZwNljhIpwJ7qch-tZksXUeJfF4PUHGmO5p0fAtUomAJ6mlO2PQL7~nvMwF-x-haWZFzAPQW2MLoMu47E9HUSKQkkYkQWCQ-chLkVyKWFplIBqcV~nShO5H~N4knTVAKzIIoE4ukzgUM0IKaURVgn1AwBj3Zivh1BqFTU99tQLPdGIj~crYdxyLy71Ychy7FGPbxEG5~Jqw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal