Abstract
Therapeutic targeting of virus-encoded proteins using cellular immunotherapy has proved successful for Epstein-Barr virus (EBV)–associated posttransplant lymphoproliferative disease. However, the more limited repertoire and immunogenicity of EBV-encoded proteins in other malignancies such as Hodgkin lymphoma and extranodal natural killer (NK)/T lymphoma has been more challenging to target. The immunosubdominant latent membrane protein 2 (LMP2) is considered the optimal target in such Latency II tumors, although data relating to its expression in T/NK malignancies are limited. In addressing the validity of LMP2 as an immunotherapeutic target we found that LMP2-specific effector CD8+ T cells recognized and killed EBV-positive NK- and T-cell tumor lines, despite an apparent absence of LMP2A protein and barely detectable levels of LMP2 transcripts from the conventional LMP2A and LMP2B promoters. We resolved this paradox by identifying in these lines a novel LMP2 mRNA, initiated from within the EBV terminal repeats and containing downstream, epitope-encoding exons. This same mRNA was also highly expressed in primary (extra-nodal) NK/T lymphoma tissue, with virtually undetectable levels of conventional LMP2A/B transcripts. Expression of this novel transcript in T/NK-cell lymphoproliferative diseases validates LMP2 as an attractive target for cellular immunotherapy and implicates this truncated LMP2 protein in NK- and T-cell lymphomagenesis. This study is registered at clinicaltrials.gov as NCT00062868.
Introduction
Expression of viral proteins in Epstein-Barr virus (EBV)–associated tumors has allowed specific therapeutic targeting of such antigens with cellular immunotherapies. This is best exemplified by the highly immunogenic lymphoproliferations arising in the T cell–compromised host after allogeneic organ or hematopoietic stem cell transplantation (posttransplant lymphoproliferative disease; PTLD),1 which express the full complement of EBV latent antigens as seen in lymphoblastoid cell lines (LCLs) generated by EBV infection of B lymphocytes in vitro.2 A more restricted pattern of EBV latent gene expression is manifest in malignancies such as Hodgkin lymphoma (HL). The so-called Latency II characteristic of these tumors confines expression of EBV-encoded proteins to Epstein-Barr virus nuclear antigen 1 (EBNA1), latent membrane protein 1 (LMP1), LMP2A, and LMP2B. This has presented a greater challenge for clinicians hoping to target such tumors with antigen-specific adoptive T-cell therapy, because these Latency II viral proteins are significantly less immunogenic than the additional Latency III viral antigens, particularly EBNA3A, EBNA3B, and EBNA3C, expressed in PTLD.2 Cytotoxic T-cell lines (CTLs) generated by in vitro stimulation with LCLs contain low frequencies of T cells specific for LMP2, LMP1, and EBNA1, with accordingly suboptimal clinical efficacy against HL.3 To address this, investigators have recently focused on skewing the EBV-specific CTL response by an in vitro system of LMP2 overexpression in antigen-presenting cells resulting in an expansion of polyclonal populations of both CD4+ and CD8+ effectors specific for LMP2.4 This approach has achieved sustained tumor responses (including some complete responses) in patients with relapsed/refractory HL, with evidence of in vivo expansion and penetration to tumor sites of LMP2-specific T cells.5
EBV-associated malignancies of natural killer (NK) and T-cell origin are also thought to display a ‘Latency II’ pattern of EBV gene expression. Extra-nodal NK/T-cell lymphoma (ENKTL) is an aggressive malignancy, occurring at a median age of 50 years and most commonly presenting in the upper-aerodigestive tract.6 EBV is invariably present within the malignant cells in all cases of ENKTL, irrespective of geographical origin.7 The clonal and episomal form of EBV in tumor biopsy material8 implicates a pathogenic role for the virus in the early stages of lymphomagenesis. Although most reports suggest that EBV antigen expression in ENKTL is of a Latency II type, it should be noted that, when expressed, LMP1 protein is usually evident in a subpopulation of malignant cells, while detection of LMP2 in tumor tissue has only been shown at the mRNA level.9 Also recognized within the spectrum of EBV-associated NK- and T-cell lymphoproliferations is chronic active EBV (CAEBV); a disease characterized by chronic infectious mononucleosis–like symptoms associated with an unusual pattern of anti-EBV antibodies and the pathognomonic presence of monoclonal EBV within NK cells or CD4+ T cells.10 The pattern of viral gene expression in CAEBV is also thought to be Latency II, at least at the mRNA level.11
ENKTL is inherently resistant to anthracycline-based chemotherapy regimens such as CHOP,12 and the outcome of extra-nasal and advanced stage disease is extremely poor.6 Even for localized disease, in spite of high rates of initial response to involved-field radiotherapy, up to 50% of such patients will relapse, usually within a year of completing first-line therapy.13 Recently published data from the retrospective International T-cell project study demonstrated a median overall survival for the whole study cohort of 7.8 months, representing the poorest survival of all T-cell lymphoma subtypes examined.6 CAEBV also confers a high mortality due to complications including hemophagocytic lymphohistiocytosis (HLH) and transformation to lymphoma.14 Allogeneic stem cell transplantation is thought to be the only potentially curative therapy for most patients with CAEBV.15 Novel therapeutic approaches are therefore urgently required for the spectrum of EBV-associated NK- and T-cell malignancies.
From the clinical experience of immunotherapeutic targeting of EBV antigens in HL, it is clear that effector T-cell responses to the immunosubdominant LMP2 protein are the most important. Although preliminary clinical data have also suggested responses in patients with ENKTL and CAEBV5 to CTL preparations containing LMP2-specific effectors, the expression of LMP2 protein has not been clearly demonstrated in NK- or T-cell malignancies, nor are in vitro data available on the validity of LMP2 as an immunologic target.
LMP2 is unusual in that it is encoded by transcripts that span the terminal repeat (TR) region of the viral genome and therefore can only be expressed after circularization of the linear genome has occurred.16 Two forms of LMP2 transcripts exist that differ by virtue of unique 5′ exons located upstream of the TR, but have in common 8 exons located downstream of the TR.17 The first exon of LMP2A (here referred to as exon 1A) encodes a 119-amino acid, N-terminal hydrophilic cytoplasmic domain, while the first exon of LMP2B (exon 1B) is noncoding. Translation of LMP2B initiates at an ATG codon close to the 5′ end of exon 2. Common to both LMP2 proteins are 12 hydrophobic transmembrane domains and a 27-amino acid cytoplasmic carboxyl-terminal domain. Monoclonal antibodies have been raised against the unique N terminus of LMP2A, allowing its detection in tissue sections and cell lines,18,19 but there are no available reagents to detect the LMP2B protein. However, reverse transcription polymerase chain reaction (RT-PCR)–based methods to distinguish between LMP2A and 2B transcripts are well established.20
Given the preliminary clinical data suggesting responses of ENKTL and CAEBV to therapeutic CTLs containing LMP2-specificities,5 we further investigated the utility of LMP2 as an immunotherapeutic target in these malignancies. We initially focused on 4 previously characterized cell lines established from patients with ENKTL and CAEBV: 2 of NK-cell origin and 2 of T-cell origin21,22 Analysis of EBV gene expression in these cells, however, revealed no detectable LMP2A protein and virtually absent or very low levels of LMP2A and LMP2B mRNA transcripts in all 4 lines. Nevertheless, we found significant recognition and killing of these T- and NK-cell lines, mediated by LMP2-specific polyclonal and monoclonal CTLs. We therefore investigated the possibility of an alternative LMP2 transcript, expressed in EBV-associated T and NK lymphoproliferative disease and encoding a target for adoptive cellular immunotherapy.
Methods
Cell lines
This research received institutional approval from the University of Birmingham and studies of clinical material received ethical approval from the West Midlands Research Ethics Committee. Cell lines SNK 6, SNT 8, SNK 10, and SNT 16, established from Japanese patients with ENKTL and CAEBV were cultured as previously described.21,22 An additional series of cell lines23 was also used for PCR studies. Human leukocyte antigen (HLA) class I and II typing of cell line-derived DNA was performed by the Anthony Nolan Trust, London United Kingdom.
ENKTL biopsies
ENKTL biopsy sections used for LMP2A immunohistochemistry fulfilled the World Health Organization (WHO) diagnostic criteria,7 including EBER-positivity, and were reviewed by 2 expert hematopathologists in the United Kingdom (S.O. and Dr David M. Clark, Nottingham University Hospitals, United Kingdom). ENKTL frozen tissue for PCR studies had undergone histology review in the United States (W.C.C.) for inclusion in a previous study.23
Flow cytometry
Cell surface expression of major histocompatibility complex (MHC) molecules was determined by staining with phycoerythrin (PE)–conjugated monoclonal antibodies (mAbs) from AbD Serotec, analyzed on a Beckman Coulter XL flow cytometer and FlowJo Version 7.6 (TreeStar). MHC class I and II were detected using W6/32 antibodies24 and YE2/36-HLK antibodies,25 respectively.
Immunoblotting
Acrylamide gel electrophoresis and immunoblotting were performed as described previously.26 The 148.3 mouse mAb to TAP1 and the 435.3 mAb to TAP2 were used at 1/100 dilution of culture supernatant, and the HC10 mAb to HLA class I heavy chains was used at 0.2 μg/mL.26 The CS1-4 mouse mAbs to LMP127 were used at 0.25 μg/mL; the PE2 mouse mAb to EBNA228 and the 14B7 rat mAb to LMP2A18 at 1 μg/mL. The human serum, AM, which has high levels of antibody to EBNA129 was used diluted 1/1000. As a loading control, rabbit antibody to calregulin (sc-11 398; Santa Cruz Biotechnology) was used at 1 μg/mL.
Immunohistochemistry
Sections of 4-μm formalin-fixed tissue were subjected to a low temperature antigen retrieval method as previously described.30 Overnight incubation at 4°C with mAb clone 15F918 diluted 1/50 (Santa Cruz Biotechnology) was used to detect LMP2A. Dako Real Envision DAB was applied for 1-2 minutes and the sections counterstained with Mayer hematoxylin.
Secreted IFN-γ ELISA
Recognition of target cells by the effector T cells was determined by enzyme-linked immunosorbent assay (ELISA) of interferon-γ (IFN-γ) release after overnight coculture (at effector:target ratios of 1:10 and 1:20), using a standard protocol described elsewhere.31
Intracellular IFN-γ assay
Effector T cells were first incubated at 37°C for 5 minutes with 0.5μM carboxyfluorescein succinimidyl ester (CFSE) then washed with ice-cold media. CFSE-labeled T cells were coincubated with target cells (effector: target ratio 1:4) and 10 μg/mL Brefeldin A in 96-well V-bottomed plates at 37°C. Staining for intracellular IFN-γ was performed as described previously.31 FlowJo software Version 7.6 was used to analyze IFN-γ production in CSFE-gated effector cells.
Chromium release assays
Retroviral-mediated expression of HLA alleles
HLA-A*1101 and DRB1*0101 genes were cloned into retroviral expression plasmid pQCXIN (Clontech) by standard methods. Vesicular stomatitis virus-pseudotyped retrovirus particles were produced in GP-293 cells cotransfected with the relevant pQCXIN vector. At 72 hours after transfection, virus was concentrated by ultracentrifugation of cell supernatant and used to infect 0.5 × 106 target cells overnight. Infected cells were selected with G418 (Invitrogen).
LMP2-specific TCR gene transfer
A CD8+ T-cell clone specific for the HLA A*1101-restricted LMP2 epitope SSCSSCPLSK was generated from a healthy EBV carrier as described.33 The TCR α and β genes were subsequently amplified from its cDNA using the BD SMART RACE cDNA Amplification kit (BD Biosciences). The TCR-α and -β genes were cloned into a single retroviral pMP71-PRE vector34 (kindly provided by C. Baum, Hannover, Germany). Retrovirus generated using the Phoenix-A packaging cell line was used to transduce peripheral blood mononuclear cells (PBMCs) that had been preactivated 48 hours earlier using anti-CD3 antibody (OKT3) at 30 ng/mL and interleukin-2 (IL-2; 600 U/mL; Chiron). T-cell receptor (TCR)–transduced T cells were cloned by limiting dilution and restimulated every 7-14 days using the autologous LCL. Further details of the cloning of this TCR and the function of the transduced T cells are being prepared for publication in a separate manuscript (Y. Zheng, G. Parsonage, L. Machado, C. James, B. Johnson, A. Salman, A. van Hasselt, L. Vlantis, E. P. Hui, K. W. Lo, A. T. C. Chan, S.P.L., manuscript in preparation, “Rapid and efficient generation of Epstein-Barr virus–specific T cells to treat nasopharyngeal carcinoma”).
QRT-PCR for EBV transcripts
Total RNA was isolated from cultured cell lines using QIAGEN RNeasy kit and treated with DNase I (Turbo DNA-free kit; Ambion). Denatured RNA (400 ng) was reverse-transcribed with AMV-RT (Roche) and random primers (Promega). Quantitative reverse-transcription polymerase chain reaction (QRT-PCR) assays for conventional LMP2A were performed as described previously.20 New PCR primer/probe combinations to amplify conventional exon 1B-exon 2 spliced LMP2B mRNA as well as LMP2 transcripts containing exon 2, exon 6, exon 5-6 splice junctions, and TR-initiated LMP2 mRNA were designed using Primer Express software Version 2.0 (Applied Biosystems): primers were synthesized by Alta Bioscience (Birmingham). TaqMan probes were synthesized by Eurogentec. Sequences are detailed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
5′ RACE
5′ Rapid amplification of cDNA ends (RACE) was performed using the manufacturer's instructions (5′RACE system v2; Invitrogen). Briefly, first strand cDNA synthesis was generated using an LMP2-specific primer (LMP2 R6; supplemental Table 1). First round PCR amplification used a combination of the Abridged Anchor Primer (AAP) and LMP2 R5 reverse primer (3′ primer sequence AGTGACGCTAGCAGTGCCAGA), followed by a second round of amplification using the Abridged Universal Amplification Primer (AUAP) and LMP2 R4 (3′ primer sequence AGAGGACGAAAGCCAGTAGCAG). PCR products were analyzed by agarose gel electrophoresis, DNA recovered from excised bands (QIAquick Gel Extraction kit; QIAGEN), and cloned in to pGEM-T Easy Vector System I (Promega) for sequencing of individual cDNA products.
Results
Characterization of ENKTL and CAEBV cell lines
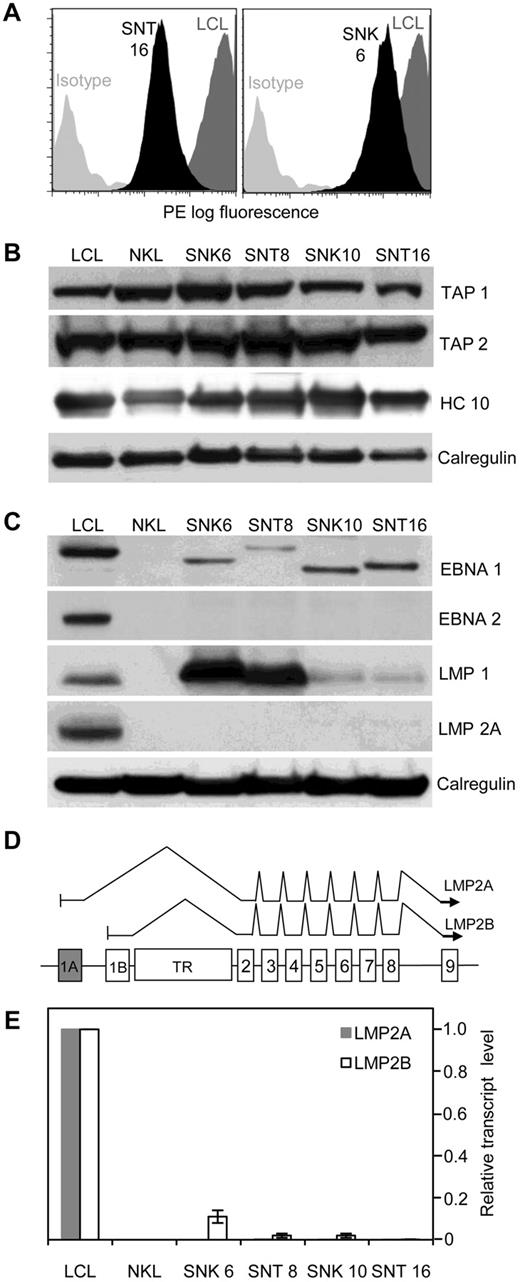
Before investigating LMP2 as a putative immunologic target in ENKTL and CAEBV, we first asked whether the target malignant cells have retained the capability to process and present antigen, because defects in the MHC class I/ transporter associated with antigen processing (TAP) pathway have been described in another EBV+ tumor, Burkitt lymphoma.35 All 4 EBV-positive NK/T tumor cell lines analyzed by flow cytometry were found to express surface HLA class I and II complexes. Figure 1A shows SNT 16 and SNK 6 as representative examples demonstrating moderate to high expression of surface HLA class I, while HLA class II expression (data not shown) was equivalent to levels in an EBV-transformed LCL. Western blot analysis of cell lysates (Figure 1B) showed total MHC class I and TAP levels to be broadly equivalent across cell lines, and comparable with a LCL.
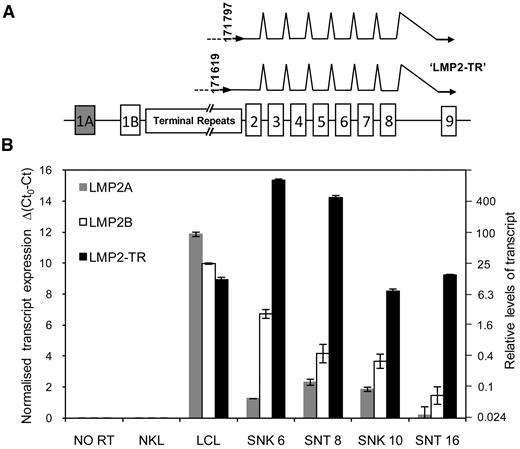
Antigen processing and presentation capability of EBV-positive T and NK tumor lines. (A) Flow cytometric analysis of cell surface HLA class I. Representative data from 2 lines (SNT 16 and SNK 6, black shading) compared with a LCL (dark gray shading) is shown. (B) Western blot of whole cell lysates probed with antibodies to transporter associated with antigen processing (TAP) 1 and 2 and total cellular class I heavy chains (HC 10) in NK- and T-cell lines, shown relative to levels in a LCL. Calregulin serves as the loading control. (C) EBV antigen expression in NK- and T-cell lines. Western blot of whole cell lysates of EBV-positive NK- and T-cell tumor lines probed with antibodies to EBNA 1, EBNA 2, LMP1, and LMP2A compared with a LCL and an EBV-negative NK line (NKL). Note the different molecular weight of EBNA 1 in the different lines is attributable to the variable size of the Gly-Ala repeat domains. (D) Schematic indicating the exon structure of the LMP2 gene and splicing patterns of conventional LMP2A and LMP2B transcripts, across the TR region of the genome. Each transcript contains a unique 5′ first exon (1A for LMP2A, shaded gray and 1B for LMP2B, shaded white). (E) Real-time PCR for LMP2A and LMP2B mRNA. Gray and white shaded bars represent quantities of LMP2A and LM2B transcripts, respectively, relative to a LCL. Error bars indicate SDs from the mean.
Antigen processing and presentation capability of EBV-positive T and NK tumor lines. (A) Flow cytometric analysis of cell surface HLA class I. Representative data from 2 lines (SNT 16 and SNK 6, black shading) compared with a LCL (dark gray shading) is shown. (B) Western blot of whole cell lysates probed with antibodies to transporter associated with antigen processing (TAP) 1 and 2 and total cellular class I heavy chains (HC 10) in NK- and T-cell lines, shown relative to levels in a LCL. Calregulin serves as the loading control. (C) EBV antigen expression in NK- and T-cell lines. Western blot of whole cell lysates of EBV-positive NK- and T-cell tumor lines probed with antibodies to EBNA 1, EBNA 2, LMP1, and LMP2A compared with a LCL and an EBV-negative NK line (NKL). Note the different molecular weight of EBNA 1 in the different lines is attributable to the variable size of the Gly-Ala repeat domains. (D) Schematic indicating the exon structure of the LMP2 gene and splicing patterns of conventional LMP2A and LMP2B transcripts, across the TR region of the genome. Each transcript contains a unique 5′ first exon (1A for LMP2A, shaded gray and 1B for LMP2B, shaded white). (E) Real-time PCR for LMP2A and LMP2B mRNA. Gray and white shaded bars represent quantities of LMP2A and LM2B transcripts, respectively, relative to a LCL. Error bars indicate SDs from the mean.
The original descriptions of the SNK and SNT cell lines, demonstrating presence of LMP1 and absence of EBNA 2 protein,22 suggested a Latency II pattern of viral gene expression. We confirmed and extended these observations by Western blot (Figure 1C) and QRT-PCR. In common with other Latency II tumors such as HL, expression of EBNA1 mRNA was initiated from the Qp promoter in the NK- and T-cell lines (data not shown) and levels of EBNA1 protein in all 4 SNK/SNT lines were lower than in LCLs (Figure 1C). As expected, EBNA2 was not detected, while LMP1 was present at variable levels. The ENKTL lines, SNK 6 and SNT 8, expressed very high levels of LMP1 protein, whereas the CAEBV-derived lines, SNK 10 and SNT 16, expressed substantially less (Figure 1C); a similar pattern of expression of LMP1 transcripts was observed by QRT-PCR (data not shown).
LMP2A protein was detected by Western blot in the LCL control but, unexpectedly, not in any of the ENKTL or CAEBV cell lines (Figure 1C). However, because the available monoclonal antibodies for detection of LMP2 protein all recognize defined epitopes within the LMP2A-unique N terminus,36 the LMP2B protein cannot be detected with this technique. A schematic of the exon structure and conventional splicing pattern, across the terminal repeat (TR) region, of LMP2A and LMP2B mRNAs is shown in Figure 1D. Consistent with the immunoblotting data, LMP2A transcripts were virtually undetectable in all cases by QRT-PCR (Figure 1E). We then used a combination of primers to amplify across the exon 1B-exon 2 junction by QRT-PCR and found that LMP2B transcripts were also expressed at extremely low levels in all 4 NK- and T-cell lines (Figure 1E). Finally, to exclude the possibility that the apparent lack of LMP2 mRNA expression by QRT-PCR was due to sequence variation within the LMP2 gene sequence, we performed genomic sequencing across LMP2 exons 1 and 2 in all 4 lines. Aside from a previously described37 single-base polymorphism detected in SNT16 compared with the prototype B95-8 EBV, corresponding to the common reverse LMP2 primer,20 there were no other sequence variations to explain the low levels of LMP2 transcripts.
Polyclonal LMP2 T-cell effectors recognize and kill ENKTL and CAEBV cell lines
Given the preliminary clinical data suggesting efficacy of polyclonal CTLs containing expanded LMP2 specificities in patients with ENKTL and CAEBV,4 we first asked whether such CTL preparations could recognize and kill the NK- and T-cell lines in spite of a virtual absence of LMP2A and LMP2B mRNA.
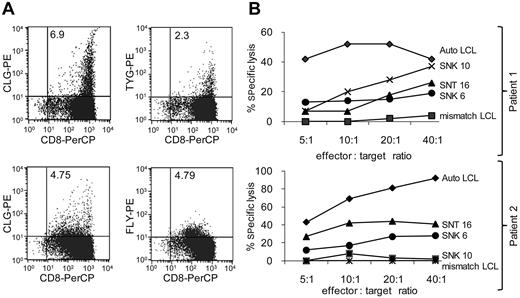
Polyclonal CTLs were produced by stimulation of PBMCs from ENKTL patients (ClinicalTrials.gov identifier: NCT00062868) with autologous antigen-presenting cells transduced with a recombinant adenovirus expressing an LMP1/LMP2A fusion protein.4,5,38 The antigen specificities of the CTLs were analyzed by peptide-loaded pentamer staining4,5 and their ability to kill HLA-compatible ENKTL and CAEBV tumor lines tested in 51Chromium-release assays. Figure 2 comprises data from 2 such analyses: CTLs from patient 1 contained 6.9% and 2.3% CD8+ cells specific for HLA-A*0201-CLGGLLTMV and HLA-A24-TYGPVFMSL, respectively (Figure 2A top plots), while CTLs from patient 2 contained 4.75% and 4.79% CD8+ cells specific for HLA-A*0201-CLGGLLTMV and HLA-A*0201-FLYALALLL, respectively (Figure 2A bottom plots). In cytotoxicity assays, CTLs from both patients killed SNT 16 and SNK 6 at significant levels compared with an autologous LCL (Figure 2B).
Specific killing of EBV+ malignant NK and T cells by polyclonal CTLs, containing LMP2-specificities, from ENKTL patients. (A) Dual-color flow cytometry of CTLs derived from 2 ENKTL patients prepared by ex vivo stimulation with LMP2/LMP1 transfected antigen-presenting cells. CTLs were stained with a PerCP (peridinin-chlorophyll protein)–conjugated anti-CD8 antibody and with the phycoerythrin-conjugated HLA-peptide pentamers: HLA-A*0201-CLGGLLTMV, HLA-A*0201-FLYALALLL, HLA-A24-TYGPVFMSL as previously described.5 Numbers within the top right quadrant of each plot indicate the percentage of viable peptide/pentamer-specific CD8+ cells. (B) The CTLs shown in panel A were tested in standard 51Chromium-release assays for their ability to kill EBV+ NK- and T-cell tumor lines. Results are expressed as the mean percentage of specific chromium release from the target cells at effector:target ratios titrated from 5:1 to 40:1, tested in triplicate. Autologous and HLA-mismatched LCLs were positive and negative controls, respectively, for each assay.
Specific killing of EBV+ malignant NK and T cells by polyclonal CTLs, containing LMP2-specificities, from ENKTL patients. (A) Dual-color flow cytometry of CTLs derived from 2 ENKTL patients prepared by ex vivo stimulation with LMP2/LMP1 transfected antigen-presenting cells. CTLs were stained with a PerCP (peridinin-chlorophyll protein)–conjugated anti-CD8 antibody and with the phycoerythrin-conjugated HLA-peptide pentamers: HLA-A*0201-CLGGLLTMV, HLA-A*0201-FLYALALLL, HLA-A24-TYGPVFMSL as previously described.5 Numbers within the top right quadrant of each plot indicate the percentage of viable peptide/pentamer-specific CD8+ cells. (B) The CTLs shown in panel A were tested in standard 51Chromium-release assays for their ability to kill EBV+ NK- and T-cell tumor lines. Results are expressed as the mean percentage of specific chromium release from the target cells at effector:target ratios titrated from 5:1 to 40:1, tested in triplicate. Autologous and HLA-mismatched LCLs were positive and negative controls, respectively, for each assay.
Recognition and killing of ENKTL and CAEBV lines by LMP2-specific CD8+ T-cell clones
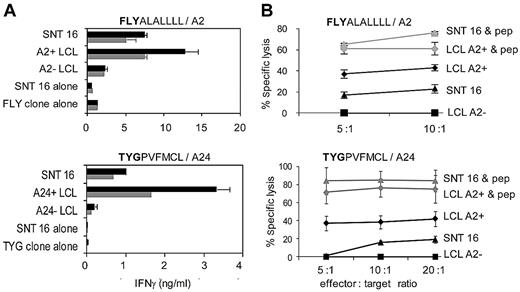
The observation that these polyclonal CTLs derived from ENKTL patients contained significant numbers of LMP2-specific T cells suggested that these cells were the dominant effectors in the cytotoxicity assays, although the presence of T cells with other specificities could not be excluded. Thus, faced with paradoxically low, or absent, levels of LMP2A and LMP2B transcripts in the T and NK lines, we further examined the specificity of this recognition. 2 previously characterized CD8+ effector clones with different LMP2-peptide specificities (FLYALALLLL, TYGPVFMCL) restricted through HLA-A2 (FLY) and -A24 (TYG)39,40 were initially tested in standard 18-hour IFN-γ–release assays. Figure 3A shows specific recognition of SNT 16 by both CD8+ clones, albeit at lower levels compared with HLA-matched LCL targets within the same assay. For these 2 effector clones, the immune recognition translated into specific killing, as demonstrated using a standard 5-hour 51Chromium-release assay (Figure 3B). Augmentation of the recognition by preloading target cells with excess exogenous cognate peptide indicated that the maximum killing of the SNT 16 targets was similar to that of HLA-matched targets, suggesting that the weaker recognition of the endogenously LMP2 antigen might reflect differences in levels of LMP2 expression or efficiency of FLY- and TYG-peptide processing. Recognition of SNT16 by CD8+ clones with specificities against LLWTLVVL39 and, to a lesser extent CLGGLLTMV,39 was also observed (data not shown).
Recognition and killing of SNT 16 by LMP2-specific CD8+ T-cell clones through HLA A2 and A24. (A) CD8+ clones specific for endogenously processed LMP2-derived peptides FLYALALLLL and TYGPVFMCL were cocultured for 18 hours with 105 target cells (gray and black bars indicate 5000 and 10 000 CD8+effectors per well, respectively). HLA-matched and -mismatched LCLs were positive and negative controls, respectively. Other controls included SNT 16 and CD8+ effectors cultured alone to assess spontaneous IFN-γ release. Error bars indicate 1 SD from the mean. Results are representative of those seen in 3 separate experiments. Supernatant IFN-γ was quantitated by ELISA. Relative to the maximal values seen using the same target cells preloaded with 5μM cognate peptide, recognition of the unmanipulated cells were as follows: FLY (SNT 16 = 14%, LCL = 26%), TYG (SNT 16 = 5%, LCL = 15%). (B) Lysis of SNT 16 and LCL targets by the LMP2-specific FLY and TYG CD8+ clones shown in panel A. Five-hour 51Chromium-release assays were conducted with SNT 16, HLA class I–matched and –mismatched LCL targets. Spontaneous release of 51Cr was < 30% of maximum release. Results are expressed as the percentage of specific chromium release from the target cells at the indicated effector-to-target ratios. Gray lines indicate results after preincubation for 1 hour with 5μM cognate peptide. Results are representative of those seen in 3 different experiments. Error bars indicate 1 SD from the mean.
Recognition and killing of SNT 16 by LMP2-specific CD8+ T-cell clones through HLA A2 and A24. (A) CD8+ clones specific for endogenously processed LMP2-derived peptides FLYALALLLL and TYGPVFMCL were cocultured for 18 hours with 105 target cells (gray and black bars indicate 5000 and 10 000 CD8+effectors per well, respectively). HLA-matched and -mismatched LCLs were positive and negative controls, respectively. Other controls included SNT 16 and CD8+ effectors cultured alone to assess spontaneous IFN-γ release. Error bars indicate 1 SD from the mean. Results are representative of those seen in 3 separate experiments. Supernatant IFN-γ was quantitated by ELISA. Relative to the maximal values seen using the same target cells preloaded with 5μM cognate peptide, recognition of the unmanipulated cells were as follows: FLY (SNT 16 = 14%, LCL = 26%), TYG (SNT 16 = 5%, LCL = 15%). (B) Lysis of SNT 16 and LCL targets by the LMP2-specific FLY and TYG CD8+ clones shown in panel A. Five-hour 51Chromium-release assays were conducted with SNT 16, HLA class I–matched and –mismatched LCL targets. Spontaneous release of 51Cr was < 30% of maximum release. Results are expressed as the percentage of specific chromium release from the target cells at the indicated effector-to-target ratios. Gray lines indicate results after preincubation for 1 hour with 5μM cognate peptide. Results are representative of those seen in 3 different experiments. Error bars indicate 1 SD from the mean.
In view of these data with the CAEBV-derived SNT16 line, we sought to broaden our investigations of LMP2-recognition to encompass other EBV-positive malignant NK and T cells. Methodologically, there were 2 obstacles to overcome: first, to broaden the HLA repertoire of potential LMP2 epitope presentation and, second, to resolve the problem of how to measure cytokine production by the effector clones in the face of significant levels of spontaneous cytokine release (including IFN-γ) from the other 3 tumor cell targets. To address the first issue, expression of a recombinant HLA-A11 molecule in SNK6 and an LCL control was achieved by infection with a retroviral construct, allowing us to test for recognition by a HLA-A*1101-restricted CD8+ LMP2 clone with specificity for SSCSSCPLSK.39 Expression of an irrelevant HLA allele, HLA-DR1, in the target cells using the same retroviral construct served as a negative control. To delineate between spontaneously produced IFN-γ from the SNK 6 target with that from the effector T cells, we prestained the effectors with CFSE enabling quantitation of IFN-γ production in CFSE+ T cells by intracellular cytokine staining. An example of such data, illustrated in Figure 4A, clearly demonstrates A11-restricted recognition of SNK6-A11 by the SSC clone. Notably, recognition of SNK6-A11 cells exceeded the recognition of an HLA-A11+ matched LCL. Finally, to confirm the LMP2 specificity of SNK6-A11 recognition, we used a recombinant TCR, recently developed in our laboratory (S.P. Lee et al, manuscript in preparation) and specific for HLA-A*1101/SSCSSCPLSK. The A11-SSC TCR, expressed in a non-SSC specific CD8+ clone derived from a healthy donor, was tested against SNK6-A11 in a CFSE/IFN-γ–release assay as before. The results (Figure 4B) confirmed the specificity and magnitude of LMP2-recognition in the ENKTL-derived cell line.
LMP2-specific T-cell recognition of SNK 6 exceeds that of a LCL. Dual-color flow cytometry: the x-axis represents fluorescence from CFSE-stained CD8+ effectors, and the y-axis denotes IFN-γ release as determined by intracellular staining. CFSE-negative cells (ie, LCL or SNK 6 targets) have been gated out of this analysis. HLA class I–mismatched LCLs and SNK 6 transduced with a control retrovirus (+ control RV) are negative controls. The top plots within panels A and B indicate T cells expressing IFN-γ in response to endogenously processed peptide, while the bottom plots of each panel show IFN-γ expression by T cells after preincubation of targets with exogenous cognate peptide (+ peptide). Numbers in the right quadrants indicate the percentage of cells positive (top quadrant) or negative (bottom quadrant) for IFN-γ release. (A) SSC-specific CD8+ clones tested against HLA-matched LCLs and SNK 6-A11. (B) Effector T cells expressing a recombinant SSC/A11 TCR tested against A11+ LCL and SNK 6-A11.
LMP2-specific T-cell recognition of SNK 6 exceeds that of a LCL. Dual-color flow cytometry: the x-axis represents fluorescence from CFSE-stained CD8+ effectors, and the y-axis denotes IFN-γ release as determined by intracellular staining. CFSE-negative cells (ie, LCL or SNK 6 targets) have been gated out of this analysis. HLA class I–mismatched LCLs and SNK 6 transduced with a control retrovirus (+ control RV) are negative controls. The top plots within panels A and B indicate T cells expressing IFN-γ in response to endogenously processed peptide, while the bottom plots of each panel show IFN-γ expression by T cells after preincubation of targets with exogenous cognate peptide (+ peptide). Numbers in the right quadrants indicate the percentage of cells positive (top quadrant) or negative (bottom quadrant) for IFN-γ release. (A) SSC-specific CD8+ clones tested against HLA-matched LCLs and SNK 6-A11. (B) Effector T cells expressing a recombinant SSC/A11 TCR tested against A11+ LCL and SNK 6-A11.
Taken together, the data in Figures 3 and 4 clearly demonstrate LMP2-specific T-cell recognition and killing of tumor cell lines derived from both CAEBV and ENKTL patients, through a number of epitopes in the context of 3 different HLA restrictions. The magnitude of T-cell recognition was comparable with an LCL in some instances, and was always greatly in excess of what might be expected from the apparent expression of LMP2A and LMP2B transcripts in the tumor lines (Figure 1D). We therefore decided to investigate further the expression of LMP2 in these cells to reconcile the immunologic data with our earlier findings of paradoxically low LMP2A and LMP2B transcripts.
Expression of a novel LMP2 transcript in ENKTL and CAEBV cell lines
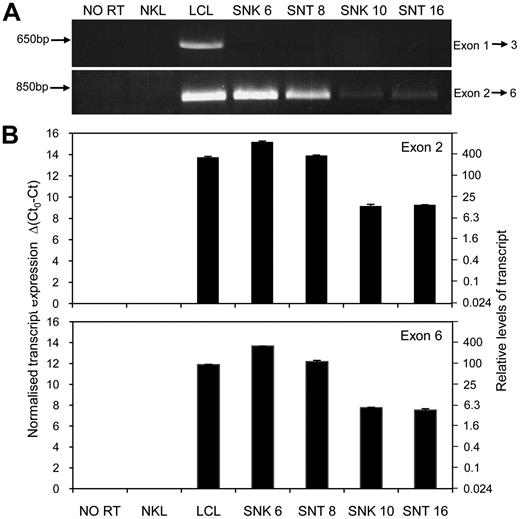
The schematic shown in Figure 1D indicates the known exon structure of conventional LMP2A and LMP2B mRNAs seen in LCLs. To address whether alternatively spliced or variant LMP2 transcripts may be expressed in our NK- and T-cell lines, we initially used conventional end point PCR to simply ask whether mRNA transcripts comprising exons 1-3 (spanning the variably sized terminal repeat region) and/or exons 2-6, (located 3′ to the terminal repeat region) were expressed in the tumor lines. While an exon 1-3 product was clearly present in the LCL control, such spliced LMP2 transcripts were undetectable in all 4 NK- and T-cell lines (Figure 5A top gel). By contrast, using primers located in exon 2 and exon 6 resulted in visible PCR products of the expected size in all 4 cases (Figure 5A bottom gel). Interestingly, similarly to LMP1 (Figure 1C, and data not shown) the 2 ENKTL lines (SNK 6 and SNT 8) apparently express these LMP2 transcripts at levels comparable with an LCL, while the 2 CAEBV lines expressed much lower levels of LMP2 exons 2-6.
Expression of a LMP2 transcript containing exons 2 to 6 in EBV positive NK- and T-cell tumor lines. (A) End point PCR products (35 cycles) visualized on a 1% agarose gel. Amplification of cDNA using a forward primer located in exon 1 and reverse primer within exon 3 (F1 and R3; see supplemental Table 1) yields a product of expected size in the LCL but no product in the NK- and T-cell lines. PCR performed with a forward primer in exon 2 and a reverse primer located within exon 6 (F2 and R6) produces a product of the anticipated size in all 4 NK- and T-cell lines. The EBV negative NK leukemia cell line, NKL, and a no reverse transcriptase (NO RT) sample from a LCL served as negative controls. (B) Quantitative RT-PCR assays, using primers and probes designed to amplify LMP2 transcripts containing exons 2 and 6, respectively (avoiding exon-exon junctions in the event of uncharacterized splice variations). Data are expressed using cycle threshold (Ct) values, where Ct0 represents the assay threshold of detection. LMP2 transcript Ct values are normalized to cellular glyceraldehyde 3-phosphate dehydrogenase (GAPDH) Ct values. The left-hand y-axis represents Δ(Ct0-Ct), while the right-hand axis shows the relative transcript levels.
Expression of a LMP2 transcript containing exons 2 to 6 in EBV positive NK- and T-cell tumor lines. (A) End point PCR products (35 cycles) visualized on a 1% agarose gel. Amplification of cDNA using a forward primer located in exon 1 and reverse primer within exon 3 (F1 and R3; see supplemental Table 1) yields a product of expected size in the LCL but no product in the NK- and T-cell lines. PCR performed with a forward primer in exon 2 and a reverse primer located within exon 6 (F2 and R6) produces a product of the anticipated size in all 4 NK- and T-cell lines. The EBV negative NK leukemia cell line, NKL, and a no reverse transcriptase (NO RT) sample from a LCL served as negative controls. (B) Quantitative RT-PCR assays, using primers and probes designed to amplify LMP2 transcripts containing exons 2 and 6, respectively (avoiding exon-exon junctions in the event of uncharacterized splice variations). Data are expressed using cycle threshold (Ct) values, where Ct0 represents the assay threshold of detection. LMP2 transcript Ct values are normalized to cellular glyceraldehyde 3-phosphate dehydrogenase (GAPDH) Ct values. The left-hand y-axis represents Δ(Ct0-Ct), while the right-hand axis shows the relative transcript levels.
In light of this finding, we designed 2 new QRT-PCR assays that amplified LMP2 sequences within exon 2 and exon 6 and quantitated their relative expression compared with that seen in a LCL. We focused on exon 2 as it represents the first transcribed exon 3′ to the TR region and exon 6 as this encodes peptides recognized in the T-cell assays described earlier. LMP2 transcripts containing exon 2 and exon 6 were highly expressed in SNK 6 and SNT 8, exceeding LCL levels, whereas lower levels corresponding to 5%-20% of LCL levels, were detected in SNK 10 and SNT 16 (Figure 5B). These results suggested expression of an alternative LMP2 transcript initiated upstream of exon 2 and pointed to an explanation for the initially paradoxical T-cell recognition data.
Identification of 5′ sequences of the novel LMP2 transcript in ENKTL and CAEBV lines
To further investigate the origin of LMP2 transcripts in the NK and T tumor cells, we subjected SNK6 RNA to RACE, using nested gene-specific primers within LMP2 exons 5 and 4. The schematic in Figure 6A demonstrates 2 novel LMP2 cDNA sequences (identified in 4 separate cDNA clones) that initiated within the last repeat of the TR region; the 5′ end of the longest cDNA is located 84 bases upstream of exon 2. Importantly, when the same RACE experiment was performed on LCL RNA, we identified 5′ cDNA ends containing exon1A or exon1B upstream of the TR, indicating the expected pattern of mRNA splicing (data not shown).
Identification of 5′ ends by RACE and quantitation of novel LMP2-TR transcripts in NK- and T-cell tumor lines. (A) The schematic represents the exon structure of the LMP2 gene spanning the EBV TR. Numbers indicate the exact 5′ ends of 4 of 7 cDNAs identified by RACE (EBV genome coordinates: NC007605). Three other 5′ cDNA ends identified in SNK 6 (coordinates 19, 80, and 83) correspond to the intervening intron and beginning of exon 2; having not reached the TR region. (B) Quantitative RT-PCR results: Ct0 represents the assay threshold of detection. LMP2 transcript Ct values are normalized to cellular GAPDH Ct values. The left y-axis represents Δ(Ct0-Ct), while the right-hand y-axis shows the equivalent fold difference in transcript levels.
Identification of 5′ ends by RACE and quantitation of novel LMP2-TR transcripts in NK- and T-cell tumor lines. (A) The schematic represents the exon structure of the LMP2 gene spanning the EBV TR. Numbers indicate the exact 5′ ends of 4 of 7 cDNAs identified by RACE (EBV genome coordinates: NC007605). Three other 5′ cDNA ends identified in SNK 6 (coordinates 19, 80, and 83) correspond to the intervening intron and beginning of exon 2; having not reached the TR region. (B) Quantitative RT-PCR results: Ct0 represents the assay threshold of detection. LMP2 transcript Ct values are normalized to cellular GAPDH Ct values. The left y-axis represents Δ(Ct0-Ct), while the right-hand y-axis shows the equivalent fold difference in transcript levels.
These results suggest that a previously uncharacterized LMP2 transcript may initiate from a novel promoter within the TR of the EBV genome and that this transcript (termed LMP2-TR), rather than the conventional LMP2A/2B mRNAs, is preferentially expressed in the context of NK- and T-cell malignancies. To confirm that this LMP2-TR mRNA is the dominant LMP2 transcript in these cells, we designed a further QRT-PCR assay using a forward primer in the intervening sequence between the TR region and exon 2; note this assay does not detect conventionally spliced LMP2A or LMP2B transcripts, both of which splice directly into exon 2. The quantitative data from this new assay are presented alongside levels of conventionally spliced LMP2A and LMP2B transcripts in Figure 6B. The novel LMP2-TR mRNA was by far the dominant LMP2 transcript in all 4 ENKTL and CAEBV cell lines. We also performed the same QRT-PCR analyses on cDNA derived from an additional series of ENKTL and CAEBV cell lines (KHYG1, YT, NKYS, HANK1, KA13, SNK1, SNT13)23 and found a similar pattern of LMP2 transcript expression in which LMP2-TR dominated in all cases (data not shown). Notably, the LMP2-TR mRNA was also transcribed in EBV-transformed B cells, albeit at 30 to 60× lower levels than in SNK 6 and SNT 8 (Figure 6B).
Expression of LMP2 in biopsy tissue from ENKTL patients
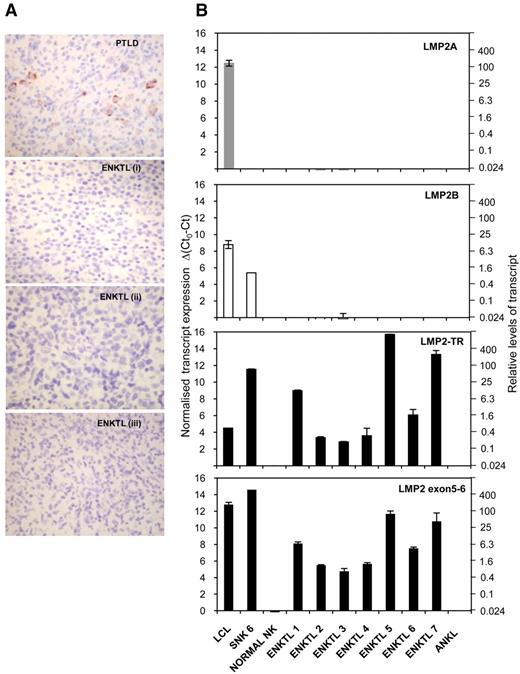
Having identified a novel LMP2-TR transcript in cell lines derived from EBV+ NK- and T-cell lymphoproliferative disease, we next examined the expression of LMP2 in primary tissue biopsies from patients with ENKTL. By means of immunohistochemical staining, we found no evidence of LMP2A protein expression on formalin-fixed biopsy sections from 7 ENKTL patients, while an EBV+ polymorphic PTLD biopsy included as a positive control demonstrated characteristic membrane/cytoplasmic staining in a subpopulation of cells (Figure 7A).
LMP2-TR, but not LMP2A or LMP2B, are expressed in primary ENKTL tissue. (A) Immunohistochemistry of formalin-fixed, paraffin-embedded tissue sections with antibody clone 15F9, specific for LMP2A. Images were captured with a Nikon Coolpix E995 digital camera, via a Nikon Eclipse E400 microscope (optical magnification × 400). A case of EBV+ B cell PTLD is a positive control. Sections from 3 representative cases ENKTL (i-iii of 7) shown. (B) Quantitative RT-PCR data using cDNA derived from 7 primary ENKTL tissue biopsies and 1 EBV+ aggressive NK leukemia sample. QRT-PCR assays were performed to detect conventional LMP2A and LMP2B, LMP2-TR and transcripts containing exon 5-6 splice; the latter representing total LMP2 mRNAs. Ct0 represents the assay threshold of detection. LMP2 transcript Ct values are normalized to cellular GAPDH Ct values. The left-hand y-axis represents Δ(Ct0-Ct), while the right-hand axis shows the equivalent fold difference in transcript levels. Note: input cDNA was 2 μL in a reaction volume of 20 μL, compared with 5 μL in 25 μL for the cell line PCR studies for Figures 5 and 6.
LMP2-TR, but not LMP2A or LMP2B, are expressed in primary ENKTL tissue. (A) Immunohistochemistry of formalin-fixed, paraffin-embedded tissue sections with antibody clone 15F9, specific for LMP2A. Images were captured with a Nikon Coolpix E995 digital camera, via a Nikon Eclipse E400 microscope (optical magnification × 400). A case of EBV+ B cell PTLD is a positive control. Sections from 3 representative cases ENKTL (i-iii of 7) shown. (B) Quantitative RT-PCR data using cDNA derived from 7 primary ENKTL tissue biopsies and 1 EBV+ aggressive NK leukemia sample. QRT-PCR assays were performed to detect conventional LMP2A and LMP2B, LMP2-TR and transcripts containing exon 5-6 splice; the latter representing total LMP2 mRNAs. Ct0 represents the assay threshold of detection. LMP2 transcript Ct values are normalized to cellular GAPDH Ct values. The left-hand y-axis represents Δ(Ct0-Ct), while the right-hand axis shows the equivalent fold difference in transcript levels. Note: input cDNA was 2 μL in a reaction volume of 20 μL, compared with 5 μL in 25 μL for the cell line PCR studies for Figures 5 and 6.
Because detection of LMP2B (and LMP2-TR) was only possible by RT-PCR, we then examined a second set of biopsies from which good quality RNA, extracted from frozen material, was available.23 While conventional LMP2A and LMP2B transcripts were absent or virtually undetectable, LMP2 transcripts were readily detected in 7/7 ENKTL biopsies using QRT-PCR assays that amplified LMP2 exons 5-6 and TR-initiated transcripts. No signals were seen in EBV− normal NK cells, or in a single case of EBV+ aggressive NK leukemia (Figure 7B).
Discussion
Immunotherapeutic targeting of viral antigens is a potentially important option for EBV-associated lymphoproliferations of NK and T cells, which are frequently resistant to conventional cytotoxic therapies, conferring a high rate of relapse and dismal prognosis for the majority of patients.6 While adoptive CTL immunotherapy has been shown to be a safe and effective treatment for PTLD, where the EBV-infected B cells express several immunogenic Latency II viral antigens, the more restricted pattern of viral gene expression in ENKTL and CAEBV could limit the success of this approach. In contrast to HL, where the most immunogenic of the available target proteins, LMP2, is demonstrably expressed,18,19 the situation in NK and T lymphoproliferations remained to be clarified. A single study previously reported the presence of conventional LMP2A and LMP2B mRNAs by nonquantitative PCR in some ENKTL biopsies, although in the majority of their cases the expression of these transcripts appeared low or absent.9
Our interest was stimulated by the unexpected observation that both conventional LMP2A and LMP2B were virtually undetectable in ENKTL and CAEBV lines. Would these tumors, therefore, be susceptible to LMP2-specific T-cell recognition? Our results showed that these malignant T and NK cells had the machinery for antigen-processing and presentation, were recognized by polyclonal effectors containing LMP2-specific CTLs as minor populations, and more importantly, by LMP2-specific CD8+ clones; efficiently recognizing multiple LMP2 epitopes across 3 different restriction elements. These data greatly strengthen the isolated reports describing killing of T and NK lines by EBNA141 or LMP142,43 clones by extending to the most immunogenic and clinically relevant of available targets, LMP2. Our data provide a rationale to pursue adoptive CTL immunotherapy for patients with ENKTL and CAEBV. Indeed, ongoing trials of adoptive transfer of LMP2-containing CTL preparations in ENKTL patients have produced encouraging results, including sustained complete responses in some cases.5,44 Given the anticipated prognosis of patients with relapsed/refractory ENKTL, whereby very few patients are salvaged even with intensive chemotherapy,6 these clinical outcome data are compelling.
Following these immunologic findings we identified a new transcript in NK- and T-cell malignancies, predicted to encode a LMP2B-like protein, but initiated from a different promoter located within the terminal repeat region of the EBV genome. This novel LMP2-TR transcript was found at high levels in ENKTL cell lines and, importantly, was the dominant LMP2 mRNA in primary ENKTL biopsy material, suggesting a role for LMP2-TR in disease pathogenesis. Interestingly, in the context of CAEBV, a recent study by Iwata et al11 demonstrated LMP2 transcripts in NK- and T-cell lines and PBMCs from CAEBV patients. Notably, in contrast to the majority of published reports, their study used a LMP2 PCR assay that targeted exon 6 and would therefore have detected all forms of LMP2 including, albeit inadvertently, the novel LMP2-TR identified in our study, although due to methods and representation of transcript quantitation, direct comparison with our data is difficult.
While the 2 viral promoters that drive transcription of LMP2A and LMP2B in B lymphocytes are dependent upon the viral transactivator protein EBNA2,45 the mechanism by which LMP2 expression is achieved in the absence of EBNA2, as in Latency II tumors, is less clear. Our data indicate that within Latency II tumors, there is an additional level of complexity, distinguishing NK and T cells from HL and nasopharyngeal carcinoma, which involves the use of alternative promoters for LMP2 expression. Although the exact transcription initiation site for the novel LMP2-TR transcript in ENKTL and CAEBV cells has not been precisely mapped, the results of the RT-PCR and RACE experiments imply that the promoter lies within the TR region. This situation is analogous to LMP1 expression in Latency II NPC cells where transcription is initiated from a promoter within the terminal repeats46 and raises the interesting possibility that similarly to the bidirectional promoter active in LCLs,47 another bidirectional promoter in the terminal repeats might give rise to both LMP1 and LMP2 transcripts. On this point, it has been reported that expression of LMP1 in ENKTL lines is reduced by depletion of IL-2 and is restored by replenishment of IL-2 or addition of IL-10 and IL-15.48 We have confirmed these data in the SNK6 line and shown that the regulation is transcriptional. Notably, we also found that the expression pattern of the LMP2-TR transcript in vitro and ex vivo mirrors that of LMP1, suggesting that these transcripts are coregulated and supporting the notion that they are expressed from a common bidirectional promoter (C.P.F. and M.R., unpublished results, May 2010). This is the subject of ongoing work in our laboratory.
To our knowledge, expression of naturally high levels of LMP2B in the absence of LMP2A has not previously been reported. The function of LMP2B in B cell and epithelial malignancies is not well understood, but it has been shown to be a negative regulator of LMP2A signaling.49 The high level of LMP2-TR transcripts (and LMP2B protein as detected by LMP2-specific effector T cells) in the absence of detectable LMP2A, in both primary ENKTL tissue and derived cell lines, suggests that the LMP2B protein is likely to have another hitherto unidentified function that might be involved in lymphomagenesis. This paper therefore has implications both for understanding the role of EBV in the development and potentiation of NK- and T-cell lymphoproliferations, and the potential effectiveness of immunotherapeutic targeting of expressed EBV antigens in these aggressive diseases.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dr Norio Shimizu, Tokyo Medical and Dental University, Japan, for kindly providing the SNK and SNT cell lines, and to Dr Souad Messahel for her expert assistance with immunohistochemistry.
This research was supported by Leukemia and Lymphoma Research, United Kingdom (through a clinical research fellowship for C.P.F.) and the Gregor MacKay Memorial Fund (a Guy's and St Thomas' Charity, London; T.A.H., M.R., and C.P.F.). Additional grant support was received from Cancer Research UK (M.R., A.I.B., C.S.-L., and S.P.L.), Medical Research Council (G.S.T. and H.M.L.), National Institutes of Health (NIH) grant P50CA136411-01 (W.C.C. and J.I.), and NIH SPORE grant P50CA126752 (C.M.B.). C.M.B. was also supported by a career development award from the Leukemia & Lymphoma Society and an award from the Gillson Longenbaugh Foundation.
National Institutes of Health
Authorship
Contribution: C.P.F. designed and performed research, analyzed data, and wrote the paper; T.A.H. performed research; G.S.T. designed research; H.M.L. performed research; S.P.L. contributed vital new reagents; C.S.-L. analyzed data; S.O., J.I., and W.C.C. contributed vital clinical samples and performed histology review; C.M.B. performed research and analyzed data; A.B.R. wrote the paper; A.I.B. designed research and analyzed data; and M.R. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martin Rowe, Professor of Tumor Virology, School of Cancer Sciences, University of Birmingham, Edgbaston, B15 2TT, United Kingdom; e-mail: m.rowe@bham.ac.uk.