Abstract
Natural killer (NK) cells are lymphocytes of the innate immune system that, following differentiation from CD56bright to CD56dim cells, have been thought to retain fixed functional and phenotypic properties throughout their lifespan. In contrast to this notion, we here show that CD56dim NK cells continue to differentiate. During this process, they lose expression of NKG2A, sequentially acquire inhibitory killer cell inhibitory immunoglobulin-like receptors and CD57, change their expression patterns of homing molecules, and display a gradual decline in proliferative capacity. All cellular intermediates of this process are represented in varying proportions at steady state and appear, over time, during the reconstitution of the immune system, as demonstrated in humanized mice and in patients undergoing hematopoietic stem cell transplantation. CD56dim NK-cell differentiation, and the associated functional imprint, occurs independently of NK-cell education by interactions with self–human leukocyte antigen class I ligands and is an essential part of the formation of human NK-cell repertoires.
Introduction
Natural killer (NK) cells participate in immune responses by the production of cytokines and chemokines, as well as by killing transformed and infected cells.1,2 In peripheral blood, mature NK cells represent 10%-15% of all lymphocytes and are commonly divided into 2 main CD3− subsets, CD56bright and CD56dim cells.3,4
In contrast to mouse NK cells,5 many details of human NK-cell development and differentiation remain undefined.6 Work by Caligiuri and colleagues have suggested that early NK-cell development in humans, from CD34+ hematopoietic stem cells (HSCs) into CD56bright NK cells, occurs in 4 discrete steps.7-9 CD56bright NK cells are then believed to be precursors of CD56dim NK cells. Such linear differentiation is supported by observations that CD56bright NK cells have longer telomeres than CD56dim NK cells,10 are more prevalent in blood early after hematopoietic stem cell transplantation (HSCT),11 and differentiate into CD56dim NK cells in humanized mice engrafted with human HSCs.12
Whereas both T and B cells undergo differentiation upon antigen encounter, CD56dim NK cells have been thought to retain a stable phenotype and function during their lifespan. This notion is supported by the observation that NK cells have a rapid turnover time in blood of approximately 2 weeks,13 and the fact that each individual has a genetically determined repertoire of killer cell immunoglobulin-like receptors (KIRs) that is stable over time.14 More recently, however, it has been shown that CD56dim NK cells are functionally fine-tuned by interactions between cell-surface receptors, including inhibitory and activating KIRs as well as NKG2A with self-human leukocyte antigen (HLA) class I ligands15-18 This process, referred to as “education” or “licensing,” (hereafter termed education) seems to be tunable and depends on the net signaling received by NK cells, thus introducing a new level of functional heterogeneity to distinct NK-cell subsets.19,20 Furthermore, in experimental models of cytomegalovirus (CMV) infection, NK cells have been shown to possess features previously attributed to cells of the adaptive immune system, including the expansion of a specific subset, followed by contraction and formation of cells with memory-like characteristics.21
The recently discovered functional heterogeneity and features shared with the adaptive immune system indicate that NK-cell differentiation and regulation of function might be more dynamic than previously appreciated. However, it is currently not known whether NK cells continue to differentiate beyond the transition from CD56bright to CD56dim cells. Furthermore, if they differentiate, it is not clear what functional consequences such differentiation would have. Discrete steps of T-cell differentiation, occurring in response to T-cell receptor (TCR)-dependent and -independent stimuli, can be distinguished by a wide array of surface molecules, including isoforms of CD45, CCR7, CD27, CD28, and CD57.22 Among these, CD57 has been suggested as a marker for replicative senescence and is associated with terminal differentiation and altered functional capacities in CD8+ T cells.23-25 Numerous studies have documented the expression of CD57 on human NK cells.23,26-30 The observations that CD57 expression is absent on fetal NK cells,31 increases with age,32,33 and is associated with replicative senescence23 suggest that CD57 may define a subpopulation of highly differentiated NK cells.
Here, we have examined how CD57 expression relates to quantitative and qualitative changes in the phenotype and function of human NK cells. We delineate a continuous differentiation process of CD56dim NK cells that ultimately ends in cells with poor responsiveness to cytokine stimulation, but retained cytolytic capacity. All cellular intermediates of this process are represented in varying proportions at steady state and appear, over time, during the development of the immune system in humanized mice as well as in patients undergoing HSCT. Furthermore, we show that differentiation of CD56dim NK cells is uncoupled from the functional tuning by self-HLA class I ligands during NK-cell education. Our data support a model in which the mature NK-cell repertoire is maintained via the continuous replenishment of NK cells undergoing a differentiation process characterized by multiple phenotypic and functional changes and gradual loss of proliferative capacity.
Methods
Human subjects and cell lines
This study was approved by the Regional Ethics Committee of Stockholm, Sweden. Buffy coats and peripheral blood mononuclear cells (PBMCs) were separated by density gravity centrifugation (Ficoll-Hypaque; GE Healthcare). PBMCs were either used directly for experiments or cryopreserved in liquid nitrogen in 10% dimethyl sulfoxide (DMSO) and 90% heat-inactivated fetal bovine serum (FBS) for later usage. KIR and HLA genotyping were performed on genomic DNA as previously described.34 The human erythroleukemia cell line, K562 (ATCC), was maintained in RPMI 1640 supplemented with 10% heat-inactivated FBS, 1mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Human cord blood was obtained from 11 healthy full-term newborns at the Karolinska University Hospital, Stockholm, Sweden, with parental informed consent in accordance with the Declaration of Helsinki, and CD34+ cells were enriched by a MACS cell-separation system using anti-CD34 microbeads, according to the manufacturer's instructions (Miltenyi Biotec).
Patients
Five patients with hematologic malignancies undergoing HSCT were sampled at weeks 4, 8, 16, 34, and 52 post-HSCT. PBMCs were isolated and cryopreserved in liquid nitrogen for later use. Pretransplantation conditioning was either fludarabine-based of reduced intensity, or myeloablative with busulphan and cyclophosphamide. Graft-versus-host disease prophylaxis consisted of rabbit antithymocyte globuline and cyclosporine A.
mAbs and flow cytometry
Monoclonal antibodies (mAbs) against the following proteins were used: CD57, CD56, CD3, NKG2A, KIR2DL1, KIR2DL1/S1, KIR2DL3, KIR2DL2/L3/S2, KIR3DL1, NKG2C, LILRB1, NKp46, NKp30, CD62L, CXCR3, CXCR4, CCR5, interleukin (IL)-2Rα, IL-2Rβ, IL-12Rβ, IL-18Rα, interferon-γ (IFN-γ), CD107a, Ki67, CD45, CD14, and CD19. Anti-KIR3DL2 mAb (Dx31) was kindly provided by Dr. J. Philips (DNAX Research Institute), and mAb against KLRG1 was kindly provided by Dr. H. Pircher (Department of Immunology, Institute of Medical Microbiology and Hygiene, University of Freiburg, Freiburg, Germany). Fluorescence-activated cell sorting (FACS) staining and analysis were performed as previously reported.34,35
FACS sorting
NK cells were freshly isolated from PBMCs of group A KIR haplotype healthy donors by negative selection using the NK-cell Isolation Kit (Miltenyi Biotec). Next, the enriched NK cells were stained with appro-priate antibody combinations and sorted using a FACSAria instrument (BD Biosciences).
Proliferation assay
For carboxyfluorescein succinimidyl ester (CFSE) labeling, NK cells were washed in phosphate-buffered saline (PBS), resuspended at 1 × 107 cells/mL in serum-free medium with 2μM of CFSE, and incubated for 7 minutes at 37°C and 5% CO2. The labeling was interrupted by adding an equal volume of medium containing 5% FBS for 10 minutes at 37°C and 5% CO2, and the cells were washed twice in medium containing 5% FBS. Labeled cells were plated in U-bottomed 96-well plates, stimulated with IL-2 (500 U/mL), IL-12 (10 ng/mL), IL-15 (10 ng/mL), and IL-18 (100 ng/mL) or combinations thereof and incubated at 37°C and 5% CO2 for 5 days.
Functional assays
PBMCs (1 × 106 cells/mL) were mixed with K562 cells at a ratio of 10:1 in V-bottomed 96-well plates, centrifuged at 300 rpm for 3 minutes, and incubated for 6 hours at 37°C and 5% CO2. After incubation, cells were harvested by centrifugation and surface stained with appropriate antibody combinations, including CD107a and IFN-γ, for functional evaluation as previously described.36
Generation of humanized mice
All animal experiments were approved by the local animal ethics committee. Humanized mice were generated as previously described.37 In brief, pregnant BALB/c Rag2−/−γcR−/− mice were injected with 100 μL busulphan (15 mg/kg; Sigma-Aldrich) at day 18 of gestation. Newborns were irradiated with 500 rad from a Cs source and injected intrahepatically with 1 × 105 CD34+ cells within 4 hours after irradiation. Six weeks later, 80 μL of blood was taken from the tail vein and analyzed for the presence of human cells (anti-CD45 and anti-CD3) by flow cytometry. Engrafted animals were used for subsequent experiments.
IL-15 treatment of humanized mice
Human IL-15 and human IL-15Rα-Fc (R&D Systems) were prepared according to the manufacturer's instructions. Before injection, human IL-15 and human IL-15Rα-Fc solutions were mixed and incubated for 20 minutes at 37°C. Humanized mice were injected intraperitoneally with 100 μL of human IL-15 (2.5 μg) mixed with human IL-15Rα-Fc (7.5 μg) or PBS once-weekly until sacrifice. All animals were killed by cervical dislocation, and spleens, thymuses, livers, BM, blood, and lymph nodes were collected for analysis. In transfer experiments, 7 × 105 NK cells of each subset were injected in the tail vein of 10-week-old mice that were treated with IL-15/IL-15Rα on days 0 and 4 before cells were recovered from the liver on day 7.
Statistical analysis
The following statistical tests were used, unless otherwise indicated in the figure legends. For multiple group comparisons, 1-way analysis of variance (ANOVA) or Kruskal-Wallis nonparametric tests were applied. For comparisons of independent groups, the Student t test or the Mann-Whitney U test were performed. For comparisons of matched groups, the paired Student t test or Wilcoxon matched test were performed. In figures, n.s. indicates not significant (***P < .001; **P < .01; and *P < .05). Statistical analyses were performed using GraphPad software Version 5.0.
Results
NKG2A, KIR, and CD57 independently correlate with the proliferative capacity of CD56dim NK cells
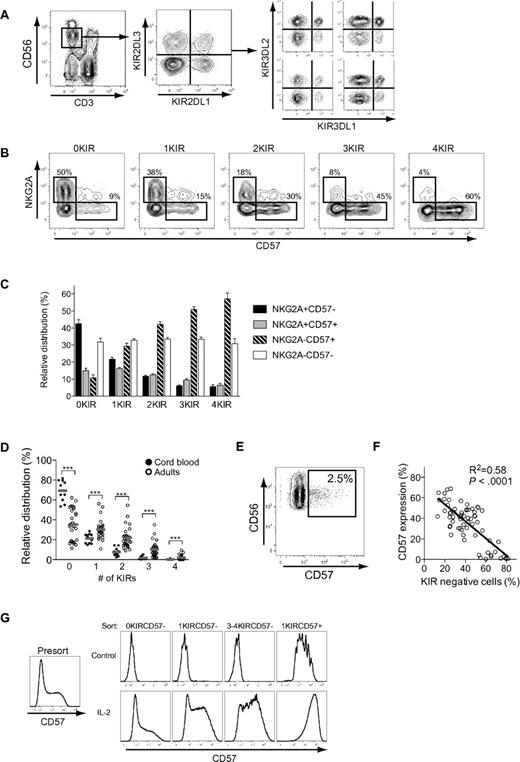
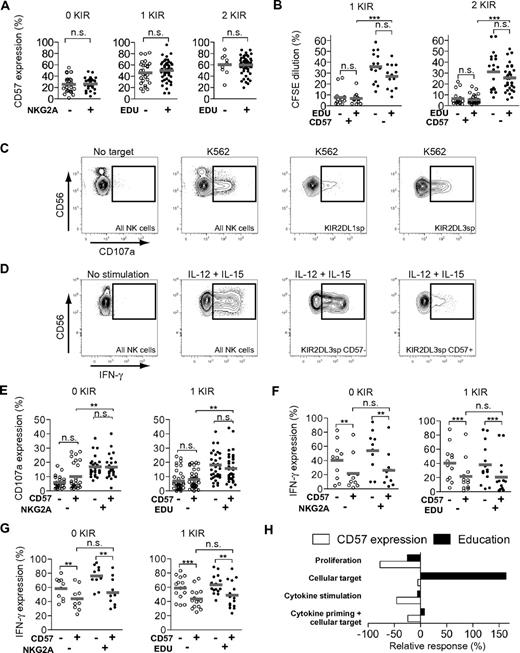
We set out to examine the possible stages of continued differentiation within the human CD56dim NK-cell subset. As a point of departure, we analyzed the expression of CD57 on human NK cells in a cohort of 26 individuals and found that, on average, 40% of the CD56dim NK cells expressed CD57, with a significant interindividual variation ranging from 5% to 70% (Figure 1A). Expression of CD57 was confined to the CD56dim NK-cell subset, and CD57+CD56dim NK cells proliferated significantly less than CD57−CD56dim NK cells upon stimulation with different combinations of cytokines (Figure 1B-C).
NKG2A, KIR, and CD57 independently correlate with the proliferative capacity of CD56dim NK cells. (A) Representative staining for CD57 on NK cells from one healthy donor. (B) Representative staining for CFSE and CD57 on NK cells from one healthy donor after 5 days of culture with the indicated cytokines. (C) Frequency of CD57− and CD57+ NK cells diluting CFSE on days 0, 3, 5, and 7 after stimulation with the indicated cytokines (n = 6; mean ± standard error of the mean [SEM]). (D) Histograms of one representative experiment in which the indicated NK-cell subsets were purified by FACS sorting and stimulated with IL-2 for 5 days. (E) Dilution of CFSE on FACS-sorted NK-cell subsets stimulated with IL-2 for 5 days (n = 8; mean). (F) Dilution of CFSE on CD57− and CD57+ NK-cell subsets expressing 0, 1, 2, 3, or 4 KIRs stimulated with IL-2 for 5 days (n = 9 0KIR, 36 1KIR, 54 2KIRs, 36 3KIRs, 9 4KIRs; mean).
NKG2A, KIR, and CD57 independently correlate with the proliferative capacity of CD56dim NK cells. (A) Representative staining for CD57 on NK cells from one healthy donor. (B) Representative staining for CFSE and CD57 on NK cells from one healthy donor after 5 days of culture with the indicated cytokines. (C) Frequency of CD57− and CD57+ NK cells diluting CFSE on days 0, 3, 5, and 7 after stimulation with the indicated cytokines (n = 6; mean ± standard error of the mean [SEM]). (D) Histograms of one representative experiment in which the indicated NK-cell subsets were purified by FACS sorting and stimulated with IL-2 for 5 days. (E) Dilution of CFSE on FACS-sorted NK-cell subsets stimulated with IL-2 for 5 days (n = 8; mean). (F) Dilution of CFSE on CD57− and CD57+ NK-cell subsets expressing 0, 1, 2, 3, or 4 KIRs stimulated with IL-2 for 5 days (n = 9 0KIR, 36 1KIR, 54 2KIRs, 36 3KIRs, 9 4KIRs; mean).
This initial observation led us to, in detail, dissect the proliferative capacity of different CD56dim NK-cell subsets. NKG2A and inhibitory KIR are major receptors that shape the functional responses of human NK cells. Therefore, we speculated that expression of these receptors could be additional determinants of proliferative capacity other than CD57. To examine this possibility, we isolated CD56bright NK cells and 4 different CD56dim NK-cell subsets: NKG2A−KIR−CD57−, NKG2A+KIR−CD57−, NKG2A− single KIR+CD57−, and NKG2A− single KIR+CD57+ cells. Sorted cells were stimulated with IL-2 for 5 days and evaluated for proliferative capacity. Lack of NKG2A and expression of KIRs independently correlated with reduced proliferation, and coexpression of CD57 was associated with a completely abolished proliferative response to cytokines (Figure 1D-E). To address whether the cellular expression of multiple KIRs affect proliferative responses, CD56dim NK cells expressing 0-4 KIRs, with or without coexpression of CD57, were analyzed. These experiments revealed a small, but significant, decrease in proliferation by CD57− NK cells expressing 2-4 KIRs, compared with those expressing only 1 KIR and KIR-negative NK cells (Figure 1F). Again, coexpression of CD57 correlated with almost completely abrogated proliferative responses, regardless of the number of KIRs expressed (Figure 1F). Loss of proliferative capacity has commonly been associated with shortening of telomeres.23 Here, we confirmed a previous report that CD56dim NK cells had shorter telomeres than CD56bright NK cells,10 but no further shortening of the telomeres was detected in NK cells that expressed multiple KIRs and/or CD57 (supplemental Figure 1; available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These results reveal that the expression of CD57, as well as NKG2A and KIRs, independently correlates with the proliferative capacity of human CD56dim NK cells.
Expression patterns of CD57, KIR, and NKG2A define different stages of on CD56dim NK-cell differentiation
Since expression of CD57, KIRs, and NKG2A all correlated with the proliferative capacity of CD56dim NK cells, we next determined how subsets expressing these receptors relate to each other. Expression patterns of NKG2A and CD57 on CD56dim NK-cell subsets expressing 0, 1, 2, 3, or 4 inhibitory KIRs (16 distinct subsets) were assessed in peripheral blood from healthy individuals (Figure 2A). When examining CD57 expression in relation to the expression of KIR and NKG2A, we found that CD57 expression increased gradually with the expression of inhibitory KIRs and correlated inversely with the expression of NKG2A (Figure 2B-C). Similar analyses were performed in 11 cord-blood specimens to assess how these subsets were distributed in naive NK-cell repertoires. CD56dim NK cells derived from cord blood expressed very few KIRs, compared with NK cells from adult individuals (Figure 2D), and almost completely lacked surface expression of CD57 (Figure 2E). The absence of CD57 expression in naïve NK-cell repertoires,31 characterized by low-KIR–expression frequencies, strengthens the correlation between KIR expression and appearance of CD57 on CD56dim NK cells. Indeed, when analyzing the expression of CD57 in relation to the total frequency of KIR-negative NK cells, we found that individuals with high frequencies of KIR− NK cells had low frequencies of CD57+ CD56dim NK cells (Figure 2F). Furthermore, CD57 expression was more easily induced by IL-2 stimulation in vitro on sorted CD57− CD56dim NK cells that expressed multiple KIRs than on subsets with few or zero KIRs (Figure 2G). Conversely, sorted CD57+ CD56dim NK cells did not lose expression of CD57 or KIRs during culture in the absence or presence of cytokines.
Expression patterns of NKG2A, CD57, and KIRs on CD56dim NK cells. (A) Identification of NK cells and representative stainings for expression of the 4 major inhibitory KIRs. (B) Representative staining for expression of CD57 and NKG2A on NK-cell subsets defined by expression of 0, 1, 2, 3, or 4 KIRs. (C) Relative distribution of CD57- and NKG2A-expressing cells within NK-cell subsets defined by expression of 0, 1, 2, 3, or 4 KIRs on CD56dim NK cells from 38 individuals homozygous for the group A KIR haplotype (n = 38; mean ± SEM). (D) Frequencies of NK cells from adult individuals (n = 29) and cord blood (n = 11) expressing 0-4 KIRs (***P < .001; mean). (E) Representative staining of CD57 on NK cells from one cord blood specimen. (F) Correlation between frequency of KIR-negative NK cells and expression of CD57 (n = 66). (G) NK cells were FACS sorted based on expression of CD57 and KIRs into the indicated subsets, stimulated for 5 days with IL-2, and then assessed for the expression of CD57 (n = 6; one representative example is shown).
Expression patterns of NKG2A, CD57, and KIRs on CD56dim NK cells. (A) Identification of NK cells and representative stainings for expression of the 4 major inhibitory KIRs. (B) Representative staining for expression of CD57 and NKG2A on NK-cell subsets defined by expression of 0, 1, 2, 3, or 4 KIRs. (C) Relative distribution of CD57- and NKG2A-expressing cells within NK-cell subsets defined by expression of 0, 1, 2, 3, or 4 KIRs on CD56dim NK cells from 38 individuals homozygous for the group A KIR haplotype (n = 38; mean ± SEM). (D) Frequencies of NK cells from adult individuals (n = 29) and cord blood (n = 11) expressing 0-4 KIRs (***P < .001; mean). (E) Representative staining of CD57 on NK cells from one cord blood specimen. (F) Correlation between frequency of KIR-negative NK cells and expression of CD57 (n = 66). (G) NK cells were FACS sorted based on expression of CD57 and KIRs into the indicated subsets, stimulated for 5 days with IL-2, and then assessed for the expression of CD57 (n = 6; one representative example is shown).
The observations that NKG2A, KIR, and CD57 independently correlated with proliferative responses, together with the distinct expression patterns of these molecules on naïve and adult human NK cells, suggest that loss of NKG2A, sequential acquisition of KIRs, and CD57 expression reflect a continued differentiation process occurring in CD56dim NK cells.
Reconstitution of human CD34+ cells in BALB/c Rag−/−γcR−/− mice reveals a gradual appearance of KIR+CD57+ NK cells
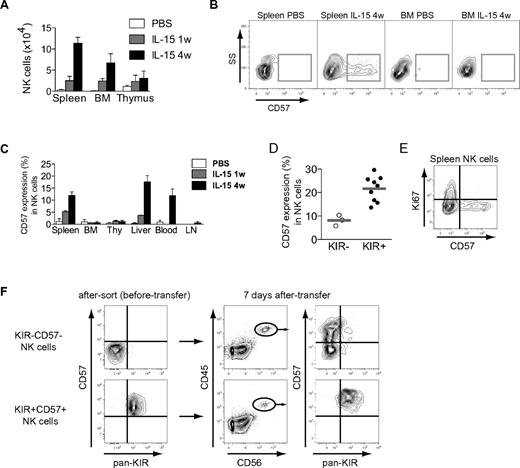
So far, data suggest that CD57 expression may define a late stage of CD56dim NK-cell differentiation. If this notion is correct, such cells should appear later than CD57− NK cells during NK-cell development. To test this prediction, we studied the development of NK cells in a humanized mouse model. BALB/c Rag2−/−γcR−/− mice were reconstituted with human cord blood–derived CD34+ HSCs. After engraftment, the mice were given weekly injections of human IL-15/IL-15Rα complexes and sacrificed after 1 or 4 injections. In agreement with previous studies,12 IL-15 treatment yielded higher NK-cell numbers in all analyzed compartments in the mice (Figure 3A, see supplemental Figure 2 for the gating strategy used to identify NK cells). CD57 expression started to appear 1 week after the first injection of IL-15/IL-15Rα and was found on a substantial fraction of splenic, peripheral blood, and intrahepatic NK cells after 4 weeks of IL-15 treatment (Figure 3B-C). Interestingly, very few CD57+ NK cells were recovered from compartments expected to harbor more naïve NK-cell subsets, such as the bone marrow, lymph node, and thymus (Figure 3B-C).6 Similar to our findings in the peripheral blood of healthy humans, CD57 expression was more common in KIR-expressing NK cells (Figure 3D). Furthermore, CD57+ NK cells were negative for Ki67, suggesting that this subset had not undergone recent proliferation in vivo (Figure 3E). To directly test the relationship between the studied subsets, we sorted and transferred CD57−KIR− and CD57+KIR+ NK cells into irradiated BALB/c Rag2−/−γcR−/− mice. Transpresentation of IL-15 induced the differentiation from CD57−KIR− into CD57+CD56dim NK cells, a proportion of which (on average, 7.4%) had acquired inhibitory KIRs after 7 days (Figure 3F). We could not observe any loss of NKG2A during the 7-day time frame (data not shown). Importantly, all CD57+KIR+ NK cells retained the expression of these molecules following IL-15 treatment in vivo. Taken together, these findings support our assumption that the expression of CD57 is a late event during differentiation of CD56dim NK cells.
Differentiation of NK cells in BALB/c Rag2−/−γcR−/− mice. (A) NK-cell number in different organs from humanized mice that have received PBS, or human IL-15 + human IL-15Rα-Fc intraperitoneally weekly for 1 or 4 weeks (w), as determined by flow cytometry (n = 3 per group; mean ± SEM) (see supplemental Figure 2 for gating algorithm used to identify human NK cells), BM (bone marrow). (B) Representative staining for CD57 on gated human NK cells from the indicated organs of PBS- or IL-15–treated mice at the indicated time point (4 weeks). (C) Frequency of CD57 expression on gated human NK cells in the indicated organs of PBS- or IL-15–treated mice at 1 and 4 weeks (n = 3 per group; mean ± SEM), Thy (thymus), LN (lymph nodes). (D) Frequency of CD57 expression within subsets of KIR− and KIR+ human NK cells from spleens of mice treated with IL-15 for 4 weeks (n = 3 KIR−, 9 KIR+; mean). (E) Representative staining for Ki67 and CD57 on gated human splenic NK cells from mice treated with IL-15 for 4 weeks. (F) The indicated subsets were sorted and transferred into irradiated BALB/c Rag2−/−γcR−/− mice. Shown are recovered NK cells at day +7 following 2 rounds of IL-15/IL-15Rα stimulation. One representative experiment of 2 is shown (n = 3 per group).
Differentiation of NK cells in BALB/c Rag2−/−γcR−/− mice. (A) NK-cell number in different organs from humanized mice that have received PBS, or human IL-15 + human IL-15Rα-Fc intraperitoneally weekly for 1 or 4 weeks (w), as determined by flow cytometry (n = 3 per group; mean ± SEM) (see supplemental Figure 2 for gating algorithm used to identify human NK cells), BM (bone marrow). (B) Representative staining for CD57 on gated human NK cells from the indicated organs of PBS- or IL-15–treated mice at the indicated time point (4 weeks). (C) Frequency of CD57 expression on gated human NK cells in the indicated organs of PBS- or IL-15–treated mice at 1 and 4 weeks (n = 3 per group; mean ± SEM), Thy (thymus), LN (lymph nodes). (D) Frequency of CD57 expression within subsets of KIR− and KIR+ human NK cells from spleens of mice treated with IL-15 for 4 weeks (n = 3 KIR−, 9 KIR+; mean). (E) Representative staining for Ki67 and CD57 on gated human splenic NK cells from mice treated with IL-15 for 4 weeks. (F) The indicated subsets were sorted and transferred into irradiated BALB/c Rag2−/−γcR−/− mice. Shown are recovered NK cells at day +7 following 2 rounds of IL-15/IL-15Rα stimulation. One representative experiment of 2 is shown (n = 3 per group).
Late appearance of KIR+CD57+ NK cells during immune reconstitution after HSCT in humans
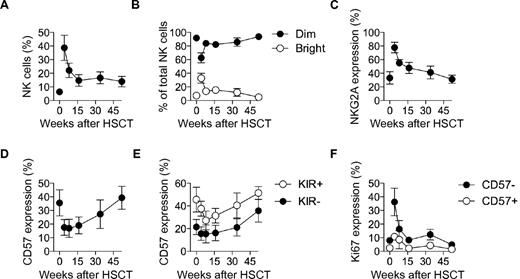
HSCT provides an opportunity for longitudinal studies of the restoration of the immune system in humans. To consolidate our studies in the humanized mouse model, we next examined NK-cell reconstitution in patients undergoing allogeneic HSCT. As shown in previous investigations, NK cells were highly overrepresented among lymphocytes early after HSCT, dominated by CD56bright and NKG2A+ CD56dim NK cells (Figures 4A-C).11,38,39 Percentages of CD57+ CD56dim NK cells were low during the first 4 months following HSCT before gradually returning to levels similar to those of the donors (Figure 4D). Corroborating our observations in the mouse model, CD57 was predominantly expressed on KIR+ NK cells (Figure 4E). Furthermore, Ki67 expression was largely confined to CD57− NK cells and peaked early after HSCT, consistent with the occurrence of homeostatic proliferation at this stage (Figure 4F). In conclusion, the late appearance of KIR+CD57+ NK cells in the periphery in immunologically reconstituted mice and in humans undergoing HSCT supports the hypothesis that these cells have reached a late differentiation stage.
Differentiation of NK cells in humans following HSCT. (A) Frequency of NK cells out of total lymphocytes in donor (time-point zero) and HSCT-recipient (longitudinally sampled at weeks 4, 8, 16, 34, and 52 post-HSCT) pairs. (B) Frequency of CD56bright and CD56dim cells out of total NK cells for donor and HSCT-recipient pairs. (C) Frequency of NKG2A expressing CD56dim NK cells out of total NK cells for donor and HSCT-recipient pairs. (D) Frequency of CD57 expressing CD56dim NK cells out of total NK cells for donor and HSCT-recipient pairs. (E) Frequency of CD57 expressing KIR− and KIR+ CD56dim NK cells out of total NK cells for donor and HSCT-recipient pairs. (F) Frequency of Ki67 expressing CD57+/−CD56dim NK cells for donor and HSCT-recipient pairs. For A-F; n = 5; mean ± SEM.
Differentiation of NK cells in humans following HSCT. (A) Frequency of NK cells out of total lymphocytes in donor (time-point zero) and HSCT-recipient (longitudinally sampled at weeks 4, 8, 16, 34, and 52 post-HSCT) pairs. (B) Frequency of CD56bright and CD56dim cells out of total NK cells for donor and HSCT-recipient pairs. (C) Frequency of NKG2A expressing CD56dim NK cells out of total NK cells for donor and HSCT-recipient pairs. (D) Frequency of CD57 expressing CD56dim NK cells out of total NK cells for donor and HSCT-recipient pairs. (E) Frequency of CD57 expressing KIR− and KIR+ CD56dim NK cells out of total NK cells for donor and HSCT-recipient pairs. (F) Frequency of Ki67 expressing CD57+/−CD56dim NK cells for donor and HSCT-recipient pairs. For A-F; n = 5; mean ± SEM.
Phenotypic changes during differentiation of CD56dim NK cells
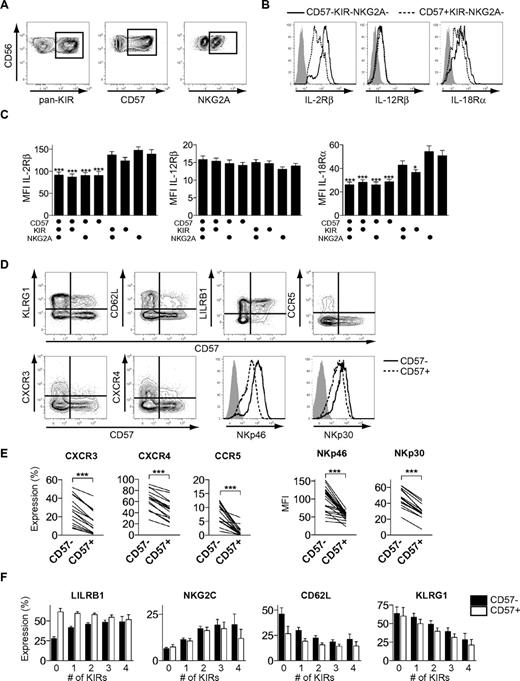
To gain further insights into the proposed differentiation process, we assessed the cell-surface expression of cytokine and chemokine receptors as well as multiple activation and inhibitory receptors on different subsets of CD56dim NK cells. Acquisition of KIRs had a small, but significant, effect on the expression of IL-18Rα (Figure 5A-C). In line with the profound effect of differentiation on cytokine-induced proliferation, the expression of CD57 was associated with significantly reduced levels of both IL-18Rα and IL-2Rβ. In contrast, expression of IL-12Rβ chain was similar on all studied NK-cell subsets and loss of NKG2A had no influence on the expression of any of the major cytokine receptors (Figure 5C). Furthermore, CD57+CD56dim NK cells expressed lower levels of the chemokine receptors, CXCR3, CXCR4, and CCR5, as well as the activation receptors, NKp46 and NKp30, compared with CD57−CD56dim NK cells (Figure 5D-E). Finally, the expression of CD57 as well as the acquisition of multiple KIRs was associated with changes in the expression of LILRB1, NKG2C, CD62L, and KLRG1 (Figure 5D-F). Together, these data show that the differentiation of CD56dim NK cells is associated with coordinated changes in the expression of cytokine receptors, activation, and inhibitory NK-cell receptors, as well as in molecules involved in homing to BM and secondary lymphoid tissues.
Phenotypic characteristics of differentiating CD56dim NK cells. (A) Boolean gating scheme to identify CD57-, pan-KIR- (KIR2DL1, KIR2DL3, KIR3DL1, and KIR3DL2), and NKG2A-expressing cells within CD56dim NK cells. (B) Representative staining for expression of IL-2Rβ, IL-12Rβ, and IL-18Rα on CD57−KIR−NKG2A− and CD57+KIR−NKG2A− CD56dim NK cells. Isotype control in gray. (C) Mean fluorescence intensity (MFI) of IL-2Rβ, IL-12Rβ, and IL-18Rα on different CD56dim NK-cell subsets (n = 10; *P < .05, ***P < .001, for indicated groups vs CD57−KIR−NKG2A+, 1-way ANOVA with Bonferroni posttest; mean ± SEM). (D) Representative costaining for expression of CD57 and the indicated receptors on CD56dim NK cells. Isotype control in gray. (E) Frequency of CXCR3-, CXCR4-, and CCR5-expressing cells (left) and MFI of NKp46 and NKp30, respectively, within the CD57− and CD57+ NK-cell population (n = 16-23; ***P < .001). (F) Expression of LILRB1, NKG2C, CD62L, and KLRG1 on CD57− and CD57+ CD56dim NK-cell subsets expressing 0-4 KIRs (n = 36-146 for LILRB1 and NKG2C and 6-36 for CD62L and KLRG1; mean ± SEM).
Phenotypic characteristics of differentiating CD56dim NK cells. (A) Boolean gating scheme to identify CD57-, pan-KIR- (KIR2DL1, KIR2DL3, KIR3DL1, and KIR3DL2), and NKG2A-expressing cells within CD56dim NK cells. (B) Representative staining for expression of IL-2Rβ, IL-12Rβ, and IL-18Rα on CD57−KIR−NKG2A− and CD57+KIR−NKG2A− CD56dim NK cells. Isotype control in gray. (C) Mean fluorescence intensity (MFI) of IL-2Rβ, IL-12Rβ, and IL-18Rα on different CD56dim NK-cell subsets (n = 10; *P < .05, ***P < .001, for indicated groups vs CD57−KIR−NKG2A+, 1-way ANOVA with Bonferroni posttest; mean ± SEM). (D) Representative costaining for expression of CD57 and the indicated receptors on CD56dim NK cells. Isotype control in gray. (E) Frequency of CXCR3-, CXCR4-, and CCR5-expressing cells (left) and MFI of NKp46 and NKp30, respectively, within the CD57− and CD57+ NK-cell population (n = 16-23; ***P < .001). (F) Expression of LILRB1, NKG2C, CD62L, and KLRG1 on CD57− and CD57+ CD56dim NK-cell subsets expressing 0-4 KIRs (n = 36-146 for LILRB1 and NKG2C and 6-36 for CD62L and KLRG1; mean ± SEM).
Differentiation and education are parallel, but uncoupled, processes
Since NK cells are functionally tuned by interactions between KIRs and/or NKG2A, with self-HLA class I molecules,15,16,18 we next asked whether NK-cell education was associated with the acquisition of CD57 and loss of proliferative capacity. First, we assessed whether education by self-KIRs and/or NKG2A influenced the expression of CD57. To this end, CD57 expression was monitored on NK-cell subsets with or without the expression of NKG2A that were single/double positive for KIR2DL1, KIR2DL3, KIR3DL1, and KIR3DL2. The education status of the respective subsets was determined through KIR and KIR-ligand genotyping. Of note, NK cells expressing only KIR3DL2 were considered to be uneducated, based on previous investigations.18,34 No difference in CD57 expression could be detected when comparing educated and uneducated subsets (Figure 6A; see supplemental Figure 3A for stratification of data into HLA-C1/C1, C2/C2, and Bw4/Bw6 donors). Hence, NK cells that acquire a self-KIR do not up- or down-regulate CD57 in this process. Since acquisition of KIR, per se, had an impact on proliferative capacity (Figure 1E-F), we studied whether education via KIR influenced the proliferative responses to cytokine stimulation. As noted before, CD57−KIR+ CD56dim NK-cell proliferated significantly better than CD57+KIR+CD56dim NK cells upon cytokine stimulation (Figure 6B). However, educated and uneducated single- or double-KIR expressing CD57− NK-cell subsets proliferated equally well (Figure 6B). Thus, whereas both expression of CD57 and KIRs were associated with reduced proliferative responses, the nature of the expressed KIR (ie, self- or nonself) had no impact on the proliferative capacity of CD56dim NK cells.
CD56dim NK-cell differentiation and education are uncoupled events. (A) Expression of CD57 on KIR−NKG2A+/− (left), singleKIR+NKG2A− educated or uneducated (middle) CD56dim NK-cell subsets, and NKG2A− subsets coexpressing 2 KIRs (right) (n = 28 left, 27 and 48 middle, 9 and 54 right; n.s., not significant; mean). (B) CFSE dilution in singleKIR+CD57+/− educated and uneducated NK-cell subsets (left) and CD57+/− subsets coexpressing 2 KIRs (right) after 5 days of IL-2 stimulation (n = 13 and 14 left, 20 and 45 right; ***P < .001; mean). (C) Representative example of an HLA C1/C1 homozygous donor showing CD107a expression of uneducated (KIR2DL1) and educated (KIR2DL3) after coculture with K562 cells for 6 hours. (D) Representative example of an HLA C1/C1 homozygous donor showing IFN-γ expression after 24 hours of stimulation with IL-12 + IL-15 of the indicated subsets. (E) Degranulation (CD107a expression) in KIR−NKG2A+/−CD57+/− (left) and singleKIR+NKG2A−CD57+/− educated and uneducated CD56dim NK-cell subsets after coculture with K562 cells for 6 hours (n = 26 left, 37 and 34 right; n.s., not significant; ** P < .01; mean). (F) Expression of IFN-γ in KIR−NKG2A+/−CD57+/− (left) and singleKIR+NKG2A−CD57+/− educated and uneducated CD56dim NK-cell subsets after 24 hours of stimulation with IL-12 + IL-15 (n = 10 left, 12 and 14 right; n.s., not significant; ** P < .01, *** P < .001; mean). (G) Expression of IFN-γ in KIR−NKG2A+/−CD57+/− (left) and singleKIR+NKG2A−CD57+/− educated and uneducated CD56dim NK-cell subsets after 18 hours of priming with IL-12 + IL-15 and 6 hours of coculture with K562 cells (n = 10 left, 15 right; **P < .01, ***P < .001; mean). (H) Relative contribution of CD57 and education to detected responses after stimulation with IL-2 for proliferation, K562 cells for degranulation, IL-12 + IL-15 for IFN-γ production, and IL-12 + 15 priming, followed by coculture with K562 cells for IFN-γ production (mean).
CD56dim NK-cell differentiation and education are uncoupled events. (A) Expression of CD57 on KIR−NKG2A+/− (left), singleKIR+NKG2A− educated or uneducated (middle) CD56dim NK-cell subsets, and NKG2A− subsets coexpressing 2 KIRs (right) (n = 28 left, 27 and 48 middle, 9 and 54 right; n.s., not significant; mean). (B) CFSE dilution in singleKIR+CD57+/− educated and uneducated NK-cell subsets (left) and CD57+/− subsets coexpressing 2 KIRs (right) after 5 days of IL-2 stimulation (n = 13 and 14 left, 20 and 45 right; ***P < .001; mean). (C) Representative example of an HLA C1/C1 homozygous donor showing CD107a expression of uneducated (KIR2DL1) and educated (KIR2DL3) after coculture with K562 cells for 6 hours. (D) Representative example of an HLA C1/C1 homozygous donor showing IFN-γ expression after 24 hours of stimulation with IL-12 + IL-15 of the indicated subsets. (E) Degranulation (CD107a expression) in KIR−NKG2A+/−CD57+/− (left) and singleKIR+NKG2A−CD57+/− educated and uneducated CD56dim NK-cell subsets after coculture with K562 cells for 6 hours (n = 26 left, 37 and 34 right; n.s., not significant; ** P < .01; mean). (F) Expression of IFN-γ in KIR−NKG2A+/−CD57+/− (left) and singleKIR+NKG2A−CD57+/− educated and uneducated CD56dim NK-cell subsets after 24 hours of stimulation with IL-12 + IL-15 (n = 10 left, 12 and 14 right; n.s., not significant; ** P < .01, *** P < .001; mean). (G) Expression of IFN-γ in KIR−NKG2A+/−CD57+/− (left) and singleKIR+NKG2A−CD57+/− educated and uneducated CD56dim NK-cell subsets after 18 hours of priming with IL-12 + IL-15 and 6 hours of coculture with K562 cells (n = 10 left, 15 right; **P < .01, ***P < .001; mean). (H) Relative contribution of CD57 and education to detected responses after stimulation with IL-2 for proliferation, K562 cells for degranulation, IL-12 + IL-15 for IFN-γ production, and IL-12 + 15 priming, followed by coculture with K562 cells for IFN-γ production (mean).
In agreement with recent literature,15 a higher frequency of educated NK cells degranulated after stimulation with K562 cells, compared with uneducated NK cells (Figure 6C-E). Interestingly, CD57+ NK cells responded equally well as CD57− NK cells when the educational status in relation to KIRs and NKG2A was taken into account (Figures 6C-E; see supplemental Figure 3B for stratification of data into HLA-C1/C1, C2/C2, and Bw4/Bw6 donors). In contrast, significantly fewer CD57+CD56dim NK cells produced IFN-γ following stimulation with IL-12 and IL-15, regardless of their educational status (Figure 6D-F; see supplemental Figure 3C for stratification of data into HLA-C1/C1, C2/C2, and Bw4/Bw6 donors). These data suggest that education by self-KIRs or NKG2A modulates cytotoxic NK-cell responses, whereas expression of CD57 correlates with poor proliferative capacity and has a negative impact on IFN-γ production in response to cytokines. These results are consistent with the finding that more highly differentiated CD57+CD56dim NK cells expressed lower levels of IL-2Rβ (Figure 5B-C). Priming of NK cells with IL-12 and IL-15 before stimulation with K562 cells revealed that fewer CD57+ cells than CD57− cells produced IFN-γ (Figure 6G). Interestingly, such cytokine priming abolished the difference in responsiveness of educated and uneducated CD56dim NK cells to K562 (Figure 6G; see supplemental Figure 3D for stratification of data into HLA-C1/C1, C2/C2, and Bw4/Bw6 donors).
In summary, our analysis demonstrates that NK-cell education regulates cytotoxic responses against cellular targets, whereas it has no significant effect on proliferation and IFN-γ secretion after cytokine stimulation (Figure 6H). In contrast, NK cells expressing CD57 respond less well to cytokine stimulation and display poor proliferative capacity. Combined, these results suggest that differentiation and education are parallel, but uncoupled, processes with distinct effects on NK-cell function.
Discussion
Compared with other lymphocyte subsets, NK cells are considered to be short-lived cells that retain relatively static phenotypic and functional properties throughout their lifespan. However, recent reports have challenged this notion by revealing a previously unappreciated functional heterogeneity within the mature human NK-cell pool and the emergence of long-lived NK cells harboring memory characteristics.15,17,19-21,40 In this context, we set out to investigate whether CD56dim NK cells are subject to differentiation. Based on a detailed analysis of NK-cell repertoires at steady state and in 2 distinct settings of immune system development, we present data suggesting that human CD56dim NK cells undergo a continuous differentiation process characterized by a gradual loss of proliferative capacity as well as coordinated changes in the surface expression of molecules involved in NK-cell activation and homing.
Previous literature have shown that surface expression of CD57 could be useful for the identification of cells with poor proliferative capacity, possibly indicating a state of cellular senescence.23-25 In this respect, one important early observation was that CD57 expression is more frequent on CD56dim NK cells that lack NKG2A and express multiple KIRs. Together, this suggested a differentiation scheme in which NKG2A+KIR−CD57− CD56dim NK cells appear earlier and are separated in a temporal fashion from NKG2A−KIR+CD57+ CD56dim NK cells. By reconstituting BALB/c Rag2−/−γcR−/− mice with human cord blood–derived CD34+ HSCs,12 we show that CD57+ NK cells appear gradually over time following IL-15 administration. Adoptive transfer experiments revealed that CD57−KIR−CD56dim cells differentiate into CD57+KIR+/−CD56dim NK cells, but not vice versa, supporting the hypothesized differentiation scheme. Differentiation of CD56dim NK cells was also examined in patients undergoing allogeneic HSCT. Previous work has shown that the appearance of CD56bright NK cells precedes that of CD56dim NK cells after transplantation.11,38 Moreover, on a receptor level, NKG2A is expressed before the acquisition of KIRs on differentiating NK cells.41 Extending these findings, our longitudinal analysis of patients undergoing HSCT shows that up to 12 months are needed for the NK-cell compartment to reach similar levels of CD57-expressing CD56dim NK cells in the recipient as in the donor. Consistent with our hypothesis, CD57+ NK cells preferentially coexpressed KIRs and showed no evidence of recent proliferation, as determined by the absence of Ki67 expression in both models of immune reconstitution.
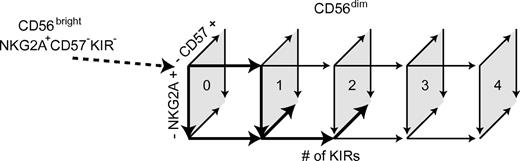
Our results suggest a unidirectional differentiation of CD56dim NK cells characterized by a gradual loss of NKG2A expression that is paralleled by the sequential acquisition of inhibitory KIRs and the appearance of CD57. Whereas loss of NKG2A may be a reversible event,42 the acquisition of KIRs and expression of CD57 were not reversible in vitro or in vivo. However, it was obvious from our studies that all CD56dim NK cells do not differentiate according to a fixed linear scheme. Rather, all possible NKG2A and KIR expression patterns are represented on CD57+ and CD57−CD56dim NK cells at steady state. Hence, some NK cells may acquire up to 4 KIRs and still express NKG2A, but lack CD57. Conversely, other cells lack KIR, but express CD57, and display an inability to proliferate. These results define a scheme of possible pathways with multiple cellular intermediates in a continuous differentiation of CD56dim NK cells from NKG2A+KIR−CD57− to NKG2A−4KIR+CD57+CD56dim cells (Figure 7). Given the uneven distribution of the different subsets and the variation among donors,43 we propose that certain pathways of differentiation are more probable than others and determine the appearance of the final steady-state NK-cell repertoires in each individual.
Schematic model of CD56dim NK-cell differentiation. Numbers within the quadrates show the number of expressed KIRs. Arrows in boldface indicate major routes for differentiation.
Schematic model of CD56dim NK-cell differentiation. Numbers within the quadrates show the number of expressed KIRs. Arrows in boldface indicate major routes for differentiation.
Yu and colleagues recently showed that CD94highCD56dim NK cells represent an intermediary differentiation step between CD94highCD56bright and CD94lowCD56dim NK cells.44 CD94 pairs with either NKG2A or NKG2C to form a heterodimer on the surface,45 and it is probable that the CD94highCD56dim NK-cell population partially overlaps with the naïve NKG2A+KIR−CD57− NK cells described here. Albeit CD94/NKG2A expression can be helpful for the discrimination of CD56dim NK cells at different levels of differentiation, we propose that the differentiation of CD56dim NK cells is nonlinear and has many phenotypically and functionally distinct intermediary subsets. Another recent paper discriminated 2 functionally distinct subsets within the CD56dim NK-cell compartment based on the expression of CD62L.46 It was proposed that CD62L+ NK cells represent a polyfunctional subset that may differentiate into CD62L− NK cells. These findings are, at least in part, compatible with our results, since we observed a gradual decrease in frequencies of CD62L expression with the acquisition of inhibitory KIRs and lower frequencies of CD62L+ NK cells among CD57+CD56dim NK cells. Consistent with our results, the immature CD62L+ NK-cell subset was more responsive to cytokine stimulation. It is possible that one may obtain a more precise view on the relationship between discrete cellular stages of CD56dim NK-cell differentiation by superimposing the 3 reported models based on CD94, CD62L, and CD57, respectively.
Since acquisition of KIRs was associated with the cellular differentiation and expression of CD57, we addressed whether the process of NK-cell education, through acquisition of one or more self-KIRs, is intertwined with the differentiation process described here. Surprisingly, we observed that the expression of CD57 was similar on educated and uneducated NK cells. Consistent with this observation, educated and uneducated single KIR+NKG2A−CD56dim NK cells proliferated equally well in response to cytokine stimulation. Furthermore, the education status of the cells did not affect cytokine production in response to IL-12/IL-15 stimulation. Instead, NK-cell differentiation, as determined by the expression of CD57, was associated with significantly reduced cytokine responses by both educated and uneducated subsets. These results suggest that NK-cell education and differentiation are parallel but uncoupled processes.
Mechanistic insights into the poor cytokine responses and reduced proliferative capacity of more differentiated NK-cell subsets was provided by the observation that IL-2Rβ, which is the major signaling unit for IL-2 and IL-15, and IL-18Rα were expressed at lower levels on KIR+CD57+CD56dim NK cells. Yu et al recently reported that differences in functionality between CD94low and CD94highCD56dim NK cells was dependent on differential IL-12–mediated phosphorylation of signal transducer and activator of transcription (STAT) 4.44 Juelke et al reported that the more mature CD62L−CD56dim NK-cell subset displayed reduced levels of STAT-5 phosphorylation.46 Our finding that CD57+ NK cells expressed lower levels of IL-2Rβ and IL-18Rα supports the notion that more differentiated cells respond poorly to cytokines, including IL-2, IL-15, and IL-18. Importantly, the reduced IFN-γ production in response to cytokines was not a consequence of an intrinsic inability to produce cytokines. In fact, Lopez-Vergès et al show in this issue of Blood that differentiation, as defined by the expression of CD57, is associated with enhanced degranulation and release of IFN-γ following stimulation with anti-CD16, linked to higher expression of CD16 on this subset.47 Notably, we have recently shown that CD56dim NK cells are more prominent cytokine and chemokine producers than CD56bright NK cells upon target cell recognition.48 Hence, the increased responsiveness to target cells (and decreased responses to cytokines) observed for CD57+ NK cells is in line with a gradual shift in functionality occurring during differentiation from CD56bright via CD57−CD56dim to CD57+CD56dim NK cells.
The observation that CD56dim NK cells continue to differentiate raises the question of whether human NK cells are as short-lived as was previously thought. The notion that NK cells may live longer is supported by data from an experimental CMV mouse model in which NK cells with adaptive memory characteristics persist for at least 90 days.21 It remains elusive which factors drive differentiation of CD56dim NK cells during normal homeostasis and whether it is influenced by acute or chronic infection. Studies examining the longevity and phenotype of human NK cells that expand as a consequence of acute viral infection are warranted. Infection and/or TLR ligation of dendritic cells induce transpresentation of IL-15, which prime and promote survival of resting NK cells.49-51 Thus, IL-15 is a central cytokine for normal homeostasis and immune responses to infection. Therefore, it is conceivable that the observed induction of CD57 on NK cells may be triggered during infection challenges. In fact, it was recently demonstrated that chronic HIV-1 infection can cause an increase of CD57+ NK cells,28 possibly reflecting the differentiation and expansion of a human “memory” NK-cell compartment. In addition, a gradual transition from naïve to more mature NK-cell repertoires must occur sometime during childhood, possibly as a result of multiple infection challenges at this point in life, since NK cells in cord blood express very few KIRs and no CD57. Interestingly, this process seems to continue slowly during life, since CD57+ NK cells accumulate with age.32,33
In conclusion, we have reported that CD56dim NK cells undergo a continuous differentiation process associated with multiple phenotypic and functional changes. This finding adds to a growing number of reports challenging the belief that NK cells are invariable cells with fixed properties.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Stella Jacobsson (Department of Medicine, Karolinska Institutet) for her help with the humanized mice, Professor Göran Roos and Dr Ulrika Svenson (Department of Medical Biosciences, Pathology, Ureå University) for their assistance in telomere assays, and Dr Stella Larsson at Transfusion Medicine, Karolinska University Hospital, for access to buffy coats.
This work was supported by grants from the Swedish Foundation for Strategic Research, the Swedish Research Council, the Swedish Cancer Society, the Swedish Children's Cancer Foundation, the Cancer Society of Stockholm, the Royal Swedish Academy of Sciences, the German Research Foundation for the Cluster of Excellence REBIRTH and the International Training Group 1273, the Tobias Foundation, the Söderberg Foundation, the Belvén Foundation, the Åke Wiberg Foundation, and the Karolinska Institutet.
Authorship
Contribution: N.K.B. designed and performed research, analyzed data, and wrote the manuscript; P.R., F.H., S.A., C.F., M.A.I., and A.T.B. designed and performed research and analyzed data; M.F.T. and M.E.R. contributed with the humanized model and analyzed data; J.M. designed research, analyzed data, and wrote the manuscript; C.A.G. analyzed data; H.G.L. analyzed data and wrote the manuscript; and K.J.M. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karl-Johan Malmberg, Center for Infectious Medicine, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, 141 86 Stockholm, Sweden; e-mail: kalle.malmberg@ki.se.

![Figure 1. NKG2A, KIR, and CD57 independently correlate with the proliferative capacity of CD56dim NK cells. (A) Representative staining for CD57 on NK cells from one healthy donor. (B) Representative staining for CFSE and CD57 on NK cells from one healthy donor after 5 days of culture with the indicated cytokines. (C) Frequency of CD57− and CD57+ NK cells diluting CFSE on days 0, 3, 5, and 7 after stimulation with the indicated cytokines (n = 6; mean ± standard error of the mean [SEM]). (D) Histograms of one representative experiment in which the indicated NK-cell subsets were purified by FACS sorting and stimulated with IL-2 for 5 days. (E) Dilution of CFSE on FACS-sorted NK-cell subsets stimulated with IL-2 for 5 days (n = 8; mean). (F) Dilution of CFSE on CD57− and CD57+ NK-cell subsets expressing 0, 1, 2, 3, or 4 KIRs stimulated with IL-2 for 5 days (n = 9 0KIR, 36 1KIR, 54 2KIRs, 36 3KIRs, 9 4KIRs; mean).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/19/10.1182_blood-2010-04-281675/5/m_zh89991060310001.jpeg?Expires=1769082656&Signature=G2d6qF2lmSZSjoG8q2QiHkBuZ-rTJ6JZ3jIU3lMLkVPxuZiC~UON-0wBk9z~SBTnHxmNMCtA~sCgv~hRZPi3qspzhQoaex6DUTK6ffLZjXCWP4nW45oUDsUBhUj1pG0ekrpFT5mbq9zPwEQJ~Af1DGpLn77DMgFA4AgZ~5fSvL7qVi9V3OvfR0RYaeYNFpKJ08vUDFbMgczpNzKDuJWzy7f4ueFxDJTqoXyfyaivGjm534mdesatwIMDLeQdyUJ6s-Vl2JB0T14OSf8qV0Q3cRAj-I5X-47ZGr1TBsk9yDooAhfE~IBZf93YxDTphlRw~aGWKjWlZncIhckaoQDQyQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal