Abstract
Recently, we showed that disturbed flow caused by a partial ligation of mouse carotid artery rapidly induces atherosclerosis. Here, we identified mechanosensitive genes in vivo through a genome-wide microarray study using mouse endothelial RNAs isolated from the flow-disturbed left and the undisturbed right common carotid artery. We found 62 and 523 genes that changed significantly by 12 hours and 48 hours after ligation, respectively. The results were validated by quantitative polymerase chain reaction for 44 of 46 tested genes. This array study discovered numerous novel mechanosensitive genes, including Lmo4, klk10, and dhh, while confirming well-known ones, such as Klf2, eNOS, and BMP4. Four genes were further validated for protein, including LMO4, which showed higher expression in mouse aortic arch and in human coronary endothelium in an asymmetric pattern. Comparison of in vivo, ex vivo, and in vitro endothelial gene expression profiles indicates that numerous in vivo mechanosensitive genes appear to be lost or dysregulated during culture. Gene ontology analyses show that disturbed flow regulates genes involved in cell proliferation and morphology by 12 hours, followed by inflammatory and immune responses by 48 hours. Determining the functional importance of these novel mechanosensitive genes may provide important insights into understanding vascular biology and atherosclerosis.
Introduction
Atherosclerosis is an inflammatory disease1,2 preferentially occurring in arterial regions exposed to disturbed flow characterized by low and oscillatory shear stress, whereas straight arterial regions exposed to table flow are protected from atherosclerosis.3,4 Despite the close association between the 2, in vivo evidence directly linking disturbed flow conditions to atherosclerosis has been scarce.
The differential mechanisms by which disturbed and stable flow promotes and inhibits atherogenesis, respectively, have been a subject of intense study, mostly using cultured endothelial cells.5-8 To define molecular mechanisms responsible for these changes, investigators have carried out DNA microarray studies using endothelial cells9-17 and have subsequently identified numerous shear-sensitive genes, such as kruppel-like factor 2 and 4 (Klf2, Klf4), endothelial nitric oxide synthase (eNOS), vascular cell adhesion molecule-1 (VCAM-1), intercellular adhesion molecule-1 (ICAM-1), bone morphogenic protein 4 (BMP-4), cathepsins, and angiopoietin-2 (Angpt2).11,14,18-29 Functional studies based on these shear-sensitive genes and their protein products have revealed the critical roles that they play in regulation of inflammation, thrombosis, vascular remodeling, angiogenesis, and arteriogenesis.11,19-22,26-30 Although these in vitro studies have provided critical insights regarding shear-sensitive mechanisms in cultured endothelial cells using modeled flow conditions, it cannot be assumed that identical mechanosensitive genes and pathways are involved in vivo regulating flow-dependent vascular responses and diseases. Therefore, it is critical to study how arterial endothelium responds to different flow conditions in vivo. However, the adequate pathophysiologic animal models enabling acute and reproducible modulation of flow conditions that rapidly lead to atherosclerosis have been lacking.
Recently, we have shown that partial ligation of mouse carotid artery causes disturbed flow with characteristic low and oscillatory wall shear stress, which in turn rapidly induces atherosclerosis, directly demonstrating the causal relationship between disturbed flow and atherosclerosis.31 In this model, disturbed flow induces endothelial dysfunction (by 1 week), robust atheroma formation (by 2 weeks), and features of advanced lesions, such as intraplaque neovascularization (by 4 weeks), in hyperlipidemic mice in the p47phox-dependent manner.31 This mouse model was also used to demonstrate how disturbed flow inhibits eNOS activity by decreasing GTP cyclohydrolase activity.32 We also showed that disturbed flow induces expression of HuR, which in turn induced inflammation by the nuclear factor-κB–dependent mechanisms in this model.33 Moreover, we developed a novel method of obtaining carotid endothelial RNA samples that are nearly free of contamination of smooth muscle cells and leukocytes from this mouse model.31 Using this method and the partially ligated mouse carotid arteries, we have shown by quantitative real-time polymerase chain reaction (PCR) studies that disturbed flow induces proinflammatory genes ICAM1, VCAM1, and BMP4, while significantly down-regulating antiatherogenic genes Klf2 and eNOS.31 These findings not only provided the proof of concept that disturbed flow rapidly regulates mechanosensitive gene expression but also demonstrated that sufficient quantity of endothelial RNA could be obtained for genome-wide microarray studies.
Here, we indeed carried out DNA microarray studies using endothelial RNAs obtained from flow-disturbed left common carotid arterial (LCA) and contralateral, undisturbed right carotid arterial (RCA) after 12- or 48-hour partial ligation in mice. These results were validated by quantitative PCR and immunostaining. Gene ontology analyses were further carried out demonstrating that disturbed flow initially regulates genes involved in cell proliferation and morphogenesis, followed by regulation of genes controlling inflammation and immune responses at a later time point.
Methods
Partial carotid ligation and flow characterization by ultrasound study
All animal studies were performed with 6- to 8-week-old male C57Bl/6 mice (The Jackson Laboratory) according to the approved Institutional Animal Care and Use Committee protocol by Emory University. Mice were partially ligated under anesthesia, and development of low and oscillatory shear in each mouse was determined by ultrasound measurements as we recently described.31 Briefly, 3 of 4 caudal branches of LCA (left external carotid, internal carotid, and occipital artery) were ligated with 6-0 silk suture, although the superior thyroid artery was left intact.
Intimal RNA isolation from carotid arteries
Total RNA from intima was separately obtained from LCA and RCA at 12, 24, and 48 hours after ligation, as we described previously.31
Microarray procedures
Total intimal RNAs were obtained from LCA and RCA at 12 and 48 hours after ligation. Intimal RNAs from 3 LCAs or RCAs were pooled to obtain approximately 30 ng of total RNA. All RNA samples used for the microarray study passed a quality control test using Agilent BioAnalyze NanoChip. Each sample was linearly amplified by WT-Ovation RNA amplification system (NuGEN) and used for the microarray study using MouseWG-6 v2 Expression BeadChip array with 45 281 probes (Illumina) at the Emory Biomarker Service Center. After hybridization, BeadChips were scanned on the Illumina BeadArray Reader to determine the probe fluorescence intensity. The raw probe intensities were then normalized by the quantile normalization algorithm34 using the GenomeStudio software from Illumina.
Microarray data analysis and bioinformatics
The normalized microarray data were statistically analyzed by Significance Analysis of Microarrays software (SAM 3.0).35 The differentially expressed genes between LCA and RCA were identified for those that showed more than 1.5-fold changes at less than 10% false discovery rate (FDR). The lists of differentially expressed genes were interrogated for statistically significant overrepresented cellular functions and disorders using DAVID analysis and Ingenuity Pathway Analysis (Ingenuity Systems).
Quantitative PCR validation
Total RNA of each sample was reverse-transcribed into cDNA using SuperScript III and random primers (Invitrogen), as we described.31 Briefly, quantitative PCR was performed on selected genes using Brilliant II SYBR Green QPCR Master Mix (Stratagene) with custom-designed primers on a Real-Time PCR System (ABI StepOne Plus). Predesigned TaqMan Gene Expression Assay probes (Applied Biosystems) were also used for some selected genes. All quantitative PCR results were normalized based on 18S RNA expression in each sample. Fold changes between LCA and RCA were determined using the ΔΔCt method.36
Immunohistochemical staining
Paraffin section immunostaining.
Mice were killed by CO2 inhalation and then were pressure-perfused at 100 mmHg with normal saline followed by pressure fixation with a 10% formalin solution. LCA and RCA were collected en bloc with the trachea and esophagus. Paraffin sections (5 μm) were then microwaved for 20 minutes in citrate buffer (0.1M, pH 6.0) for BMP4 and LMO4 staining or in Tris buffer (0.1M, pH 9.0) for Angpt2 and Jam2 staining. Sections were blocked with 10% donkey serum for 1 hour at room temperature and incubated with primary antibodies specific to BMP4 (5 μg/mL; Biovision), Lmo4 (5 μg/mL37-39 ), Jam2 (2 μg/mL; R&D Systems), and Angpt2 (0.4 μg/mL; Santa Cruz Biotechnology) overnight at 4°C in a humidified chamber.18 To visualize primary antibodies, rhodamine-conjugated secondary antibodies (donkey anti–goat, anti–rat IgG; Jackson ImmunoResearch Laboratories) were used for 1 hour at room temperature. Nuclei were counterstained with Hoechst 33258. For microscopic image acquisition, stained slides were mounted with Dako fluorescent mounting medium. Fluorescent images were taken using an Axiocam MRm camera (Zeiss) and an Axiovert 200M inverted microscope (Zeiss) with 20× (Plan-Achromat, NA 0.75) and 5× (Plan-Neofluar, NA 0.15) objective lenses. Axiovision 3.1 software (Zeiss) was used for image acquisition and processing. Paraffin sections of human coronary arteries from patients undergoing heart transplantations were obtained with the patients' consent obtained in accordance with the Declaration of Helsinki, according to the Institutional Review Board protocol approved at Emory as described previously.18 The same staining method used for mouse carotids as described above in this section was used for Lmo4 staining.
En face staining.
Ex vivo tissue culture
Mice were killed by CO2 inhalation and then pressure-perfused with heparinized normal saline. Under sterile conditions, common carotid arteries were harvested and carefully cleaned of perivascular fat. Carotid artery rings (∼ 3 mm) were incubated for 3 to 5 days at 37°C and 5% CO2 in Dulbecco modified Eagle medium supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin and 10% of heat-inactivated fetal bovine serum.
Statistical analysis
Data are mean plus or minus SEM. Paired Student t test was carried out for all quantitative PCR results of each gene to compare LCA versus RCA, and P value less than .05 (n = 3-5) was considered statistically significant.
Results
Discovery of mechanosensitive genes regulated by disturbed flow in mouse carotid endothelium in vivo
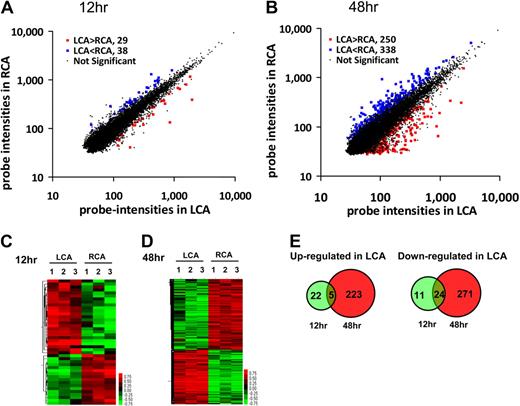
We carried out a DNA microarray study using Illumina BeadChip array containing 45 281 mouse gene probes and endothelial RNAs obtained from the flow-disturbed LCA and contralateral RCA at 12 and 48 hours after the partial ligation of LCA of C57BL/6 mice. The array results of detected probes were analyzed by the Significance Analysis of Microarrays analysis. We found that 67 (29 up-regulated and 38 down-regulated) of 45 281 gene probes were significantly altered by more than 50% in flow-disturbed LCA endothelium compared with the RCA by 12 hours after surgery at 10% FDR (Figure 1A). By 48 hours, 588 gene probes (250 up-regulated and 338 down-regulated) were regulated in LCA endothelium compared with the RCA at 10% FDR (Figure 1B). The detailed list of the significantly changed probes is shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article), and the array data were deposited to Gene Expression Omnibus (GSE 20741). Next, the significantly altered gene probes in individual samples were analyzed by hierarchical clustering to examine the intragroup and intergroup variations. As shown in the heat maps (Figure 1C-D), the results showed remarkably low variations within each group at both time points, demonstrating the reproducibility of the data. Because a single gene can be represented by multiple gene probes, the number of mechanosensitive genes is smaller than the number of detected probes. For example, 4 different probes representing the eNOS gene were identified as significantly changed by our microarray result. By 12 hours, 27 genes were up-regulated, whereas 35 genes were down-regulated in LCA endothelium (Figure 1E). In contrast, by 48 hours, 228 genes were up-regulated and 295 genes were down-regulated in LCA endothelium (Figure 1E). As the Venn diagrams show (Figure 1E), 5 of the 27 genes that were up-regulated in the LCA endothelium at the early time point (12 hours) continued to be up-regulated at the later time point (48 hours). These 5 genes are Ctgf, Ctps, Fosl2, Got2, and Lmo4. In contrast, 24 of 35 genes that were down-regulated at the early time point continued to be down-regulated at the later time point. These genes (supplemental Table 2) include some of the well-known shear-sensitive genes, such as Klf2 and Klf4, although the majority of them have never been reported previously as mechanosensitive genes.11,20,26-29 These results demonstrated that in vivo microarray study not only confirmed some of the well-known shear-sensitive genes reported previously but also discovered numerous novel mechanosensitive genes.
Global gene expression profiles in response to disturbed flow in mouse carotid artery endothelium in vivo. Total RNAs were obtained from intima of mouse left carotid (flow-disturbed, LCA) and right carotid (contralateral control, RCA) 12 and 48 hours after ligation. Illumina BeadChips containing 45 281 mouse genome-wide probes were used for the array study. Scatter plots show normalized intensities of each probe under 2 experimental conditions: LCA versus RCA at 12 hours (A) and 48 hours (B) after ligation. Genes that were up-regulated (red) or down-regulated (blue) (≥ 1.5-fold) at the FDR (≤ 10%) in LCA compared with RCA. (C-D) Hierarchical clustering analyses of mechanosensitive genes found in LCA endothelium compared with that of RCA are shown as heat maps. Each column represents a single sample pooled from 3 different LCAs or RCAs, and each row represents a single gene probe. (E) Venn diagrams show the temporal effects of disturbed flow on the number of up- or down-regulated mechanosensitive genes.
Global gene expression profiles in response to disturbed flow in mouse carotid artery endothelium in vivo. Total RNAs were obtained from intima of mouse left carotid (flow-disturbed, LCA) and right carotid (contralateral control, RCA) 12 and 48 hours after ligation. Illumina BeadChips containing 45 281 mouse genome-wide probes were used for the array study. Scatter plots show normalized intensities of each probe under 2 experimental conditions: LCA versus RCA at 12 hours (A) and 48 hours (B) after ligation. Genes that were up-regulated (red) or down-regulated (blue) (≥ 1.5-fold) at the FDR (≤ 10%) in LCA compared with RCA. (C-D) Hierarchical clustering analyses of mechanosensitive genes found in LCA endothelium compared with that of RCA are shown as heat maps. Each column represents a single sample pooled from 3 different LCAs or RCAs, and each row represents a single gene probe. (E) Venn diagrams show the temporal effects of disturbed flow on the number of up- or down-regulated mechanosensitive genes.
Validation of mechanosensitive genes by quantitative PCR
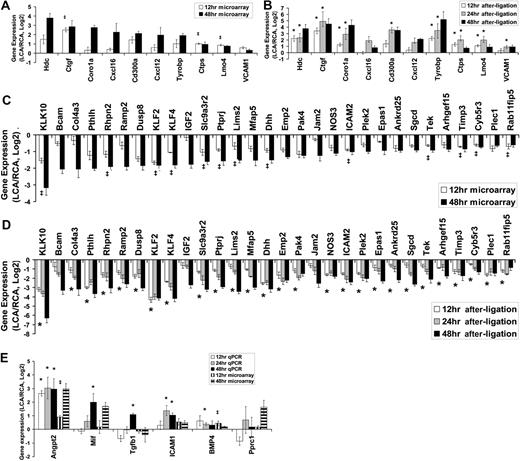
Next, we validated the microarray results by 2 different methods of quantitative PCR. We selected 46 genes (10 up-regulated, 30 down-regulated genes plus 6 additional genes of interests, although they were not significantly changed at 48 hours after ligation) and tested by quantitative PCR assay with either the TaqMan (28 genes, supplemental Table 3) or SYBR Green (18 genes, supplemental Table 4) method using total endothelial RNAs obtained from LCAs or RCAs collected at 3 different time points (12, 24, and 48 hours after ligation). These RNAs used for quantitative PCR validation were entirely independent from those used in the microarray study. First, our quantitative PCR results validated the microarray results for the 48-hour time point for the 10 up-regulated and 30-down regulated genes (Figure 2, A vs B and C vs D). For the 12-hour time point, 3 significantly up-regulated genes (Ctgf, Ctps, and Lmo4) were all confirmed by quantitative PCR assays (Figure 2A-B). In addition, 13 genes that were significantly down-regulated at 12 hours in the microarray study were also validated by the quantitative PCR analysis (Figure 2C-D). These results demonstrate the superb accuracy of microarray results with few false positives. There were, however, some genes at the 12-hour time point that were not shown to be significantly changed according to the array results, but quantitative PCR results showed that they were significantly changed. These include Hdc, Coro1a, and Tyrobp (Figure 2B) and those marked with an asterisk, including Col4a3, Pthlh, and Ramp2 (Figure 2D). This suggests that our microarray result using 10% FDR underestimated the number of mechanosensitive genes that changed significantly. Therefore, we selected 6 additional genes of interest, which did not change significantly according to the microarray result. For example, Angpt2, BMP4, and ICAM1 were previously identified as mechanosensitive genes 19,21,22,30 but were not included in the list of the significantly changed genes by the microarray (Figure 2E). Quantitative PCR results showed that Angpt2, BMP4, and ICAM1 were all significantly up-regulated at 24 hours after ligation and showed a similar trend at 48 hours (Figure 2E). Overall, to our complete but pleasant surprise, we were able to validate 16 of 16 genes at 12 hours and 40 of 40 genes at 48 hours.
Validation of mechanosensitive genes by quantitative PCR. Total RNAs from intima of LCA or RCA at different time points (12, 24, and 48 hours) after ligation were collected. Differentially expressed genes were selected for quantitative PCR analyses based on 48-hour microarray results. Each RNA sample at each time was pooled from 3 different mouse carotids, representing a total of 9 (n = 3) to 15 (n = 5) mice. Microarray results for 12- and 48-hour time points are shown as fold increase (A) or fold decrease (C) of genes expressed in LCA over RCA in log2 scale (mean ± SEM; n = 3). All genes shown in the graphs were significant at FDR less than 10% at 48 hours. ‡Genes were also statistically significant (FDR < 10%) at 12 hours. Quantitative PCR validation results for 12-, 24-, and 48-hour time points are shown as fold increase (B) or fold decrease (D) of genes expressed in LCA over RCA in log2 scale (mean ± SEM; n = 3-5). All genes shown in the graphs were significant (P < .05) at 48 hours. *Genes were also statistically significant (P < .05) at 12 and 48 hours. (E) Six genes of interests that did not reach statistical significance (> 10% FDR) were examined by quantitative PCR. Data are mean ± SEM (n = 3). ‡< 10% FDR (LCA vs RCA). * < .05 (LCA vs RCA).
Validation of mechanosensitive genes by quantitative PCR. Total RNAs from intima of LCA or RCA at different time points (12, 24, and 48 hours) after ligation were collected. Differentially expressed genes were selected for quantitative PCR analyses based on 48-hour microarray results. Each RNA sample at each time was pooled from 3 different mouse carotids, representing a total of 9 (n = 3) to 15 (n = 5) mice. Microarray results for 12- and 48-hour time points are shown as fold increase (A) or fold decrease (C) of genes expressed in LCA over RCA in log2 scale (mean ± SEM; n = 3). All genes shown in the graphs were significant at FDR less than 10% at 48 hours. ‡Genes were also statistically significant (FDR < 10%) at 12 hours. Quantitative PCR validation results for 12-, 24-, and 48-hour time points are shown as fold increase (B) or fold decrease (D) of genes expressed in LCA over RCA in log2 scale (mean ± SEM; n = 3-5). All genes shown in the graphs were significant (P < .05) at 48 hours. *Genes were also statistically significant (P < .05) at 12 and 48 hours. (E) Six genes of interests that did not reach statistical significance (> 10% FDR) were examined by quantitative PCR. Data are mean ± SEM (n = 3). ‡< 10% FDR (LCA vs RCA). * < .05 (LCA vs RCA).
Functional annotation and categorization of mechanosensitive genes
To understand the potential functional importance of the mechanosensitive genes that changed in response to disturbed flow in mouse carotid endothelium, we used the list of the significantly changed genes at the 12- and 48-hour groups from the microarray result (supplemental Table 1) for functional annotation analysis. Ingenuity Pathway Analysis showed that disturbed flow for 12 hours regulated genes that are involved in the disease processes, such as developmental disorder, cancer, immunologic and cardiovascular diseases that are mediated through changes in cell growth, proliferation, development, and morphology (supplemental Table 5). By 48 hours, disturbed flow induced genes that are involved in inflammatory and immunologic diseases while regulating cellular responses, such as antigen presentation, cellular movement, and cell-cell signaling (supplemental Table 5). Top mechanosensitive genes that are involved in inflammation and cell growth and proliferation are listed in Table 1. These include inflammatory cytokines (Ccl11, Ccl4, Cxcl12, and Cxcl16), adhesion molecules (SELL and VCAM1), and transcription factors (Klf2, Klf4, and Fosl2) and morphogens (Id1 and BMP4). These results suggested that disturbed flow initially induces genes that regulate cell morphogenesis and proliferation, followed by those that regulate inflammatory and immune responses at the later time point.
Flow-regulated genes involved in inflammation and cell growth and proliferation in mouse carotid endothelium
| Gene symbol . | 12-hour (LCA/RCA) . | FDR . | 48-hour (LCA/RCA) . | FDR . |
|---|---|---|---|---|
| Inflammation | ||||
| Down-regulated | ||||
| Col4a3 | 0.71 | > 10 | 0.22 | 8.1 |
| CCl11 | 1.41 | > 10 | 0.64 | 8.4 |
| CD40 | 0.88 | > 10 | 0.43 | 8.4 |
| CD59a | 0.93 | > 10 | 0.45 | 9.9 |
| Up-regulated | ||||
| Tyrobp | 2.93 | > 10 | 3.64 | 0.0 |
| CCl4 | 1.78 | > 10 | 2.41 | 3.1 |
| SELL | 1.47 | > 10 | 2.82 | 3.4 |
| Coro1a | 2.03 | > 10 | 6.70 | 7.5 |
| CD74 | 1.41 | > 10 | 3.80 | 8.6 |
| CD300a | 2.98 | > 10 | 4.31 | 9.5 |
| IL17RA | 2.01 | > 10 | 3.69 | 9.5 |
| Cxcl12 | 0.96 | > 10 | 3.43 | 9.5 |
| Cxcl16 | 1.32 | > 10 | 2.20 | 9.5 |
| SPP1 | ND | NA | 2.11 | 9.5 |
| ITGB2 | 1.15 | > 10 | 1.89 | 9.5 |
| TNFRSF1B | 1.46 | > 10 | 1.75 | 9.5 |
| IL1RN | ND | NA | 1.67 | 9.5 |
| VCAM1 | 1.47 | > 10 | 1.53 | 9.5 |
| Cell growth and proliferation | ||||
| Down-regulated | ||||
| KLF2 | 0.30 | 0.0 | 0.27 | 9.1 |
| ELN | 0.56 | 0.0 | 0.57 | 6.0 |
| Id1 | 0.59 | 0.0 | 0.45 | 0.0 |
| GNAQ | 0.66 | 0.0 | 0.76 | > 10 |
| PDGFA | 0.54 | 4.8 | 0.87 | > 10 |
| KLF4 | 0.48 | 7.2 | 0.28 | 8.1 |
| KRAS | 0.54 | 7.2 | 0.63 | 8.6 |
| CDKN1A | 0.54 | 7.2 | 1.31 | > 10 |
| F2RL1 | 0.56 | 7.2 | 0.60 | 6.9 |
| Pthlh | 0.42 | > 10 | 0.24 | 0.0 |
| IGF2 | 0.85 | > 10 | 0.29 | 9.1 |
| MAP3K1 | 1.03 | > 10 | 0.60 | 8.1 |
| Up-regulated | ||||
| BMP4 | 1.54 | 0.0 | 1.37 | > 10 |
| FOSL2 | 1.79 | 9.6 | 1.71 | 3.4 |
| BCL2L11 | 1.44 | > 10 | 1.84 | 9.5 |
| Gene symbol . | 12-hour (LCA/RCA) . | FDR . | 48-hour (LCA/RCA) . | FDR . |
|---|---|---|---|---|
| Inflammation | ||||
| Down-regulated | ||||
| Col4a3 | 0.71 | > 10 | 0.22 | 8.1 |
| CCl11 | 1.41 | > 10 | 0.64 | 8.4 |
| CD40 | 0.88 | > 10 | 0.43 | 8.4 |
| CD59a | 0.93 | > 10 | 0.45 | 9.9 |
| Up-regulated | ||||
| Tyrobp | 2.93 | > 10 | 3.64 | 0.0 |
| CCl4 | 1.78 | > 10 | 2.41 | 3.1 |
| SELL | 1.47 | > 10 | 2.82 | 3.4 |
| Coro1a | 2.03 | > 10 | 6.70 | 7.5 |
| CD74 | 1.41 | > 10 | 3.80 | 8.6 |
| CD300a | 2.98 | > 10 | 4.31 | 9.5 |
| IL17RA | 2.01 | > 10 | 3.69 | 9.5 |
| Cxcl12 | 0.96 | > 10 | 3.43 | 9.5 |
| Cxcl16 | 1.32 | > 10 | 2.20 | 9.5 |
| SPP1 | ND | NA | 2.11 | 9.5 |
| ITGB2 | 1.15 | > 10 | 1.89 | 9.5 |
| TNFRSF1B | 1.46 | > 10 | 1.75 | 9.5 |
| IL1RN | ND | NA | 1.67 | 9.5 |
| VCAM1 | 1.47 | > 10 | 1.53 | 9.5 |
| Cell growth and proliferation | ||||
| Down-regulated | ||||
| KLF2 | 0.30 | 0.0 | 0.27 | 9.1 |
| ELN | 0.56 | 0.0 | 0.57 | 6.0 |
| Id1 | 0.59 | 0.0 | 0.45 | 0.0 |
| GNAQ | 0.66 | 0.0 | 0.76 | > 10 |
| PDGFA | 0.54 | 4.8 | 0.87 | > 10 |
| KLF4 | 0.48 | 7.2 | 0.28 | 8.1 |
| KRAS | 0.54 | 7.2 | 0.63 | 8.6 |
| CDKN1A | 0.54 | 7.2 | 1.31 | > 10 |
| F2RL1 | 0.56 | 7.2 | 0.60 | 6.9 |
| Pthlh | 0.42 | > 10 | 0.24 | 0.0 |
| IGF2 | 0.85 | > 10 | 0.29 | 9.1 |
| MAP3K1 | 1.03 | > 10 | 0.60 | 8.1 |
| Up-regulated | ||||
| BMP4 | 1.54 | 0.0 | 1.37 | > 10 |
| FOSL2 | 1.79 | 9.6 | 1.71 | 3.4 |
| BCL2L11 | 1.44 | > 10 | 1.84 | 9.5 |
Comparison of microarray data between the in vivo mouse carotid endothelium and in vitro cultured endothelial cells
We next determined whether the 42 validated mechanosensitive genes from mouse carotid endothelium in vivo behaved similarly in cultured endothelium in vitro. For this study, we compared the in vivo mouse microarray and quantitative PCR results to the microarray results of human umbilical vein endothelial cells (HUVECs) exposed to oscillatory shear as opposed to laminar shear for 1 day. Because it is well known that microarray results obtained by different laboratories significantly vary,40 we first compared our mouse array result obtained using the mouse Illumina BeadChip array with that of HUVEC microarray using the human Illumina BeadChip array, as we recently reported (GSE20739). As shown in supplemental Table 6, several mechanosensitive genes (eg, Klf2, Klf4, NOS3, VCAM1, Ctgf, Angpt2, and BMP4) in mouse carotid endothelium were also found in the HUVEC microarray results. Of 42 mechanosensitive genes compared here, 23 genes (55%) showed similar responses between the mouse LCA endothelium and OS-exposed HUVECs. Of the remaining 19, 6 genes (14%) were not detected in cultured HUVECs, whereas 13 genes (31%) showed either no change or opposite trends. We hypothesized that those 6 undetectable genes in HUVECs were lost during culture under static condition. To test this hypothesis, we incubated mouse carotid arteries ex vivo for 0, 3, and 5 days under sterile conditions and examined endothelial mRNA levels by quantitative PCR. Klk10 (down-regulated gene in LCA) became undetectable by 3 days of culture. In contrast, Lmo4 (up-regulated in LCA) levels did not change significantly in the same samples, whereas Klf2 and Dhh (down-regulated in LCA) were decreased but still detectable at 3 and 5 days during culture (supplemental Figure 1). Additional quantitative PCR results using cultured HUVECs further confirmed that Klk10 and Col4a3 genes were not detectable even under shear conditions (supplemental Figure 2). These results are consistent with the notion that expression of some mechanosensitive genes became low or undetectable in cultured endothelial cells, at least in part, because of their no-flow culture conditions.
Validation of mechanosensitive genes at the protein level in mouse and human arterial endothelium
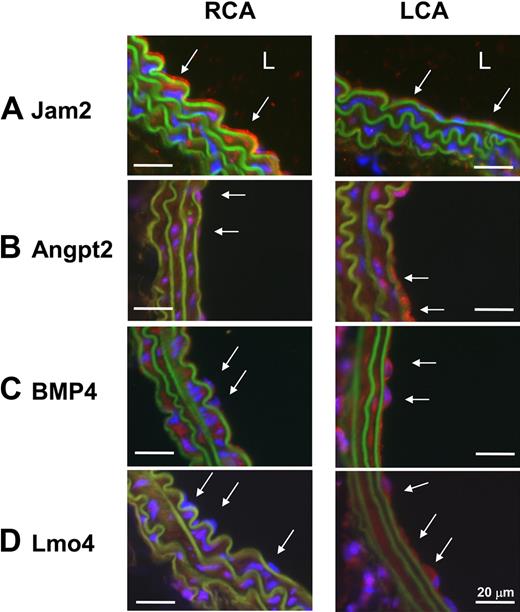
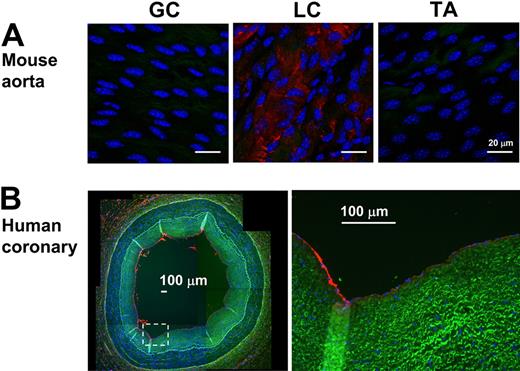
To further examine the validity of the mechanosensitive genes discovered in vivo, we examined protein expression levels of 2 newly identified mechanosensitive genes (Jam2 and Lmo4) and 2 previously known ones (Angpt2 and BMP4). Immunohistochemical staining confirmed that all 4 protein expression levels changed consistent to the mRNA results (Figure 3). Disturbed flow in LCA increased expression of Angpt2, BMP4, and Lmo4 proteins in endothelium while decreasing Jam2 (Figure 3). In addition, Lmo4 expression was easily detected in the flow-disturbed lesser curvature region of aortic arch, but not in stable flow regions in the greater curvature and thoracic aorta in C57BL/6 mice (Figure 4A). Moreover, Lmo4 was specifically expressed in human coronary artery endothelium in an asymmetric pattern, as shown in the compositive microphotograph (Figure 4B; and also see supplemental Figure 3 for identification of individual microphotographs used for Figure 4B). These results not only confirm that disturbed flow induced in our mouse carotid model changes protein expression of some of the expected mechanosensitive genes but also provide further evidence supporting the validity of the newly discovered mechanosensitive genes.
Disturbed flow in LCA decreases protein expression of Jam2 while up-regulating Angpt2, BMP4, and Lmo4. C57BL/6 mice underwent partial ligation, and LCA and RCA were collected 2 days after ligation. Paraffin sections were stained with specific antibodies for Jam2 (A), Angpt2 (B), BMP4 (C), and Lmo4 (D). Nuclei were counterstained with Hoechst (blue). Arrows indicate the protein expression in endothelial cells. L indicates lumen. Images are representative of n = 4 mice.
Disturbed flow in LCA decreases protein expression of Jam2 while up-regulating Angpt2, BMP4, and Lmo4. C57BL/6 mice underwent partial ligation, and LCA and RCA were collected 2 days after ligation. Paraffin sections were stained with specific antibodies for Jam2 (A), Angpt2 (B), BMP4 (C), and Lmo4 (D). Nuclei were counterstained with Hoechst (blue). Arrows indicate the protein expression in endothelial cells. L indicates lumen. Images are representative of n = 4 mice.
Lmo4 is differentially expressed in mouse aortic arch and human coronary artery. (A) En face staining of greater curvature (GC), lesser curvature (LC) of the arch, and the thoracic aorta (TA) was performed with Lmo4 antibody (red). Blue signal indicates nuclei stained with 4,6-diamidino-2-phenylindole; green signal indicates elastic laminae detected by autofluorescence. Shown are representative images of 7 different mice. (B) Paraffin sections of human left anterior descending coronary artery were stained for Lmo4 protein expression. Overall staining patterns were shown at low magnification (original magnification ×5) as a composite figure and zoomed views (original magnification ×20) of the indicated areas (broken box). The raw images of the composite figure are shown in supplemental Figure 3.
Lmo4 is differentially expressed in mouse aortic arch and human coronary artery. (A) En face staining of greater curvature (GC), lesser curvature (LC) of the arch, and the thoracic aorta (TA) was performed with Lmo4 antibody (red). Blue signal indicates nuclei stained with 4,6-diamidino-2-phenylindole; green signal indicates elastic laminae detected by autofluorescence. Shown are representative images of 7 different mice. (B) Paraffin sections of human left anterior descending coronary artery were stained for Lmo4 protein expression. Overall staining patterns were shown at low magnification (original magnification ×5) as a composite figure and zoomed views (original magnification ×20) of the indicated areas (broken box). The raw images of the composite figure are shown in supplemental Figure 3.
Discussion
Although the association between localization of atherosclerotic lesions and local hemodynamics has been recognized for more than several decades, compelling evidence directly demonstrating the cause-and-effect relationship between disturbed flow and atherosclerosis has been scarce, largely because of a lack of adequate animal models to test the hypothesis directly. We recently provided evidence directly demonstrating that disturbed flow acutely caused by partial ligation of carotid artery rapidly induces endothelial dysfunction in 1 week and atherosclerosis in 2 weeks.31 We further developed a novel method of endothelial RNA extraction from mouse carotid intima.31 The availability of sufficient quantity of endothelial RNA from mouse carotid intima enabled us to carry out genome-wide high-throughput screening studies to identify mechanosensitive genes in the mouse model. Using this novel method and mouse model, we identified 62 (27 up-regulated and 35 down-regulated) genes at 12 hours and 523 (228 up-regulated and 295 down-regulated) genes at 48 hours after the partial ligation that changed significantly in the flow-disturbed LCA endothelium compared with the contralateral RCA (Figure 1).
The microarray results were further validated for 46 selected genes by quantitative PCR (Figure 2). To our great surprise, all 40 up- or down-regulated genes tested were validated by quantitative PCR. Four of those genes (2 previously known mechanosensitive genes: BMP4 and Angpt2; 2 novel mechanosensitive genes: Lmo4 and Jam2) were further validated by immunostaining of mouse carotid artery (Figure 3). In particular, expression of Lmo4 was validated by immunostaining of mouse aortic arch and human coronary artery (Figure 4). Gene ontology analyses using the mechanosensitive genes suggest that disturbed flow rapidly controls expression of endothelial genes involved in cell morphology and proliferation pathways, followed by additional genes regulating inflammatory and immune responses by 48 hours after ligation (supplemental Table 5). These secondary responses involving inflammation and immune responses may lead to subsequent endothelial dysfunction by 1 week and atherosclerosis by 2 weeks. This is the first in vivo genome-wide DNA microarray study revealing the gene expression profiles in response to acute exposure to flow disturbance using mouse carotid endothelium. Although confirming some of the previously known mechanosensitive genes, this study reports numerous novel mechanosensitive genes that have never been reported previously to our knowledge.
Our study uncovers one interesting group of mechanosensitive genes as immediate and persistent responders that were up-regulated or down-regulated by disturbed flow in LCA endothelium at both the 12- and 48-hour time points (supplemental Table 2). These genes include some of the well-known mechanosensitive genes, such as Klf2 and Klf4,11,20,26-29 although the majority of them have never been reported previously as mechanosensitive genes, such as transcription regulators Lmo4, Fosl2, and Id1. These early and persistent responders could represent the primary mechanosensitive genes that respond immediately to disturbed flow in endothelium, potentially playing a key role in vascular biology and atherosclerosis.
Lmo4 (LIM-only protein 4) is a potential oncogene and associated with growth, migration, and invasion of breast cancer cells.37-39 In our study, we found that Lmo4 expression is up-regulated in disturbed flow regions, including mouse LCA endothelium and aortic arch. Interestingly, Lmo4 expression in human coronary artery was found specifically in endothelial cells in an asymmetric manner, consistent with the idea of its flow-dependent expression. Interestingly, oscillatory shear stress stimulates endothelial cell proliferation,41-44 suggesting a potential role for Lmo4 overexpressed in flow-disturbed regions in the pro-atherogenic response.
Previously, several DNA microarray studies have been reported generating the lists of potential mechanosensitive genes using cultured endothelial cells exposed to various shear stress conditions, laminar, pulsatile laminar, oscillatory shear, and turbulent flow. In most studies, gene expression profiles in endothelial cells exposed to laminar shear were compared with that of static culture conditions,9-17 whereas a few compared laminar shear with that of oscillatory or turbulent shear,14,43,45 better simulating pathophysiologic conditions. These microarray studies have identified many mechanosensitive genes, such as Klf2, Klf4, BMP-4, cathepsins, and Angpt2, and subsequent studies have revealed functional significance of these mechanosensitive genes in regulation of inflammation, thrombosis, vascular remodeling, angiogenesis, and arteriogenesis,11,19-22,26-30 demonstrating the critical use of these microarray studies in studying vascular biology and diseases. Because cultured endothelial cells are prone to phenotypic changes during extended culture (no flow condition) and in vitro shear conditions cannot exactly replicate in vivo conditions, we wanted to examine whether the mechanosensitive genes found in our mouse endothelium in vivo were similar or different from those found in vitro. Our initial comparison was carried out between the 42 confirmed mechanosensitive genes in vivo and our HUVEC array result using the same microarray platform. These comparisons showed that approximately 55% mechanosensitive genes examined here were conserved, whereas the remaining approximately 45% were either dysregulated or lost. Similarly, we found that approximately 50% of the in vivo genes were also conserved, whereas approximately 50% were not when our in vivo data were compared with another independent study recently reported by Conway et al (supplemental Table 7).14 These results demonstrate that approximately half of the mechanosensitive genes found in vivo can be confirmed in vitro, whereas the other half may not be found in vitro because of phenotypic changes in cultured cells. These findings clearly demonstrate the critical need of in vivo models in studying flow-dependent vascular responses and diseases.
Recently, Davies et al conducted in vivo DNA microarray studies using endothelial RNAs obtained directly from the flow-disturbed inner aortic arch and undisturbed flow region of normal pig aorta.40,46,47 When we compared our list of 42 confirmed mechanosensitive genes found in vivo with that of pig endothelial array result (supplemental Table 7), we found only 2 (Klf4 and eNOS) were found in their list. This discrepancy could be the result of one or more of the following causes: First, this may represent the difference in the acute mouse model versus chronic pig aorta model. Because pig aortic arch is exposed to chronic changes, including flow disturbance for many months from birth, the observed gene profile changes may be complex and may not be solely attributed to flow disturbance. On the other hand, we isolated endothelial RNA samples within 12 to 48 hours after partial ligation, enabling us to study direct effect of flow disturbance on endothelial gene expression in vivo. Second, although the inner curvature of pig aortic arch is a well-known naturally occurring flow-disturbed region, it may be difficult to identify a distinct region exposed to disturbed flow and to obtain RNA samples from the small area only. In contrast, our mouse carotid artery (LCA) is exposed to flow disturbance occurring nearly homogeneously along the length of the common carotid artery, as we recently demonstrated.31 Third, unlike our mouse array study using the mouse genome-wide probes, the pig array was carried out against human probes because of the lack of porcine specific arrays. This could have resulted in underestimation of mechanosensitive genes in the pig array study. One advantage of our study using the mouse model is that the identified mechanosensitive genes could easily be further examined for their pathophysiologic importance in transgenic or knockout mice.
One caveat of in vivo studies such as ours is the potential contamination of RNAs originating from leukocytes accumulated in the carotid intima or medial smooth muscle cells. However, as we have recently demonstrated, our intimal RNA isolation method is free of markers of smooth muscle cells (α-SMA) and leukocytes (CD11b) as determined by quantitative PCR.31 In addition, we did not find discernible CD11b-positive staining in the intima of LCA and RCA within 2 days of partial ligation,31 although we found them in the adventitia.31 This was the reason that we limited our experimental time points to 12 and 48 hours to prevent potential contamination of infiltrating cells in the LCA intima. Furthermore, we examined whether additional markers of infiltrating leukocytes in our microarray results. CD3, CD4, CD28, CD11b, CD43, CD16, and CD56 are either not detectable or not significantly different between LCA and RCA, suggesting that there is no obvious contamination of T cells, B cells, or macrophages in our RNA samples. Although we cannot completely rule out the possibility that infiltrating cells affect our gene lists, we are especially confident for those mechanosensitive genes, such as well-known Klf2, Klf4, eNOS, and the novel Jam2, Klk10, and Dhh, which are highly expressed in the contralateral RCA but are decreased in flow-disturbed LCA.
In conclusion, we have carried out in vivo genome-wide microarray studies using mouse carotid endothelium exposed to disturbed flow. From this study, we identified more than 500 mechanosensitive genes that change in response to disturbed flow within 2 days. Based on our analysis of confirmed 42 mechanosensitive genes identified in mouse carotid endothelium, we estimate approximately 50% of the in vivo mechanosensitive genes are novel, whereas the rest confirms the previous results reported in cultured endothelial cells. These findings suggest that, although the in vitro flow studies are valid and play important roles in studying detailed mechanistic studies, it highlights the critical and unique need of in vivo models to study vascular biology and diseases because many of the mechanosensitive genes are lost or dysregulated during culture. These novel mechanosensitive genes identified in this study need to be further studied to determine their functional importance in cells and animal models in the future. They may provide novel therapeutic and diagnostic targets of vascular diseases, such as atherosclerosis.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr W. Robert Taylor, Daiana Weiss, and David Vega for providing human coronary sections.
This work was supported in part by the Emory Biomarker Service Center and by the National Institutes of Health (grants HL75209, HL87012, and HL80711; H.J.) and the World Class University Project of Korea and Ada Lee and Pete Correll Professorship (H.J.).
National Institutes of Health
Authorship
Contribution: C.-W.N. designed and performed research, analyzed data, and wrote the manuscript; H.Q., A.R., K.K., D.N., and D.J.S. performed research and analyzed data; J.E.V. provided a vital reagent; and H.J. oversaw the entire project, designed research, analyzed data, secured funding, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hanjoong Jo, Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University, 2005 WMB, Atlanta, GA 30322; e-mail: hjo@bme.gatech.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal