Abstract
Heat shock protein 27 (HSP27) is a chaperone whose cellular expression increases in response to various stresses and protects the cell either by inhibiting apoptotic cell death or by promoting the ubiquitination and proteasomal degradation of specific proteins. Here, we show that globin transcription factor 1 (GATA-1) is a client protein of HSP27. In 2 models of erythroid differentiation; that is, in the human erythroleukemia cell line, K562 induced to differentiate into erythroid cells on hemin exposure and CD34+ human cells ex vivo driven to erythroid differentiation in liquid culture, depletion of HSP27 provokes an accumulation of GATA-1 and impairs terminal maturation. More specifically, we demonstrate that, in the late stages of the erythroid differentiation program, HSP27 is phosphorylated in a p38-dependent manner, enters the nucleus, binds to GATA-1, and induces its ubiquitination and proteasomal degradation, provided that the transcription factor is acetylated. We conclude that HSP27 plays a role in the fine-tuning of terminal erythroid differentiation through regulation of GATA-1 content and activity.
Introduction
Terminal erythroid differentiation is driven by the glycoprotein hormone erythropoietin (Epo) and involves the sequential formation of proerythroblasts and basophilic, polychromatic, and orthochromatic erythroblasts in the bone marrow. This differentiation program is under control of the transcription factor GATA-1 (globin transcription factor 1),1,2 which induces the expression of erythroid genes such as glycophorin A, Epo receptor, and hemoglobin.3 GATA-1 also cooperates with Epo to promote erythroid precursor survival by positively regulating the bcl-xL gene.3-5 On Epo starvation or engagement of the death receptor Fas (CD95/APO-1), caspases are activated and GATA-1 is cleaved. These events arrest erythroid precursor maturation and provoke cell death.6,7 On Epo stimulation of erythroid precursors, caspase-3 is also transiently activated, and this activation is required for terminal erythroblast maturation, but GATA-1 remains uncleaved and erythroid cells do not die.8-10 We have demonstrated that, in Epo-stimulated erythroid precursors, GATA-1 was protected from caspase-3–mediated cleavage by the stress-inducible heat shock protein 70 (HSP70). At the onset of caspase-3 activation, HSP70 translocates from the cytoplasm to the nucleus and interacts with GATA-1. Epo deprivation disrupts the GATA-1/HSP70 interaction and exposes GATA-1 to the proteolytic effect of caspase-3.11
Another stress-inducible HSP that is expressed in erythroid cells undergoing differentiation is HSP27.11 This small stress protein has shown prosurvival functions through interaction with proteins such as cytochrome c, caspase-3, death domain–associated protein, and actin (for a review, see Lanneau et al12 ). HSP27 is also an adenosine triphosphate (ATP)–independent chaperone that binds ubiquitin, with a higher affinity for long chains of ubiquitin than for mono-ubiquitin, and orchestrates the degradation of proteins such as the nuclear factor κB inhibitor13 and the cell cycle inhibitory protein, p27kip1.14 This confers a function to this chaperone in the so-called “protein triage” that occurs in stressful conditions.15 The role of HSP27 in erythroid cell differentiation remained unexplored.
In the present study, we demonstrate that HSP27 depletion in red cell precursors provokes an increase in GATA-1 content and prevents terminal erythroid differentiation. A strict regulation of GATA-1 expression level and activity might be critical for a fine-tuning of terminal erythroid differentiation; that is, murine erythroleukemic cells overexpressing GATA-1 fail to undergo terminal differentiation in response to chemical inducers,16 and embryonic stem cells overexpressing GATA-1 generate erythroid colonies whose terminal differentiation is inhibited.17 The transcription factor can be acetylated, which, on one hand, stimulates its DNA binding capability and enhances its transcriptional activity and, on the other hand, targets its ubiquitinylation and degradation by the proteasome. Likewise, mutation of the main sites of acetylation prevents GATA-1 from inducing erythroid differentiation.18 We demonstrate that, in late stages of erythroid differentiation, a phosphorylated form of HSP27 generated in a p38-dependent manner binds to acetylated GATA-1 to promote its ubiquitination and proteasomal degradation. These results identify a function for HSP27 in terminal red cell differentiation through regulating GATA-1 expression level.
Methods
Cell culture, reagents, and transfections
Human erythroleukemia K562 cells were cultured in RPMI medium (BioWittaker); HeLa and COS cells were cultured in Dulbecco minimal essential medium (BioWittaker). Each cell line was supplemented with 10% (vol/vol) fetal bovine serum (Gibco BRL). Transfection was performed with the Jetpei reagent according to the manufacturer's instruction (Polyplustransfection; Ozyme). Plasmids used were HA-HSP27-Wt, HSP27-Ala (kindly provided by M. Gaestel, Institut fur biochemie) and GATA-Myc and AcMut-GATA-1.19 Empty plasmids were used as transfection controls. K562 cells and primary erythroblasts were also transfected with scramble and HSP27 small interfering RNA (siRNA; 20nM; Ambion) with the use of INTERFERin according to the manufacturer's instructions (Polyplustransfection). The protein synthesis inhibitor cycloheximide (CHX; Sigma-Aldrich) was used at a final concentration of 5μM for 8 hours, the proteasome inhibitor MG132 (Euromedex) at 20μM for 5 hours, trichostatin A at 300nM for 15 hours, hemin at 40μM, SB203580 20μM (added 8 hours before differentiation and then each 24 hours) and G418 at 800 μg/mL (all from Sigma-Aldrich).
Primary erythroblast culture
Umbilical cord blood units from normal full-term deliveries were obtained, with the agreement of mothers, from the Obstetrics Unit of Hôpital Necker-Enfants Malades. Erythroid cells were generated with a 2-step amplification culture system as previously described.8 Briefly, CD36+ erythroid progenitors were generated from CD34+ progenitors, isolated from cord blood (CD34 Progenitor Cell Isolation Kit; Miltenyi Biotec), after 6 days of treatment with a cocktail of cytokines (interleukin-6 [IL-6; 100 ng/mL], IL-3 [10 ng/mL], stem cell factor [SCF; 100 ng/mL]) from Diaclone. Sorted CD36+ erythroid progenitors were further cultured in the presence of IL-3 (10 ng/mL), SCF (100 ng/mL), Epo (Sigma-Aldrich; 2 U/mL) in Iscove modified Dulbecco medium (Gibco BRL) supplemented with 15% BIT 9500 (StemCell Technologies). When indicated, 5 × 106 CD34+ progenitor cells treated for 5 days with IL-6 (100 ng/mL), IL-3 (10 ng/mL), SCF (100 ng/mL) were transfected with 3 μg of HSP27 specific or scramble siRNA in a human CD34 Cell Nucleofector buffer using a Nucleofector (Amaxa Biosystems) according to the manufacturer's protocol. Cells were further cultured for 1 day with IL-6, IL-3, and SCF, then with Epo (2 U/mL), IL-3 (10 ng/mL), and, when indicated, transforming growth factor-β (TGFβ; 2.5 ng/mL; R&D Systems) in serum-free medium.
Primary erythroblast infection
CD34+ cells isolated from cord blood were cultured during 5 days as described in “Primary erythroblast culture,” then infected by adding a mix of 3 different short hairpin RNA (shRNA) HSP27 lentiviral particles (SH-005269-01, SH-005269-02, SH-005269-03; Thermo Scientific, Dharmacon) or shRNA lentiviral control particles (Thermo Scientific, Dharmacon). For GATA-1 overexpression, GATA-1 lentiviral particles were produced as previously described.11 A second round of infection was performed 24 hours later by changing fresh medium with cytokines. After 24 hours, cells were extensively washed in phosphate-buffered saline (PBS) and stained with anti–CD36-allophycocyanin monoclonal antibody. The CD36+ green fluorescent protein–positive cell population was purified by cell sorting and cultured in serum-free media in the presence of IL-3, SCF, and Epo.
Erythroid cell differentiation
Differentiation of primary cells in liquid culture was assessed morphologically on May-Grünwald-Giemsa–stained smears. Primary erythroblasts and K562 cells containing heme or hemoglobin were detected by a specific reaction with a benzidine/hydrogen peroxide solution as described.20,21 The final concentration of benzidine was 0.2% in 0.5M glacial acetic acid, 3% H2O2 (Sigma-Aldrich). To detect the formation of reticulocytes, CD36+ green fluorescent protein–positive erythroid progenitor sorted cells (described in “Primary erythroblast transfections”) were cultured with Epo, IL-3, or TGFβ for 6 days as described in “Primary erythroblast culture,” or in the presence of 30% fetal calf serum (HyClone; Thermo Scientific) for 18 days. These culture conditions allowed the production of around 3% to 5% and 30% to 35% enucleated cells, respectively. After May-Grünwald-Giemsa staining,8 the production of reticulocytes was assessed by microscopy (Leica DMRB microscope with a PLFluotar 40× oil objective), and the percentage was determined on 300 cells randomly chosen in different microscopic fields. Vectashield was used as a mounting medium (Cliniscience). For image acquisition, Ziess LSM5 Pascal software was used (Carl Ziess Nv-SA), and for image processing, Adobe Photoshop 7 was used.
Immunoblotting and antibodies
Proteins were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) 10% to 8% and transferred to nitrocellulose membrane with the use of wet transfer apparatus (Bio-Rad). After blocking the nonspecific binding sites with 5% nonfat dry milk or 5% bovine serum albumin, the membranes were probed overnight with primary antibodies and then incubated for 1 hour with the appropriate secondary antibody coupled to horseradish peroxidase (Dako). Proteins were visualized with the use of the enhanced chemiluminescence Western blotting kit (Santa Cruz Biotechnology). Goat anti–GATA-1, -lamin B, -actin, rabbit anti-HSP90, 14-3-3, mouse anti-ubiquitin, -HSC70, nonrelevant immunoglobulin G were purchased from Santa Cruz Biotechnology; rabbit anti-HSP27 and anti-phospho Ser15 and 78 HSP27 were purchased from Stressgen; Myc-tag was purchased from Millipore; and hemagglutinin (HA)–tag was purchased from Biomol.
Immunoprecipitation
Cells were lysed in immunoprecipitation buffer [50mM HEPES (N-2-hydroyethylpiperazine-N′-2-ethanesulfonic acid) pH 7.4, 140mM NaCl, 5mM EDTA (ethylenediaminetetraacetic acid), 0.2% NP40, protease inhibitor cocktail (Roche)] for 30 minutes on ice followed by centrifugation at 12 000g for 10 minutes at 4°C. The protein concentration was evaluated (DC protein assay, Bio-Rad). Except for the immunoprecipitation of endogenous GATA-1/HSP27 in K562 cells, which was performed with the Exactacruz system (Santa Cruz Biotechnology; sc-45039), 800 μg of each lysate was precleared with 40 μL of sepharose beads for 45 minutes and incubated with 3 μg of GATA-1 (Santa Cruz Biotechnology), HSP27 (Stressgen), Myc-tag (Millipore), or HA-tag (Biomol) antibodies with constant agitation at 4°C. Then, the immunocomplexes were precipitated with 25 μL of protein G-Sepharose (Amersham Bioscience, GE Healthcare), and the beads were washed 4 times in 20mM HEPES, pH 7.4, EDTA 5mM, NaCl 150mM, NP40 0.2%, and resuspended in Laemmli buffer. The fraction of each cell lysate used in the inputs was 40 μg.
For ubiquitination studies in vivo, immunoprecipitation was performed in the presence of 5μM ubiquitin-aldehyde (Boston Biochem) to block the de-ubiquitination process, and an agarose-conjugated anti-multi Ubiquitin (MBL) was used.
In vitro ubiquitination assay
Ubiquitination reactions were performed with Ubiquitin-Protein Conjugation Kit (Boston Biochem, Euromedex). The manufacturer's protocol was slightly modified as follows: the 55-μL final volume reactions were performed in a buffer containing 30 μg of ubiquitin, 14.5 μg of fraction A, 14.5 μg of fraction B, 1× energy solution, 5μM ubiquitin-aldehyde (Boston Biochem), and 2μM MG132. In vitro–translated human GATA-1 protein was used as substrate in the presence or absence of in vitro–translated human HSP27. Both proteins were produced with TNT Quick Coupled Transcription/Transcription System (Promega) as follows: 1 μg of template plasmid DNA was added to the reaction mixture that was afterward incubated at 30°C for 90 minutes. The in vitro–translated proteins (3 μL) were preincubated with ubiquitin-aldehyde at 37°C for 6 minutes before adding the ubiquitinylation reaction mix. Samples were incubated at 37°C for 3.5 hours, and the reaction was stopped by adding Laemmli buffer. The samples were boiled 5 minutes and then resolved on sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Nucleus/cytoplasm extraction
Cytoplasm and nucleus extracts were obtained after lysis of 5 to 10 × 106 cells for 10 minutes on ice in lysis buffer (20mM HEPES pH 7.4, 10mM KCl, 1mM EDTA, 10% glycerol) with 0.2% NP-40 in the presence of protease inhibitors. Cell lysates were centrifuged at 14 000 rpm for 10 minutes, and the supernatant was carefully collected (cytoplasm fraction). The pellet was washed once and resuspended in lysis buffer (20mM HEPES pH 7.4, 10mM KCl, 1mM EDTA, 20% glycerol, 350mM NaCl, protease inhibitors), and nuclear fractions were harvested after centrifugation (16 000g, 10 minutes).
Proteasome activity
Proteasome activity was determined as described.13,14 Briefly, 4 × 106 cells in 200 μL of PBS (pH 7.4) were incubated for 30 minutes at 37°C with 100μM of the cell-permeant fluorogenic substrate N-succinyl-L-leucyl-L-leucyl-L-tyrosine-7-amido-4-methyl coumarin (Bachem). Fluorescence generated by the substrate cleavage was quantified with the use of a Kontron SFM 25 spectrofluorometer (Kontron AG). When needed, proteasome activity was inhibited by exposure of the cells to MG132 (Euromedex).
Yeast 2 hybrid assay
The bait vector used was pGBKT7 and pGADT7, the prey vector (Clontech). We cloned human wild-type (WT)–GATA-1 full-length coding sequence, and the deletion mutants GATA-1Δ1-83 and GATA-1Δ84-413, into ClaI/XhoI sites of the pGADT7. Human WT-HSP27 coding sequence was cloned into the EcoRI/BamHI sites of the pGBKT7. As positive control bait plasmid we use pGBKT7-p53. This plasmid encodes the GAL4 DNA-BD fused with murine p53. As positive control prey plasmid we used pGADT7-SV40 that encodes the GAL4 activation domain fused with SV40 large T-antigen. The empty vector pGADT7 was used as a negative control prey plasmid. All pGADT7- and pGBKT7-derived vectors were transformed into Y187 and Y2HGold yeast strains, respectively (Clontech Yeastmaker Yeast Transformation System 2). The transformants were, respectively, selected on Leu− and Trp− minimal media plates after growth at 30°C for 3 to 5 days. Each prey strain was mating with the bait strain to generate diploid yeast cells (Clontech Matchmaker Gold Yeast 2-Hybrid system). Diploid yeast cells selected on Leu−Trp− minimal media plates were then patched onto Leu−Trp− minimal media plates with X-α-Galactosidase (40 μg/mL) and Aureobasidin A (70 ng/mL). Blue diploid cells appear after 3 to 5 days at 30°C, indicating the interaction between the bait protein and the prey one. To confirm the result, the diploid yeast cells were then patch onto a higher stringency His−Ade−Leu−Trp− minimal media plates supplemented with X-α-Galactosidase (40 μg/mL) and Aureobasidin A (70 ng/mL).22,23
Immunofluorescence staining
Cells were fixed in PBS-paraformaldehyde 4% during 15 minutes and permeabilized by incubation with PBS-Triton 0.1% for 3 minutes. After washing with PBS (Cambrex), samples were saturated with PBS–bovine serum albumin 3% (Sigma-Aldrich) during 30 minutes before incubation overnight at 4°C with anti-HSP27 (Stressgen), anti–phospho-Ser15-HSP27 (Stressgen), or anti–GATA-1 (Santa Cruz Biotechnology). After 4 washes in PBS, appropriate secondary antibodies coupled with fluorochromes (Alexa 486 and 568 nm; Molecular Probe) were added during 1 hour at room temperature in the dark. The nucleus was labeled with Hoescht 33342. Aquatex was used as a mounting medium (Merck). For image acquisition, LASAF software was used (Leica Microsystèmes SAS), and for image processing, Adobe Photoshop 7 was used.
Statistics
Quantitative experiments were analyzed with the use of Student t test. All P values resulted from the use of 2-sided tests.
Results
Expression of GATA-1 and HSP27 during terminal erythroid differentiation
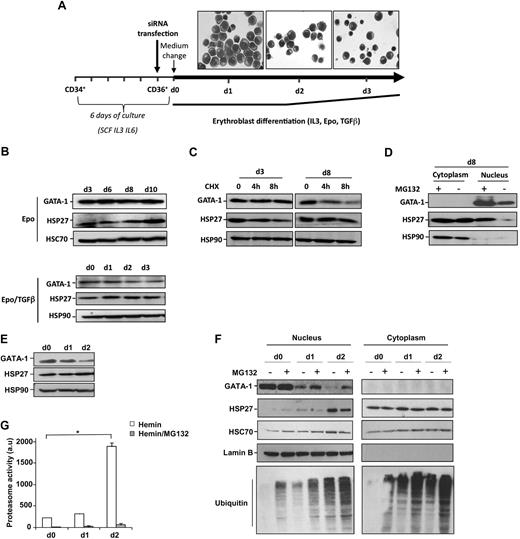
The expression of GATA-1 and HSP27 expression was studied in 2 models of erythroid cell differentiation. CD34+ cells isolated from human cord blood were cultured in the presence of IL-6 (100 ng/mL), IL-3 (10 ng/mL), and SCF (100 ng/mL) for 5 days before sorting CD36+ cells. The following day, the medium was changed, and CD36+ cells were cultured for the indicated additional days with Epo (2 U/mL) and IL-3 (10 ng/mL; Figure 1A). When indicated, TGFβ (2.5 ng/mL) was added to accelerate the differentiation process.24 In this way, days 1, 2, and 3 in the presence of TGFβ, shown in Figure 1A, correspond to days 2, 4, and 6 in the absence of TGFβ. GATA-1 levels were quite constant during erythroblast differentiation (Figure 1B). The addition of CHX (5μM) for 4 hours before collecting the cells indicated that GATA-1 turnover was faster at day 8 compared with day 3 (Figure 1C). At this concentration and time of treatment, CHX did not have any significant effect in cell survival (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Inhibition of the proteasome machinery at day 8 with either MG132 (20μM, 5 hours; Figure 1D) or lactacystin (25μM, 4 hours, not shown) induced an accumulation of the protein in the nucleus (Figure 1D), indicating that the proteasomal degradation of GATA-1 was involved in its turnover. In human erythroid K562 cells induced to differentiate in the presence of hemin (40μM), the expression of GATA-1 decreased during erythroid differentiation (Figure 1E). This decrease was partially prevented by MG132 (Figure 1F) and lactacystin (not shown). GATA-1 decrease correlated with an increase in the proteasome activity measured in cell lysates (Figure 1G) and ubiquitinated proteins in the nucleus of differentiated cells in the presence of MG132 (Figure 1F). Because HSP27 is expressed throughout the differentiation process in both models (Figure 1B-C,E-F) and partially accumulates in the nucleus of differentiating cells (Figure 1D,F), we explored whether HSP27 was involved in GATA-1 protein level regulation.13,14
HSP27 and GATA-1 expression during erythroblast differentiation. (A) Scheme of the model used for human primary erythroblast differentiation. A representative microscopic image shows the structure of the differentiating erythroblasts at the indicated time points. (B) Human primary erythroblasts were induced to differentiate in the presence of IL-3, Epo with or without TGFβ. At the indicated times, the level of GATA-1 and HSP27 was determined by Western blot. HSC70 an HSP90 serve as loading control. (C) Differentiating cells were treated or not with the protein synthesis inhibitor CHX (5μM, 8 hours). Cells were harvested at indicated days, and lysates were blotted with the indicated antibodies. The data are representative of 3 independent experiments. (D) Human primary erythroblasts induced to differentiate in the presence of IL-3 and Epo were, at day 8, treated or not with the proteasome inhibitor MG132 (20μM, 5 hours), and nuclear/cytosolic GATA-1 and HSP27 expression was assessed by Western blotting. (E) Human K562 cells were induced to differentiate with hemin (40μM). At the indicated days, GATA-1 and HSP27 levels were assessed by Western blot. HSP90 serves as a loading control. (F) When indicated, K562 cells induced to differentiate by the presence of hemin were treated for 5 hours with the proteasome inhibitor MG132 (20μM). Nuclear/cytosolic GATA-1 and HSP27 expression was assessed by Western blot. HSC70 and lamin B serve as loading controls. (G) The proteasome activity was determined by the measurement of Suc-LLVY-AMC cleavage in the control lysates from hemin-treated K562 cells in the absence ( ) or presence (
) or presence ( ) of MG132 (20μM, 5 hours). a.u. indicates arbitrary units; bars, SD; n = 3.
) of MG132 (20μM, 5 hours). a.u. indicates arbitrary units; bars, SD; n = 3.
HSP27 and GATA-1 expression during erythroblast differentiation. (A) Scheme of the model used for human primary erythroblast differentiation. A representative microscopic image shows the structure of the differentiating erythroblasts at the indicated time points. (B) Human primary erythroblasts were induced to differentiate in the presence of IL-3, Epo with or without TGFβ. At the indicated times, the level of GATA-1 and HSP27 was determined by Western blot. HSC70 an HSP90 serve as loading control. (C) Differentiating cells were treated or not with the protein synthesis inhibitor CHX (5μM, 8 hours). Cells were harvested at indicated days, and lysates were blotted with the indicated antibodies. The data are representative of 3 independent experiments. (D) Human primary erythroblasts induced to differentiate in the presence of IL-3 and Epo were, at day 8, treated or not with the proteasome inhibitor MG132 (20μM, 5 hours), and nuclear/cytosolic GATA-1 and HSP27 expression was assessed by Western blotting. (E) Human K562 cells were induced to differentiate with hemin (40μM). At the indicated days, GATA-1 and HSP27 levels were assessed by Western blot. HSP90 serves as a loading control. (F) When indicated, K562 cells induced to differentiate by the presence of hemin were treated for 5 hours with the proteasome inhibitor MG132 (20μM). Nuclear/cytosolic GATA-1 and HSP27 expression was assessed by Western blot. HSC70 and lamin B serve as loading controls. (G) The proteasome activity was determined by the measurement of Suc-LLVY-AMC cleavage in the control lysates from hemin-treated K562 cells in the absence ( ) or presence (
) or presence ( ) of MG132 (20μM, 5 hours). a.u. indicates arbitrary units; bars, SD; n = 3.
) of MG132 (20μM, 5 hours). a.u. indicates arbitrary units; bars, SD; n = 3.
HSP27 depletion inhibits terminal erythroid differentiation
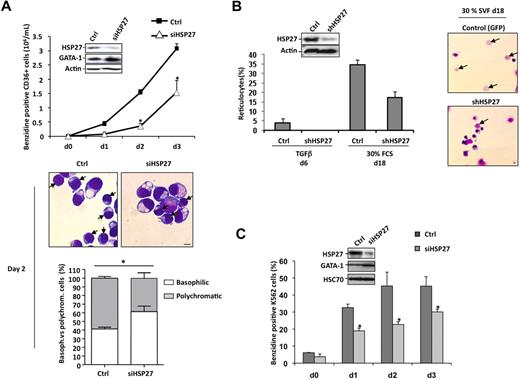
HSP27 was depleted from sorted CD36+ cells by transfection with a specific siRNA (siHSP27). As described in Figure 1A, the medium was changed after 24 hours, and the cells were cultured with Epo and IL-3 in the presence of TGFβ to accelerate and synchronize the differentiation process. The efficacy of HSP27 siRNA was checked by immunoblot at day 2 (Figure 2A insert). The decrease in HSP27 protein expression was associated with a delay in the appearance of benzidine-positive cells (Figure 2A) and in the morphologic maturation of erythroblasts; for example, at day 2 the percentage of polychromatic (mature) erythroblasts decreased from 59% plus or minus 2% in control siRNA to 37% plus or minus 6.2% in HSP27-siRNA–transfected cells (P < .05; Figure 2A bottom panels). Confirming the negative effect of HSP27 depletion on erythroid cell differentiation, HSP27-targeting siRNA induced a decrease in the appearance of cells expressing the erythroblast differentiation markers glycophorin A (GPA) and transferrin receptor CD71 (supplemental Figure 2). This decrease was not related to an increase in mature cell death rate when HSP27 was down-regulated (supplemental Figure 3A-B).
HSP27 depletion delays erythroid differentiation of human primary erythroblasts and K562 cells. (A) Human primary erythroblasts were induced to differentiate with Epo, IL-3,and TGFβ (2.5 ng/mL) after 24 hours of transfection with HSP27 siRNA (siHSP27) or control siRNA (Ctrl,) (20nM). Percentages of cell transfection and survival were approximately 60% to 70% and 80%, respectively. At the days indicated (day 0 to day 3) the percentage of differentiated cells was evaluated by benzidine assay. Insert, cell lysate from day 2 was resolved on a gel and blotted with the indicated antibodies. (Middle) Differentiating cells at day 2 were visualized by microscopy after cell fixation and staining with May-Grünwald-Giemsa (magnification ×40; bar, 10μm). → indicates polychromatic. One representative image is shown. (Bottom) Percentages of basophilic and polychromatic cells. Quantification, at day 2, from a total number of 300 cells in randomly chosen microscopic fields. (B) CD34+ cells, growing in presence of cytokines (IL-3, Epo), were transduced with shRNA specific for HSP27 or shRNA control (Ctrl). At day 5, CD36+ green fluorescent protein–positive (GFP+) cells were sorted and differentiated in the presence of 30% SVF or TGFβ (2.5 ng/μL) to allow the production of reticulocytes. (Insert) Western blot analysis of HSP27 expression after shRNA transduction. Actin serves as a loading control. Reticulocytes were quantified at the indicated times as the ratio of their number to a total number of 300 cells chosen randomly in different microscopic fields. (Right) Reticulocytes visualized by microscopy after cell fixation and staining with May-Grünwald-Giemsa (magnification ×20; bar, 10 μm). → indicates reticulocytes. Note that HSP27 siRNA and shRNA target different sequences of HSP27 mRNA. (C) Human K562 cells were induced to differentiate with hemin (40μM) after 24 hours of transfection with HSP27 siRNA (siHSP27;  ) or scrambled (Ctrl;
) or scrambled (Ctrl;  ). The percentage of differentiated cells was determined by benzidine assay. (Insert) Cell lysate from days 0 and 2 was resolved on a gel and blotted with the indicated antibodies.
). The percentage of differentiated cells was determined by benzidine assay. (Insert) Cell lysate from days 0 and 2 was resolved on a gel and blotted with the indicated antibodies.
HSP27 depletion delays erythroid differentiation of human primary erythroblasts and K562 cells. (A) Human primary erythroblasts were induced to differentiate with Epo, IL-3,and TGFβ (2.5 ng/mL) after 24 hours of transfection with HSP27 siRNA (siHSP27) or control siRNA (Ctrl,) (20nM). Percentages of cell transfection and survival were approximately 60% to 70% and 80%, respectively. At the days indicated (day 0 to day 3) the percentage of differentiated cells was evaluated by benzidine assay. Insert, cell lysate from day 2 was resolved on a gel and blotted with the indicated antibodies. (Middle) Differentiating cells at day 2 were visualized by microscopy after cell fixation and staining with May-Grünwald-Giemsa (magnification ×40; bar, 10μm). → indicates polychromatic. One representative image is shown. (Bottom) Percentages of basophilic and polychromatic cells. Quantification, at day 2, from a total number of 300 cells in randomly chosen microscopic fields. (B) CD34+ cells, growing in presence of cytokines (IL-3, Epo), were transduced with shRNA specific for HSP27 or shRNA control (Ctrl). At day 5, CD36+ green fluorescent protein–positive (GFP+) cells were sorted and differentiated in the presence of 30% SVF or TGFβ (2.5 ng/μL) to allow the production of reticulocytes. (Insert) Western blot analysis of HSP27 expression after shRNA transduction. Actin serves as a loading control. Reticulocytes were quantified at the indicated times as the ratio of their number to a total number of 300 cells chosen randomly in different microscopic fields. (Right) Reticulocytes visualized by microscopy after cell fixation and staining with May-Grünwald-Giemsa (magnification ×20; bar, 10 μm). → indicates reticulocytes. Note that HSP27 siRNA and shRNA target different sequences of HSP27 mRNA. (C) Human K562 cells were induced to differentiate with hemin (40μM) after 24 hours of transfection with HSP27 siRNA (siHSP27;  ) or scrambled (Ctrl;
) or scrambled (Ctrl;  ). The percentage of differentiated cells was determined by benzidine assay. (Insert) Cell lysate from days 0 and 2 was resolved on a gel and blotted with the indicated antibodies.
). The percentage of differentiated cells was determined by benzidine assay. (Insert) Cell lysate from days 0 and 2 was resolved on a gel and blotted with the indicated antibodies.
CD36+-sorted cells were also transduced with a lentivirus encoding either a HSP27-specific (shHSP27) or a scrambled shRNA (Figure 2B insert) and cultured with Epo, IL-3, or TGFβ for 6 days as described in the previous paragraph, or in the presence of 30% fetal calf serum for 18 days, which led to the production of around 3% to 5% and 30% to 35% enucleated cells, respectively. In both conditions, HSP27 depletion induced a significant decrease in the formation of reticulocytes (Figures 2B) without affecting erythroblast cell death and proliferation (not shown). The siRNA HSP27 was also used to decrease HSP27 expression in K562 cells, which delayed the hemin-induced erythroid differentiation (Figure 2C).
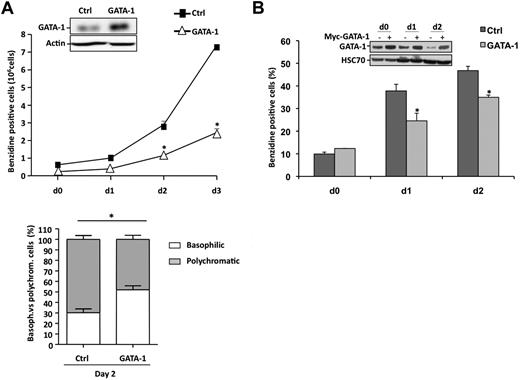
In both, CD36+-sorted cells (Figure 2A) and K562 cells (Figure 2C), HSP27 depletion correlated with an increase in GATA-1 protein level. As previously reported in mouse models,16,17 lentivirus-mediated GATA-1 overexpression in sorted human CD36+ cells (Figure 3A insert) significantly delayed the appearance of benzidine-positive cells (Figure 3A) and GPA-positive cells (supplemental Figure 4) in a TGFβ-containing medium. GATA-1 overexpression delayed the structural maturation of erythroblasts; for example, at day 2 the percentage of polychromatic (mature) erythroblasts decreased from 70% plus or minus 3.7% in control to 48% plus or minus 3.9% in GATA-1–transduced cells (P < .05; Figure 3A bottom panel) and impaired the formation of reticulocytes (not shown). As for HSP27 depletion, GATA-1 overexpression affected erythroblast maturation without significantly altering cell proliferation or survival (supplemental Figure 3C-D). Interestingly, transient overexpression of HSP27 in the GATA-1–overexpressing cells could rescue the erythroblasts, at least partially, from their impaired differentiation as shown by the restored appearance of GPA- and benzidine-positive cells (supplemental Figure 4).
GATA-1 overexpression impairs erythroid differentiation. (A) Primary erythroblasts were transduced 3 times with an empty lentivirus construct (■) or one encoding GATA-1 (▵). Then, differentiation was induced as in Figure 1A. The percentage of differentiated cells was evaluated by benzidine assay at the indicated times. (Bottom) The percentages of basophilic and polychromatic cells was quantified at day 2 as in Figure 2A. (B) K562 cells were transiently transfected with empty plasmid ( ) or with Myc-GATA-1 (
) or with Myc-GATA-1 ( ). After 24 hours, differentiation was induced with hemin (40μM). The percentage of differentiated cells was evaluated by benzidine assay at the indicated times. GATA-1 expression was monitored by Western blotting. HSC70 and actin are the loading controls. Data are expressed as the mean ± SE (n = 3; *P < .05).
). After 24 hours, differentiation was induced with hemin (40μM). The percentage of differentiated cells was evaluated by benzidine assay at the indicated times. GATA-1 expression was monitored by Western blotting. HSC70 and actin are the loading controls. Data are expressed as the mean ± SE (n = 3; *P < .05).
GATA-1 overexpression impairs erythroid differentiation. (A) Primary erythroblasts were transduced 3 times with an empty lentivirus construct (■) or one encoding GATA-1 (▵). Then, differentiation was induced as in Figure 1A. The percentage of differentiated cells was evaluated by benzidine assay at the indicated times. (Bottom) The percentages of basophilic and polychromatic cells was quantified at day 2 as in Figure 2A. (B) K562 cells were transiently transfected with empty plasmid ( ) or with Myc-GATA-1 (
) or with Myc-GATA-1 ( ). After 24 hours, differentiation was induced with hemin (40μM). The percentage of differentiated cells was evaluated by benzidine assay at the indicated times. GATA-1 expression was monitored by Western blotting. HSC70 and actin are the loading controls. Data are expressed as the mean ± SE (n = 3; *P < .05).
). After 24 hours, differentiation was induced with hemin (40μM). The percentage of differentiated cells was evaluated by benzidine assay at the indicated times. GATA-1 expression was monitored by Western blotting. HSC70 and actin are the loading controls. Data are expressed as the mean ± SE (n = 3; *P < .05).
We also overexpressed GATA-1 in K562 cells by transient transfection with a Myc-tagged GATA-1 construct (Figure 3B insert). As shown in Figure 3B, overexpression of the transcription factor delayed the appearance of benzidine-positive cells on exposure to hemin. Altogether, HSP27 depletion was associated with GATA-1 accumulation, and both HSP27 depletion and GATA-1 overexpression delayed erythroid cell differentiation.
HSP27 favors GATA-1 degradation by the proteasome
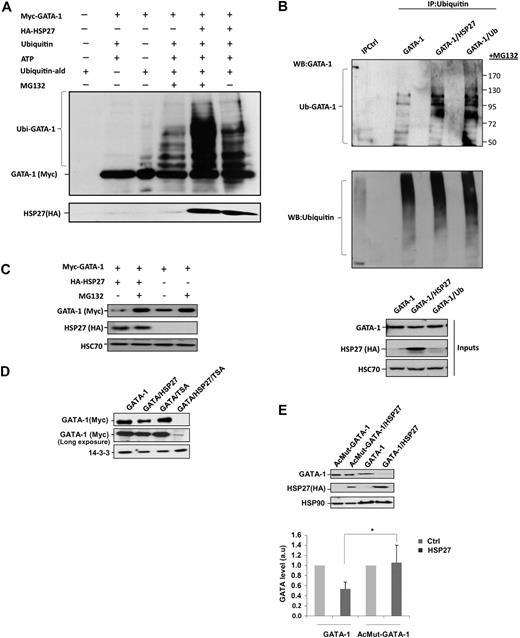
We have shown previously that HSP27 was an ubiquitin-binding protein involved in the proteasomal degradation of certain proteins under stress conditions.13,14 To explore whether HSP27 could regulate GATA-1 degradation, we performed an in vitro assay. The addition of purified HSP27 to Myc-tagged GATA-1 protein in the presence of ubiquitin favored the appearance of ubiquitinated forms of GATA-1, which was better evidenced when the proteasome was inhibited by MG132 (Figure 4A). Ubiquitinated forms of GATA-1 also accumulated in vivo, as observed in K562 cells transiently transfected with GATA-1 and HSP27 or an ubiquitin construct, in the presence of the proteasome inhibitor MG132 (Figure 4B). Overexpression of HSP27 or ubiquitin had a similar effect in the appearance of GATA-1–ubiquitinated forms (Figure 4B). In the absence of MG132, these HSP27-induced ubiquitinated GATA-1 forms were degraded (supplemental Figure 5). On line with this observation, cotransfection of a Myc-tagged GATA-1 and a HA-tagged HSP27 in HeLa cells induced the degradation of GATA-1, which was prevented by MG132 (Figure 4C). Similar results were obtained in transiently transfected COS cells (not shown). Acetylation of GATA-1 was shown to precede its ubiquitination.19 The histone deacetylase inhibitor trichostatin A, which enhances the amount of acetylated proteins in the cells, strongly stimulated HSP27-induced degradation of GATA-1 (Figure 4D). Transfection experiments in HeLa cells showed that HSP27 failed to induce the degradation of AcMut–GATA-1, a GATA-1 mutant in which the acetylation sites have been mutated (Figure 4E). These results suggested that HSP27 could promote the ubiquitination of acetylated GATA-1 and its degradation by the proteasome.
HSP27 induces GATA-1 ubiquitination and proteasomal degradation. (A) Recombinant GATA-1 protein, generated with TNT T7-coupled reticulocyte lysate system, was incubated in the presence or absence of recombinants ubiquitin (Ub) and HSP27 for 40 minutes in an ubiquitin buffer, as described in “In vitro ubiquitination assay.” The reaction was stopped with Laemmli buffer, run on a gel, and blotted with the Myc-tag (GATA-1) antibody. The smear corresponds to the different forms of ubiquitinated GATA-1. (B) K562 cells were transiently transfected with GATA-1 alone or GATA-1 to together with a HSP27 or an ubiquitin construct. Cells were treated 5 hours with MG132, and then lysates were immunoprecipitated with an agarose-conjugated multiubiquitin antibody followed by a Western blot with a GATA-1 antibody. Control for immunoprecipitation (IPCtl) corresponds to a mix of nonrelevant immunoglobulin G and protein G in the presence of lysate. Inputs: protein expression in total extracts (40 μg). (C) HeLa cells were transiently transfected with HA-HSP27 and Myc-GATA-1 plasmids and treated or not with MG132 (20μM, 5 hours). GATA-1 content was analyzed by immunoblot. (D) HeLa cells were transiently transfected with Myc-GATA-1 or with both Myc-GATA-1 and HA-HSP27. Cell extracts from cells either left untreated or treated with trichostatin A (300nM, 16 hours) were resolved on a gel and immunoblotted with Myc-tag (GATA-1). (E) HeLa cells were transiently transfected with Myc–GATA-1 or the acetyl mutant GATA-1 (AcMut-GATA-1) in the presence or absence of HA-HSP27. After 24 hours of transfection, expression of GATA-1 and HSP27 was assessed by Western blot. (Bottom) Densitometry analysis to quantify HSP27-induced degradation of GATA-1. One representative blot of 3 performed is shown. HSP90, HSC70, and 14-3-3 are loading controls.
HSP27 induces GATA-1 ubiquitination and proteasomal degradation. (A) Recombinant GATA-1 protein, generated with TNT T7-coupled reticulocyte lysate system, was incubated in the presence or absence of recombinants ubiquitin (Ub) and HSP27 for 40 minutes in an ubiquitin buffer, as described in “In vitro ubiquitination assay.” The reaction was stopped with Laemmli buffer, run on a gel, and blotted with the Myc-tag (GATA-1) antibody. The smear corresponds to the different forms of ubiquitinated GATA-1. (B) K562 cells were transiently transfected with GATA-1 alone or GATA-1 to together with a HSP27 or an ubiquitin construct. Cells were treated 5 hours with MG132, and then lysates were immunoprecipitated with an agarose-conjugated multiubiquitin antibody followed by a Western blot with a GATA-1 antibody. Control for immunoprecipitation (IPCtl) corresponds to a mix of nonrelevant immunoglobulin G and protein G in the presence of lysate. Inputs: protein expression in total extracts (40 μg). (C) HeLa cells were transiently transfected with HA-HSP27 and Myc-GATA-1 plasmids and treated or not with MG132 (20μM, 5 hours). GATA-1 content was analyzed by immunoblot. (D) HeLa cells were transiently transfected with Myc-GATA-1 or with both Myc-GATA-1 and HA-HSP27. Cell extracts from cells either left untreated or treated with trichostatin A (300nM, 16 hours) were resolved on a gel and immunoblotted with Myc-tag (GATA-1). (E) HeLa cells were transiently transfected with Myc–GATA-1 or the acetyl mutant GATA-1 (AcMut-GATA-1) in the presence or absence of HA-HSP27. After 24 hours of transfection, expression of GATA-1 and HSP27 was assessed by Western blot. (Bottom) Densitometry analysis to quantify HSP27-induced degradation of GATA-1. One representative blot of 3 performed is shown. HSP90, HSC70, and 14-3-3 are loading controls.
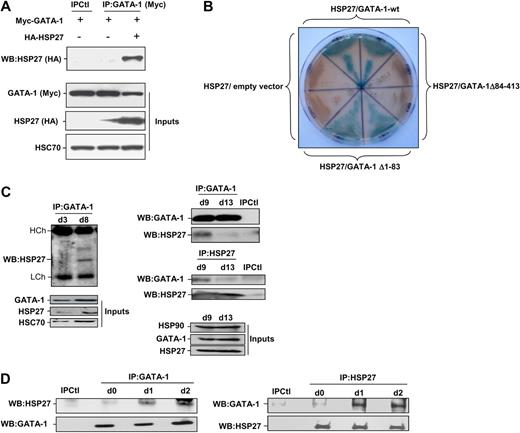
Phosphorylated HSP27 localizes in erythroid cell nucleus to interact with GATA-1
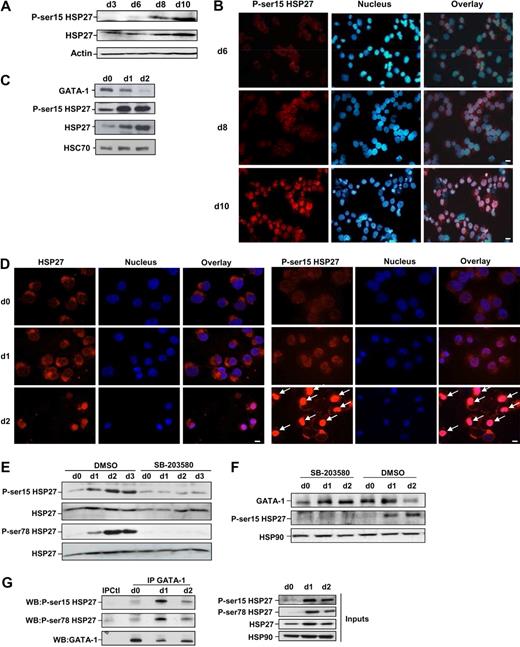
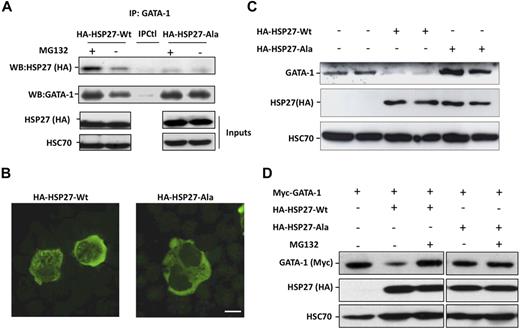
Coimmunoprecipitation experiments performed in COS cells expressing Myc-tagged GATA-1 and HA-tagged HSP27 showed an interaction between the 2 proteins (Figure 5A). To further characterize the interaction GATA-1/HSP27, we used a yeast 2 hybrid assay. As shown in Figure 5B, HSP27 was able to interact with GATA-1 wild type as well as with a GATA-1 deletion mutant in which we have deleted from amino acid 1 to 83 (GATA-1Δ1-83). In contrast, no interaction was observed with a GATA-1 mutant in which we deleted the domain containing the zinc fingers of the protein (GATA-1Δ84-413). Altogether, we conclude that GATA-1 interacts directly with HSP27 and, although a finer mapping is required, it is probably that the zinc finger domain of GATA-1 is involved in the interaction. We then studied the interaction between the endogenous proteins in our erythroid cell models. An interaction between the 2 proteins, GATA-1 and HSP27, could be detected in nuclear extracts of CD36+ primary cells exposed for 8 to 9 days to IL-3 and Epo (Figure 5C) and in hemin-treated K562 cells by day 1 to 2 (Figure 5D). Human HSP27 can be reversibly phosphorylated at serine S15, S78, and S82 by kinases of the p38 mitogen-activated protein kinase (MAPK) family, more specifically by MAPKAPK2/3, downstream of p38α kinase.15,25-27 This phosphorylating event was observed during erythroid differentiation.28 By using antibodies that recognize HSP27 when phosphorylated on S15 or S78, we confirmed the accumulation of the phosphorylated small stress protein in CD36+ exposed for 8 to 10 days to IL-3 and Epo (Figure 6A) and in hemin-treated K562 cells by day 1 to 2 (Figure 6C). Therefore, HSP27 phosphorylation was coincidental with the apparition of the interaction between HSP27 and GATA-1 in vivo (Figure 5C-D). In both cellular models, phosphorylated HSP27 localized in the nucleus (Figure 6B,D; cell fractionation studies in supplemental Figure 6). An inhibitor of p38MAPK pathway (SB-203580) prevented the phosphorylation of HSP27 (Figure 6E) and the consequent degradation of GATA-1 (Figure 6F) in hemin-treated K562 cells, in which the phosphorylated HSP27 was shown to interact with GATA-1 (Figure 6G). Expression of WT HSP27 or a nonphosphorylatable HSP27 mutant (HA-HSP27-Ala) in K562 cells showed that the HSP27-Ala mutant did not significantly interact with the transcription factor (Figure 7A). Interestingly, the HSP27-Ala mutant did not localize within the nucleus during erythroid differentiation (Figure 7B) and did not induce GATA-1 degradation (Figure 7A,C), which was further confirmed in HeLa cells stably overexpressing HSP27-Ala and transiently transfected with Myc–GATA-1 (Figure 7D). Altogether, phosphorylation of HSP27 that is inhibited by SB-203580 is required for modulation of acetylated GATA-1 level in the nucleus of erythroid cells (Figure 8).
HSP27 interacts with GATA-1. (A) COS cells were cotransfected or not with Myc–GATA-1 and HA-HSP27. GATA-1 was immunoprecipitated from cell extracts, subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis migration and immunoblotted with HA-tag (HSP27) antibody. Control for immunoprecipitation (IPCtl) corresponds to a mix of nonrelevant immunoglobulin G and protein G in the presence of lysate. Inputs: protein expression in total extracts. (B) Yeast 2 hybrid assay that detect a direct interaction of HSP27 with GATA-1 wild type (GATA-1-wt) and GATA-1 Δ1-83 (blue staining) but not with GATA-1 Δ84-413. Each section contains diploid yeast cells resulting from one independent yeast mating experience with the corresponding bait protein and a prey protein. The empty vector pGADT7 is used as a negative control. (C) Nuclear extracts from differentiating progenitors CD36+ cells were subjected to immunoprecipitation with GATA-1 antibody and blotted with HSP27 antibody. (D) Immunoprecipitation of endogenous GATA-1 (left) or HSP27 (right) from K562 cell lysates at indicated times of differentiation was followed by HSP27 and GATA-1 immunoblotting. IPCtl, immunoprecipitation in lysates from differentiating cells with a nonrelevant antibody and G protein. Inputs: protein expression in total extracts (40 μg).
HSP27 interacts with GATA-1. (A) COS cells were cotransfected or not with Myc–GATA-1 and HA-HSP27. GATA-1 was immunoprecipitated from cell extracts, subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis migration and immunoblotted with HA-tag (HSP27) antibody. Control for immunoprecipitation (IPCtl) corresponds to a mix of nonrelevant immunoglobulin G and protein G in the presence of lysate. Inputs: protein expression in total extracts. (B) Yeast 2 hybrid assay that detect a direct interaction of HSP27 with GATA-1 wild type (GATA-1-wt) and GATA-1 Δ1-83 (blue staining) but not with GATA-1 Δ84-413. Each section contains diploid yeast cells resulting from one independent yeast mating experience with the corresponding bait protein and a prey protein. The empty vector pGADT7 is used as a negative control. (C) Nuclear extracts from differentiating progenitors CD36+ cells were subjected to immunoprecipitation with GATA-1 antibody and blotted with HSP27 antibody. (D) Immunoprecipitation of endogenous GATA-1 (left) or HSP27 (right) from K562 cell lysates at indicated times of differentiation was followed by HSP27 and GATA-1 immunoblotting. IPCtl, immunoprecipitation in lysates from differentiating cells with a nonrelevant antibody and G protein. Inputs: protein expression in total extracts (40 μg).
HSP27 phosphorylation is required for its association with GATA-1 and to induce GATA-1 degradation. (A) Human primary CD36+ erythroblasts were induced to differentiate in the presence of IL-3, and Epo. Phosphorylated HSP27 expression (on S15) was assessed at the indicated days by Western blotting. Actin serves as a loading control. (B) Localization of S15-phosphorylated HSP27 during Epo-induced CD36+ differentiation. Immunofluorescence was performed as described in “Methods.” Nuclei were stained with Hoechst 33342 (1 μg/mL). Magnification ×40; bar, 10 μm. (C) Phosphorylated HSP27 (on S15) was determined by Western blot in K562 erythroid cells treated with hemin. HSC70 serves as loading control. (D) Localization by immunofluorescence studies of S15-phosphorylated HSP27 during hemin-induced K562 differentiation (days 0-2). Magnification ×40; bar, 10 μm. Arrows indicate nuclear localization of phosphorylated HSP27 (E) Phosphorylated HSP27 (both on S15 and S78) was determined in lysates from K562 cells induced to differentiate with hemin (day 0 to day 3) in the presence or absence of the p38MAPK pathway inhibitor SB-203580 (20μM). (F) GATA-1 level was determined in lysates from K562 cells induced to differentiate with hemin (day 0 to day 2) in the presence or absence of the p38MAPK pathway inhibitor SB-203580 (20μM). (G) At the indicated days after hemin treatment of K562 cells, GATA-1 was immunoprecipitated followed by immunoblotting with P-Ser15-HSP27 and P-Ser78-HSP27 antibodies. IPCtl, immunoprecipitation with a nonrelevant antibody and G protein. HSP90 serves as a loading control. Inputs: protein expression in total extracts (40 μg). DMSO indicates dimethyl sulfoxide.
HSP27 phosphorylation is required for its association with GATA-1 and to induce GATA-1 degradation. (A) Human primary CD36+ erythroblasts were induced to differentiate in the presence of IL-3, and Epo. Phosphorylated HSP27 expression (on S15) was assessed at the indicated days by Western blotting. Actin serves as a loading control. (B) Localization of S15-phosphorylated HSP27 during Epo-induced CD36+ differentiation. Immunofluorescence was performed as described in “Methods.” Nuclei were stained with Hoechst 33342 (1 μg/mL). Magnification ×40; bar, 10 μm. (C) Phosphorylated HSP27 (on S15) was determined by Western blot in K562 erythroid cells treated with hemin. HSC70 serves as loading control. (D) Localization by immunofluorescence studies of S15-phosphorylated HSP27 during hemin-induced K562 differentiation (days 0-2). Magnification ×40; bar, 10 μm. Arrows indicate nuclear localization of phosphorylated HSP27 (E) Phosphorylated HSP27 (both on S15 and S78) was determined in lysates from K562 cells induced to differentiate with hemin (day 0 to day 3) in the presence or absence of the p38MAPK pathway inhibitor SB-203580 (20μM). (F) GATA-1 level was determined in lysates from K562 cells induced to differentiate with hemin (day 0 to day 2) in the presence or absence of the p38MAPK pathway inhibitor SB-203580 (20μM). (G) At the indicated days after hemin treatment of K562 cells, GATA-1 was immunoprecipitated followed by immunoblotting with P-Ser15-HSP27 and P-Ser78-HSP27 antibodies. IPCtl, immunoprecipitation with a nonrelevant antibody and G protein. HSP90 serves as a loading control. Inputs: protein expression in total extracts (40 μg). DMSO indicates dimethyl sulfoxide.
A nonphosphorylatable mutant of HSP27 does not localize in the nucleus and does not bind to GATA-1 to induce its degradation. (A) K562 cells transiently transfected with HA-HSP27-Wt, HA-HSP27-Ala, or a control vector, treated or not with MG132 (20μM, 5 hours), were immunoprecipitated with GATA-1 before immunoblotting with HSP27 (HA) and GATA-1 antibodies. IPCtl, immunoprecipitation with a nonrelevant antibody and G protein. Inputs: protein expression in total extracts (40 μg). (B) K562 cells were transiently transfected with HA-HSP27-Wt and HA-HSP27-Ala. Localization of the transfected HSP27 forms was determined by immunofluorescence after 24 hours of hemin treatment. (Magnification ×100; bar, 10 μm). (C) GATA-1 content was determined by Western blot in duplicate in K562 cells expressing a control vector, HA-HSP27-Wt, or HA-HSP27-Ala. (D) GATA-1 content was determined by Western blot in HeLa cells stably overexpressing HA-HSP27-Wt, HA-HSP27-Ala, or a control vector and transiently transfected with Myc-GATA-1. When indicated, cells were treated with MG132 (20μM, 5 hours). HSC70 was used as a loading control.
A nonphosphorylatable mutant of HSP27 does not localize in the nucleus and does not bind to GATA-1 to induce its degradation. (A) K562 cells transiently transfected with HA-HSP27-Wt, HA-HSP27-Ala, or a control vector, treated or not with MG132 (20μM, 5 hours), were immunoprecipitated with GATA-1 before immunoblotting with HSP27 (HA) and GATA-1 antibodies. IPCtl, immunoprecipitation with a nonrelevant antibody and G protein. Inputs: protein expression in total extracts (40 μg). (B) K562 cells were transiently transfected with HA-HSP27-Wt and HA-HSP27-Ala. Localization of the transfected HSP27 forms was determined by immunofluorescence after 24 hours of hemin treatment. (Magnification ×100; bar, 10 μm). (C) GATA-1 content was determined by Western blot in duplicate in K562 cells expressing a control vector, HA-HSP27-Wt, or HA-HSP27-Ala. (D) GATA-1 content was determined by Western blot in HeLa cells stably overexpressing HA-HSP27-Wt, HA-HSP27-Ala, or a control vector and transiently transfected with Myc-GATA-1. When indicated, cells were treated with MG132 (20μM, 5 hours). HSC70 was used as a loading control.
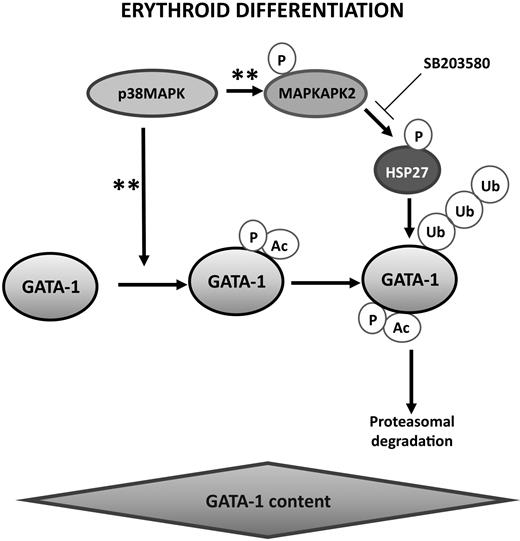
Proposed model for the coordinated molecular events that may control GATA-1 level and erythroblast differentiation. Activation of p38MAPK during erythropoiesis was described previously to promote GATA-1 phosphorylation and acetylation28 and to phosphorylate HSP27 through MAPKAPK2 activation.26,28 The present study indicates that phosphorylated HSP27 accumulates in the nucleus and interacts with GATA-1 to induce its ubiquitination and proteasomal degradation. GATA-1 acetylation precedes its degradation by the proteasomal machinery. This model provides new evidence of the major role of chaperones in erythropoiesis.29 **Reference to data from the literature as mentioned above.
Proposed model for the coordinated molecular events that may control GATA-1 level and erythroblast differentiation. Activation of p38MAPK during erythropoiesis was described previously to promote GATA-1 phosphorylation and acetylation28 and to phosphorylate HSP27 through MAPKAPK2 activation.26,28 The present study indicates that phosphorylated HSP27 accumulates in the nucleus and interacts with GATA-1 to induce its ubiquitination and proteasomal degradation. GATA-1 acetylation precedes its degradation by the proteasomal machinery. This model provides new evidence of the major role of chaperones in erythropoiesis.29 **Reference to data from the literature as mentioned above.
Discussion
GATA-1 levels and activity must be tightly regulated for proper erythroblast differentiation; that is, gata-1 gene deletion is responsible for embryonic lethality through erythroid differentiation arrest and apoptosis of erythroid progenitors30 and GATA-1 overexpression arrests cell differentiation and causes mouse embryonic lethality as well.17 The present study identifies HSP27 as one of the proteins that regulate the GATA-1 expression level and activity along erythroid cell differentiation. More specifically, our results suggest that HSP27 promotes the ubiquitinylation and proteosomal degradation of the transcription factor when acetylated.19 This biologic effect of HSP27 requires its phosphorylation on serine residues, probably by a p38-dependent mechanism (Figure 8). The small stress protein HSP27 was previously shown to favor the ubiquitination and proteasomal degradation of the nuclear factor κB inhibitor13 and the cell cycle protein p27kip1,14 which suggested that driving the selective degradation of proteins was part of its protective role in stressed cells. Here, we demonstrate that HSP27-driven ubiquitination and proteasomal degradation of the transcription factor GATA-1 contributes to the normal differentiation of erythroid cells. This effect is rather specific because growth factor independent 1b levels, another transcription factor necessary for erythroid cell differentiation, is not affected by HSP27 (supplemental Figure 7).
Ubiquitination requires an enzyme cascade that includes an E1-activating enzyme, an E2-conjugating enzyme, an E3-ligase, and, at least in some situations, E4 factors that increase ubiquitination efficiency in a substrate-specific manner, either by accelerating the transfer of ubiquitin chains to the substrate or merely by providing a scaffold.31 Our data suggest that HSP27 is a scaffold protein for GATA-1 and triggers its ubiquitination and degradation when acetylated, which, in human primary erythroblasts, does not affect GATA-1 expression but affects its turnover. Because HSP27 can bind to a GATA-1 mutant that cannot be acetylated (supplemental Figure 8), we suggest that HSP27 binds to GATA-1 before its acetylation that could be the triggering signal for HSP27 to induce a rapid degradation of the transcription factor. This result confirms previous data indicating that acetylation was needed for GATA-1 ubiquitination and degradation.19 Thus, although the reason why the GATA-1 turnover is increased remains elusive, our results indicate that a tightly regulated balance between HSP27 and deacetylases/acetylases may regulate the erythroid differentiation program through modulation of GATA-1 cellular content.
Phosphorylation of GATA-1 by MAP kinases precedes the transcription factor acetylation and participates to the control of its expression.19 Human HSP27 is also phosphorylatable on 3 serine residues (S15, S78, and S82), and this reversible posttranslational modification is mediated by p38 MAP kinases (Figure 8). In cells exposed to a variety of stimuli, this process was previously shown to be catalyzed mainly by MAPKAP kinase-2, a downstream target of p38α (Figure 8).32,33 HSP27 phosphorylation modulates the protein oligomerization by provoking a shift toward small oligomers.15 Nonphosphorylatable (HSP27-Ala) mutants of the protein protects the cells from caspase-mediated apoptosis34 and binds to actin,35 whereas a constitutively phosphorylated (HSP27-Asp) mutant interacts with the death-domain associated protein.36 The phosphorylated HSP27 also displays higher affinity for ubiquitin chains than WT protein.13,14 The present study suggests that HSP27 phosphorylation is required for the protein to localize in the nucleus, to interact with GATA-1, and to trigger its proteosomal degradation. The HSP27 phosphorylation may account for the function of the p38 MAPK cascade in erythroid cell differentiation.28 It may be of interest to determine whether overactivation of the p38 MAPK pathway, which has been associated with altered erythropoiesis in some myelodysplastic syndromes, could affect GATA-1 turnover and expression level.37 Another interesting point to determine is whether the HSP27 effect controlling erythropoiesis through GATA-1 stability is conserved through the species. HSP27 effect in the ubiquitination process has only been studied in human and rodent cells, and the relevance of GATA-1 degradation by the proteasome in its stability has not been established in other species. However, the fact that HSP27 phosphorylation sites as well as GATA-1 acetylation sites are very well conserved through evolution suggests that the effect of HSP27 might also be conserved. Further, HSP27, in Drosophila, has been shown to associate to Ubc9, an ubiquitin conjugating enzyme in flies.38
The stress-inducible HSP27 and HSP70 are abundantly expressed during erythroblast differentiation. We previously demonstrated that HSP70 protected GATA-1 from caspase-mediated cleavage when caspase-3 was transiently activated along the differentiation process.11 We show here that HSP27 is involved in the increased GATA-1 turnover that precedes the terminal stages of erythroid cell differentiation. HSP70 is an ATP-dependent chaperone, whereas HSP27 does not require ATP for inhibiting the aggregation of misfolded proteins.39 The 2 stress proteins cooperate to modulate the stability of AU-rich labile mRNAs40 and to trigger resistance of cancer cells to DNA-damaging inducing agents.41,42 On the basis of their role in the regulation of GATA-1 expression and function, a deregulation of either HSP27 or HSP70 or both could possibly play a role in the pathogenesis of some congenital or acquired erythroid disorders.43,44
Altogether, our data suggest that the coordinated molecular events that tightly control the expression of GATA-1 along erythroid cell differentiation could include the MAPK-mediated phosphorylation of GATA-1, which provokes its acetylation leading to an increased transcriptional activity. Meanwhile, MAPK-mediated phosphorylation of HSP27 favors its nuclear localization. In the nucleus, interaction between phosphorylated HSP27 and acetylated GATA-1 may lead to GATA-1 ubiquitination and subsequent proteosomal degradation (Figure 8). Chaperones play several key functions in the control of normal erythropoiesis that include the control of globin folding and expression29 as well as GATA-1 integrity.11 The present study adds a new function for a chaperone in GATA-1 turnover. Proteins from an additional stress-inducible protein subfamily, HSP90, are expressed in erythroid cells undergoing differentiation, and their function in the process remains to be explored, which may be of importance because HSP90 inhibitors are now developed in cancer treatment.45,46
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank A. Bouchot for excellent technical assistance and P. Vyas for critically reading the manuscript.
This work was supported by grants from the Agence National de la Recherche and Institut National pour le Cancer. C.G. and E.S. lead teams Labellisées from the Ligue Nationale contre le Cancer. A.d.T. is recipient of a postdoctoral fellowship from Association pour la Recherche sur le Cancer (ARC). S.S. has a postdoctoral fellowship from the Conseil Regional de Bourgogne. M.B. is recipient of a doctoral fellowship from the Ligue Nationale contre le Cancer. J.V., D.L., and S.M. have fellowships from the Ministère de l'Education de France.
Authorship
Contribution: A.d.T. and J.V. designed experiments, performed research, and analyzed data; D.L., S.S., A. Hazoume, A. Hammann, M.B., G.C., S.M., A.S.G., Y.Z., and J.A.R. performed research and analyzed data; J.B. provided mutant GATA plasmid constructs and analyzed data; C.G. and O.H. supervised the experiments; C.G., O.H., and E.S. directed the work and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carmen Garrido, Inserm U866, Faculty of Medicine & Pharmacy, 7, boulevard Jeanne d'Arc, 21033 Dijon, France; e-mail: cgarrido@u-bourgogne.fr.
References
Author notes
A.d.T. and J.V. contributed equally to this study.
O.H. and C.G. shared the direction of the work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal