Abstract
Genomic instability (GI) of cells may lead to their malignant transformation. Carcinoma after hematopoietic cell transplantation (HCT) frequently involves some (eg, oral) but not other (eg, nasal) epithelia. We examined GI in oral and nasal mucosal specimens from 105 subjects, including short-term (7-98 days, n = 32) and long-term (4-22 yrs, n = 25) allogeneic HCT survivors. Controls included autologous HCT survivors (n = 11), patients treated with chemotherapy without HCT (n = 9) and healthy controls (n = 27). GI was detected in 60% oral versus only 4% nasal specimens in long-term allogeneic HCT survivors (P < .001). None of the controls showed GI. In oral specimens, GI was significantly associated with history of oral chronic graft-versus-host disease (cGVHD). We conclude that GI after HCT is frequent in some (oral) but rare in other (nasal) epithelia. This may explain why some epithelia (especially those involved with cGVHD) are prone to develop cancer.
Introduction
Hematopoietic cell transplantation (HCT) recipients have 2- to 4-fold higher likelihood of developing a solid cancer compared with the general population.1-4 However, cancers of some organs are frequent (eg, skin, mouth, esophagus, thyroid, liver, bones, brain) whereas cancers of other organs are rare (eg, nasal mucosa, small intestine) or do not appear to have a higher incidence compared with general population (eg, lung, colon, testis).1,3 The cause of this discrepancy is unknown.
Genomic instability (GI) is characteristically a precancerous or cancerous state.5 It has also been reported in non-neoplastic, chronically inflamed tissues.6,7 GI refers to a set of somatic alterations within the genome, and is classified into chromosomal and microsatellite instabilities. Chromosomal instability refers to a gain or loss of chromosomes, caused by failures in mitotic chromosome separation.8 On the contrary, microsatellite instability (MSI) leads to expansions or contractions of short tandem repeats (STR), resulting from replication slippage, mismatch repair (MMR) or homologous recombination.8
Here we evaluated MSI as a marker of GI to determine whether after allogeneic HCT (allo-HCT), GI occurs more frequently in oral epithelium (representing epithelium frequently involved with cancer) than nasal epithelium (representing epithelium rarely, if ever, involved with cancer). We also evaluated whether chronic graft-versus-host disease (cGVHD) can contribute to GI development, as cGVHD appears associated with increased likelihood of developing cancer, in particular squamous cell cancer of skin or oral cavity.1-4,9,10
Methods
A total of 105 subjects were accrued (Table 1). The Research Ethics Board of the University of Calgary approved the study and all subjects signed a written consent in accordance with the Declaration of Helsinki.
Demographic and clinical information on subjects
| Characteristics . | Distribution and range . | Testing variables . | P . | |
|---|---|---|---|---|
| Long-term allo HCT survivors . | Short-term allo HCT survivors* . | |||
| N | 25 | 32 | NA | |
| Time after transplantation | Median = 10 y; range =6-22 y | Median = 56 d; range = 7-98 d | NA | |
| Sex | Male = 15; female = 10 | Male = 14; female = 18 | Male, Female | .28 |
| Age at transplantation | Median = 42 y; range = 18-59 y | Median = 53 y; range = 19-65 y | ≤ 40 y, older | .29 |
| Age at time of sample collection | Median = 51 y; range = 31-68 y | Median = 53 y; range = 20-65 y | ≤ 40 y, older | .39 |
| Source of graft | Bone marrow = 18; blood stem cells = 7 | Blood stem cells = 30; cord blood = 2 | BM, PBSC | .001‖ |
| Disease | AML = 4; CML = 8; ALL = 6; CLL = 2; NHL = 3; MM = 1; SAA = 1 | AML = 13; CML/CMML = 2; ALL = 5; CLL/lymphoma = 11; MF = 1 | Meyloid, lymphoid | .92 |
| Type of transplantation | Related = 19; unrelated = 6 | Related = 16; unrelated = 16 | Related, unrelated | .06 |
| Conditioning† | Cy+TBI = 6, Cy+Bu = 7, Cy = 1, Flu+Bu = 9, VP16+TBI = 2 | Flu+Bu+TBI = 19, Flu+Bu = 12, VP16+TBI = 1 | TBI, no TBI | .07 |
| GVHD prophylaxis | CSA+MTX = 15, MTX+steroid = 2, CSA+MTX+ATG = 8 | CSA+MTX+ATG = 32 | ATG, no ATG | > .001‖ |
| Acute GVHD | Yes = 9; No = 16 | Yes = 13; No = 19 | Yes, No | −.78 |
| Chronic GVHD‡ | Yes = 18; No = 7 | NA | NA | |
| Oral chronic GVHD‡ | Yes = 14; No = 11 | NA | NA | |
| Time from oral cGVHD diagnosis to sampling | Median = 9 y; range = 4-21 y | NA | NA | |
| Active oral cGVHD at time of sampling | Yes = 2; N = 23 | NA | NA | |
| Patients on immunosuppression§ at time of sampling | Yes = 2; No = 23 | |||
| Characteristics . | Distribution and range . | Testing variables . | P . | |
|---|---|---|---|---|
| Long-term allo HCT survivors . | Short-term allo HCT survivors* . | |||
| N | 25 | 32 | NA | |
| Time after transplantation | Median = 10 y; range =6-22 y | Median = 56 d; range = 7-98 d | NA | |
| Sex | Male = 15; female = 10 | Male = 14; female = 18 | Male, Female | .28 |
| Age at transplantation | Median = 42 y; range = 18-59 y | Median = 53 y; range = 19-65 y | ≤ 40 y, older | .29 |
| Age at time of sample collection | Median = 51 y; range = 31-68 y | Median = 53 y; range = 20-65 y | ≤ 40 y, older | .39 |
| Source of graft | Bone marrow = 18; blood stem cells = 7 | Blood stem cells = 30; cord blood = 2 | BM, PBSC | .001‖ |
| Disease | AML = 4; CML = 8; ALL = 6; CLL = 2; NHL = 3; MM = 1; SAA = 1 | AML = 13; CML/CMML = 2; ALL = 5; CLL/lymphoma = 11; MF = 1 | Meyloid, lymphoid | .92 |
| Type of transplantation | Related = 19; unrelated = 6 | Related = 16; unrelated = 16 | Related, unrelated | .06 |
| Conditioning† | Cy+TBI = 6, Cy+Bu = 7, Cy = 1, Flu+Bu = 9, VP16+TBI = 2 | Flu+Bu+TBI = 19, Flu+Bu = 12, VP16+TBI = 1 | TBI, no TBI | .07 |
| GVHD prophylaxis | CSA+MTX = 15, MTX+steroid = 2, CSA+MTX+ATG = 8 | CSA+MTX+ATG = 32 | ATG, no ATG | > .001‖ |
| Acute GVHD | Yes = 9; No = 16 | Yes = 13; No = 19 | Yes, No | −.78 |
| Chronic GVHD‡ | Yes = 18; No = 7 | NA | NA | |
| Oral chronic GVHD‡ | Yes = 14; No = 11 | NA | NA | |
| Time from oral cGVHD diagnosis to sampling | Median = 9 y; range = 4-21 y | NA | NA | |
| Active oral cGVHD at time of sampling | Yes = 2; N = 23 | NA | NA | |
| Patients on immunosuppression§ at time of sampling | Yes = 2; No = 23 | |||
| Controls: autologous HCT survivors (controls for alloreactive milieu and effect of chemotherapy) . | |
|---|---|
| Characteristics . | Distribution and range . |
| N | 11 |
| Sex | Male = 7; female = 4 |
| Age at transplantation | Median = 54 y; range = 28-63 y |
| Age at time of sample collection | Median = 57 y; range = 50-68 y |
| Time after transplantation | Median = 5 y; range = 3 mo-8 y |
| Source of graft | All blood stem cells |
| Disease | Lymphoma = 10; amyloidosis = 1 |
| Conditioning* | BEAM = 7, Flu+Bu = 2, Mel+TBI = 1, Mel = 1 |
| Controls: autologous HCT survivors (controls for alloreactive milieu and effect of chemotherapy) . | |
|---|---|
| Characteristics . | Distribution and range . |
| N | 11 |
| Sex | Male = 7; female = 4 |
| Age at transplantation | Median = 54 y; range = 28-63 y |
| Age at time of sample collection | Median = 57 y; range = 50-68 y |
| Time after transplantation | Median = 5 y; range = 3 mo-8 y |
| Source of graft | All blood stem cells |
| Disease | Lymphoma = 10; amyloidosis = 1 |
| Conditioning* | BEAM = 7, Flu+Bu = 2, Mel+TBI = 1, Mel = 1 |
| Controls: chemotherapy recipients without HCT (controls for effect of chemotherapy) . | |
|---|---|
| N | 9 |
| Sex | Male = 2; female = 7 |
| Age at time of sample collection | Median = 52 y; range = 31-60 y |
| Time before transplantation | Median = 11 d; range = day 3-36 |
| Disease, stage | AML in 1st remission = 3; AML in/beyond 2nd remission = 4; CLL/lymphoma in/beyond 2nd remission = 2 |
| Controls: healthy controls | |
| N | 27 |
| Sex | Male = 10; female = 17 |
| Age at time of sample collection | Median = 38 y; range = 18-54 y |
| Controls: chemotherapy recipients without HCT (controls for effect of chemotherapy) . | |
|---|---|
| N | 9 |
| Sex | Male = 2; female = 7 |
| Age at time of sample collection | Median = 52 y; range = 31-60 y |
| Time before transplantation | Median = 11 d; range = day 3-36 |
| Disease, stage | AML in 1st remission = 3; AML in/beyond 2nd remission = 4; CLL/lymphoma in/beyond 2nd remission = 2 |
| Controls: healthy controls | |
| N | 27 |
| Sex | Male = 10; female = 17 |
| Age at time of sample collection | Median = 38 y; range = 18-54 y |
HCT indicates hematopoietic cell transplantation; GVHD, graft-versus-host disease; AML, acute myeloid leukemia; Allo, allogeneic HCT; ALL, acute lymphoid leukemia; CML, chronic myeloid leukemia; CLL, chronic lymphocytic leukemia; MM, multiple myeloma; NHL, non-Hodgkin lymphoma; SAA, severe aplastic anemia; MDS, myelodysplastic syndrome; CMML, chronic myelomonocytic leukemia; MF, myelofibrosis; BEAM, BCNU+etoposide+cytarabine+melphalan; Flu, fludarabine; Bu, busulfan; TBI, total body irradiation; Cy, cyclophosphamide; VP16, etoposide; Mel, mephalan; CSA, cyclosporine A (given for the first 3-6 months after transplantation, longer in case of chronic GVHD); MTX, methotrexate (given on days 1, 3, 6, 11); ATG, antithymocyte globulin (given on days −2, −1, 0); BM, bone marrow; and PBSC, peripheral blood stem cells (filgrastim-mobilized).
Controls for short-term effect of MTX or CSA.
Myeloablative in all patients, except possibly the one patient with severe aplastic anemia who received 200 mg/kg cyclophosphamide.
At any time between transplant and sample collection. In most patients, cGVHD was inactive at the time of sample collection.
Mycophenolate mofetil.
Significant P value.
Buccal and nasal swabs were collected by twirling a sterile swab on the inside surface of each cheek or inferior turbinate of nose. Blood was drawn within 1 week of swabs. DNA from blood leukocytes and buccal and nasal swabs was extracted using the QIAGEN QIAamp DNA Micro Kit. DNA was PCR amplified for 15 autosomal tetra-nucleotide microsatellites (Identifiler; Applied Biosystems) using manufacturer's recommended operating procedures. The microsatellites are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The fluorochrome-labeled amplicons were size-fractionated by capillary electrophoresis on ABI-3130 genetic analyzer (Applied Biosystems). The allele analysis was done by Genemapper-v2 software (Applied Biosystems). Stutter peaks, dye-associated peaks, blobs and spikes were excluded. MSI was defined as previously described.11,12 Briefly, only peak shifts or gains and not peak intensity changes were counted for MSI. At least 3 successful PCRs were performed for each specimen to designate microsatellite as stable or unstable.
Results and discussion
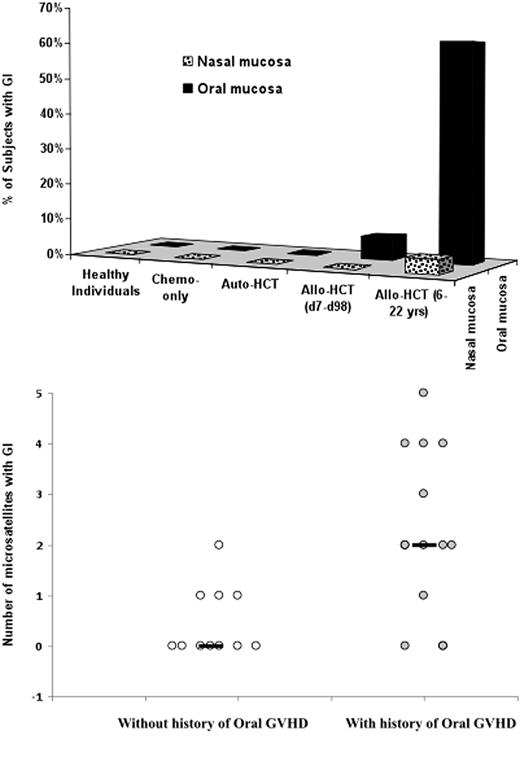
Genomic instability (defined here as MSI) was detected exclusively in the allo-HCT recipients. GI typically occurred late after transplantation and was frequently observed in the oral mucosal cells (60% long-term and 6% short-term allo-HCT survivors) and rarely in the nasal mucosal cells (4% long-term and 0% short-term allo-HCT survivors; Figure 1 top). The difference between the short-term and the long-term survivors in the occurrence of GI in oral mucosa was significant (P < .001, Fisher exact test), as was the difference between oral and nasal mucosal GI occurrence in long-term survivors (P < .001, Fisher exact test). GI occurred in mucosal cells (presumably epithelial cells) and not in leukocytes infiltrating the mucosa, as no GI was detected in blood leukocytes of any subject and was detected in the nasal mucosa of only 1 of 57 allo-HCT recipients.
Genomic instability (based on microsatellite analysis) in oral and nasal mucosa. (Top) Percent of subjects with genomic instability detected in oral and nasal specimens among the following subjects: Healthy individuals, Chemotherapy-only controls (received chemotherapy but no transplantation), Autologous HCT controls, Short-term survivors of allogeneic HCT (7-98 days), and Long-term survivors of allogeneic HCT (4-22 years). (Bottom) Number of microsatellite markers showing instability in long-term allogeneic HCT survivors with and without history of oral cGVHD. Black horizontal lines represent the median number of microsatellite markers showing instability. Significance of difference calculated for the 2 groups was P = .007 in univariate analysis (Mann-Whitney-Wilcoxon rank sum test) and P = .005 in multivariate analysis (multinomial logistic regression).
Genomic instability (based on microsatellite analysis) in oral and nasal mucosa. (Top) Percent of subjects with genomic instability detected in oral and nasal specimens among the following subjects: Healthy individuals, Chemotherapy-only controls (received chemotherapy but no transplantation), Autologous HCT controls, Short-term survivors of allogeneic HCT (7-98 days), and Long-term survivors of allogeneic HCT (4-22 years). (Bottom) Number of microsatellite markers showing instability in long-term allogeneic HCT survivors with and without history of oral cGVHD. Black horizontal lines represent the median number of microsatellite markers showing instability. Significance of difference calculated for the 2 groups was P = .007 in univariate analysis (Mann-Whitney-Wilcoxon rank sum test) and P = .005 in multivariate analysis (multinomial logistic regression).
To determine which factors are associated with the development of oral GI, we searched for associations between oral GI and clinical/demographic characteristics of the long-term allo-HCT recipients including age at time of transplantation and sampling, time from oral cGVHD diagnosis to sampling, sex, underlying disease (myeloid vs lymphoid), type of conditioning (TBI vs no TBI), type of the graft (bone marrow vs peripheral blood stem cells), immunosuppression at the time of sampling (yes vs no), history of mucositis (presence vs absence), history of cGVHD of organs other than oral mucosa (presence vs absence) and history of oral cGVHD (presence vs absence). The only significant association was observed with the history (P = .05, Fisher exact test) of oral cGvHD (supplemental Table 2). Despite this was only marginally significant, the association between the history of oral cGVHD and the number of microsatellites showing GI in the oral mucosal specimens (Figure 1 bottom) was highly significant (P = .007, Mann-Whitney-Wilcoxon rank sum test). This association was also significant in a multivariate analysis considering patient age at the time of sampling and sex as a covariates (P = .005, multinomial logistic regression), because age and sex are known to be associated with GI.13 GI did not appear to be a result of cytotoxic therapy as none of the recipients receiving intensive chemotherapy with or without autologous HCT were positive for GI (Figure 1 top). Likewise, Faber et al did not find GI in recipients receiving chemotherapy without allo-HCT,12 even though chemotherapeutic agents can induce GI in vitro.14 Also, GI did not appear to be a result of GVHD prophylaxis as GI was rarely observed in the short-term allo-HCT survivors (at time of or soon after administration of methotrexate and cyclosporine). However, we could not completely rule out this possibility as methotrexate or cyclosporine might have a late effect. It may also not be true that GI resulted from inadequately treated cGVHD as cGVHD was inactive in 12 of 14 patients (supplemental Table 2). It is also unlikely that the observed GI was a result of technical artifacts such as stutter peaks, contamination with exogenous DNA or differences in PCR efficiency, as we followed a rigorous protocol and used multiple controls. Therefore, it would be extremely unlikely to have an identical pattern of MSI peaks in multiple PCR experiments, if the MSI was not true. Moreover, examination of oral mucosal specimens from controls performed under similar conditions did not result in MSI detection.
A limitation of our study is that we analyzed only tetra-nucleotide STRs. Thus, though unlikely, it is theoretically possible that instability of mono/bi/tri-nucleotide STRs or other markers of GI were present with similar frequency in oral and nasal mucosa, early and late after transplantation, in allogeneic and autologous transplantation recipients and in allograft recipients with and without history of oral cGVHD. However, Faber et al12 found MSI only at tetranucleotide STRs and not at any of the 3 mononucleotide STRs tested in colonic and oral epithelium. A second limitation of the study is that although we were able to demonstrate the association of GI with the history of oral cGVHD, we did not determine the precise mechanism. A recent report showed that alloreactive microenvironment after HCT induces GI in epithelium through a reactive oxygen species (ROS)–mediated mechanism supports our results.15 Finally, we have not studied any patients with secondary oral squamous cell malignancy. Faber et al12 documented GI in 3 of 3 patients with secondary squamous cell malignancy and Themeli et al15 reported GI in 5 of 6 patients with secondary epithelial cell malignancy.
The most important finding of our study is that instability of tetranucleotide STRs (and possibly GI in general) occurs frequently in oral and rarely in nasal mucosal cells, typically late after transplantation and typically in patients with history of oral cGVHD. We speculate that the chronic inflammation associated with cGVHD may induce GI,16-19 as GI was not detected in auto-HCT recipients and was more frequently detected in allo-HCT recipients with history of oral cGVHD. Faber et al also found GI in oral mucosa in only allograft and not autograft recipients, however, they did not find an association between GI and history of oral GVHD.12 This may be because Faber et al analyzed oral specimens from only 14 allo-HCT recipients surviving more than 1 year after transplantation, and used 3 tetra-nucleotide microsatellites spanning 3 chromosomes. In contrast, we analyzed 15 microsatellites spanning 13 chromosomes. This is critical as the occurrence of GI may be limited to some chromosomes.8
Even though we speculate that the pathogenesis of oral cancer after transplantation may be “GVHD→GI→oral cancer,” we have not evaluated the hypothesis that GI leads to posttransplantation oral cancer. An argument against this hypothesis is that even though posttransplantation GI is frequently detected in colonic epithelium,12 colonic carcinoma incidence in transplant recipients is not higher than in the general population.1,3 Conversely, an argument in favor of the above hypothesis is that the type of GI (tetra-nucleotide MSI) observed in our study is frequently detected in sporadic carcinomas.20 Larger studies with longer follow-up are needed to evaluate the “GVHD→GI→oral cancer” hypothesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded by the Alberta Heritage Foundation for Medical Research, the Canada Research Chair Program, the Canada Foundation for Innovation, and the Alberta Cancer Foundation. J.S. is a recipient of Canada Research Chair and Alberta Heritage Foundation Clinical Scholar awards. The work was selected for the travel award at the annual tandem meeting of the Center for International Blood and Marrow Transplantation/American Society for Blood and Marrow Transplantation–2008.
Authorship
Contribution: F.M.K. performed experiments, analyzed data, and generated the initial and final drafts of the manuscript; S.S. performed experiments; P.L. recruited subjects and collected specimens; A.U.-T. performed data analysis; N.B. and G.D.S. provided pretransplantation specimens of donors and recipients and edited the final draft of the manuscript; D.A.S. and J.A.R. recruited allogeneic and autologous HCT patients, respectively; and J.S. designed the study and edited the draft and the final versions of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Faisal M. Khan, PhD, AE-412, 9, 3535 Research Rd NW, Calgary, Alberta, Canada T2L 2K8; e-mail: fkhan@ucalgary.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal