Abstract
Five spontaneous, allelic mutations in the α-spectrin gene, Spna1, have been identified in mice (spherocytosis [sph], sph1J, sph2J, sph2BC, sphDem). All cause severe hemolytic anemia. Here, analysis of 3 new alleles reveals previously unknown consequences of red blood cell (RBC) spectrin deficiency. In sph3J, a missense mutation (H2012Y) in repeat 19 introduces a cryptic splice site resulting in premature termination of translation. In sphIhj, a premature stop codon occurs (Q1853Stop) in repeat 18. Both mutations result in markedly reduced RBC membrane spectrin content, decreased band 3, and absent β-adducin. Reevaluation of available, previously described sph alleles reveals band 3 and adducin deficiency as well. In sph4J, a missense mutation occurs in the C-terminal EF hand domain (C2384Y). Notably, an equally severe hemolytic anemia occurs despite minimally decreased membrane spectrin with normal band 3 levels and present, although reduced, β-adducin. The severity of anemia in sph4J indicates that the highly conserved cysteine residue at the C-terminus of α-spectrin participates in interactions critical to membrane stability. The data reinforce the notion that a membrane bridge in addition to the classic protein 4.1-p55-glycophorin C linkage exists at the RBC junctional complex that involves interactions between spectrin, adducin, and band 3.
Introduction
The membrane skeleton, a multiprotein complex attached to the cytoplasmic surface of the red blood cell (RBC) lipid bilayer, is critical to RBC strength and deformability.1 The major component of the skeleton, spectrin, consists of α and β subunits, encoded in mice by the Spna1 and Spnb1 genes, respectively.2 Spectrin tetramers are cross-linked into a 2-dimensional hexagonal array by short actin protofilaments at “junctional complexes,” which also include protein 4.1, dematin (protein 4.9), adducin, tropomyosin, tropomodulin, and p55.3 The spectrin array is attached to the overlying plasma membrane by ankyrin, which binds β-spectrin to the integral membrane protein band 3 (anion exchanger, SLC4A1), and by protein 4.1-p55-glycophorin C interactions at the junctional complexes. The structural components and function of the RBC membrane skeleton have been comprehensively reviewed elsewhere.2,4-7
Each spectrin molecule is composed of a series of approximately 106 amino acid repeats that fold into triple helices; α-spectrin contains 21 numbered repeats, although repeat 10 is actually an SH3 domain, and β-spectrin contains 16 repeats.6 α- and β-spectrin align side-to-side (“nucleate”) in an antiparallel manner to form heterodimers.8 Repeats 18 to 21 at the C-terminus of α-spectrin and repeats 1 to 4 at the N-terminus of β-spectrin form the nucleation site that initiates “zippering” of the 2 subunits together via electrostatic interactions.6,9,10 Heterodimers self-associate to form heterotetramers, the major form of spectrin in RBCs. The self-association site is at opposite ends of the respective nucleation sites.11,12 The C-terminus of α-spectrin contains 4 EF-hand motifs. The 2 most N-terminal EF hands 1 and 2 are homologous to those in calmodulin.13 They bind calcium with low affinity, and calcium binding induces a conformational change in the domain.14 The more C-terminal EF hands 3 and 4 are calcium independent and structurally similar to those in α-actinin.15 The EF hand domains have long been considered nonfunctional, although recent evidence indicates that protein 4.2 binds to α-spectrin within the EF hand domain in a calcium- and calmodulin-dependent manner.16 However, the overall function of the EF hand domain is poorly understood with regard to spectrin and the membrane skeleton as a whole.
Genetic defects in membrane skeleton components composing vertical interactions (spectrin, ankyrin, band 3) result in hereditary spherocytosis (HS), whereas defects in horizontal interactions affecting spectrin dimer-dimer interactions or protein 4.1 cause hereditary elliptocytosis (HE).1,6,7,17-19 In mice 5 allelic, recessive mutations in α-spectrin have been identified (spherocytosis [sph], sph1J, sph2J, sph2BC, sphDem) and 1 in β-spectrin (ja); all result in severe HS except sphDem, which is a model of severe HE.2,20,21 Here, we describe novel mouse α-spectrin spherocytosis mutations that arose at The Jackson Laboratory, sph3J and sph4J. While studies of sph3J and sph4J were in progress, we imported a mutant strain with features of HS, designated Ihj (Iasi hereditary jaundice),22,23 and confirmed that Ihj is also an sph allele. Analyses of these mutant strains reveal unique, not previously described, protein changes within the spectrin-deficient RBC membrane in vivo, including loss of band 3 and adducin. Furthermore, our studies indicate that a novel, critical function(s) is mediated by a highly conserved cysteine residue within the α-spectrin EF hand domain and provide evidence for an additional vertical bridge critical to RBC membrane stability.
Methods
Animals
Mice were bred at The Jackson Laboratory in climate-controlled rooms (12-hour light/dark cycle) and provided acidified water and chow (NIH 5K52) ad libitum. The sph3J and sph4J mutations were first identified by the appearance of severe pallor in neonates. The sph3J mutation arose spontaneously on the NOD/LtJ strain and was backcrossed onto the congenic, nondiabetic, NOD.B10Sn-H2b/J (NOD) background.24 The sph4J mutation arose in a third-generation offspring of an N-ethyl-N-nitrosourea-mutagenized C57BL/6J (B6) male mated to a wild-type B6 female. The sphIhj mutation arose spontaneously in the mouse colony of Petre Andrei University from a cross between strains HRS/J and LAH/Pas and was imported to The Jackson Laboratory while studies of sph3J and sph4J were in progress. All studies were performed on adult mice 6 weeks of age or older. The Jackson Laboratory Animal Care and Use Committee approved all protocols.
Blood analyses
Whole blood was collected from the retro-orbital sinus by the use of EDTA (ethylenediaminetetraacetic acid)-coated (complete blood counts) or heparinized (plasma iron and bilirubin) microhematocrit tubes. Complete blood counts were obtained by the use of an ADVIA 120 Hematology System (Bayer Healthcare). Iron and bilirubin levels were determined with the use of a CX5 Delta (Beckman Coulter Inc) chemistry analyzer. Peripheral blood smears were Wright-Giemsa (Sigma-Aldrich) stained. Reticulocytes were enumerated after staining with new methylene blue.
Scanning electron microscopy
Heparinized RBCs were washed in mouse phosphate-buffered saline (10mM NaCl, 155mM KCl, 10mM glucose, 1mM MgCl2, 2.5mM KHPO4, pH 7.4), fixed in 2% gluteraldehyde in cacodylate buffer (pH 7.4, 4°C), and processed for examination with a Hitachi 3000N VP (Hitachi High Technologies America Inc) scanning electron microscope as previously described.25
Histology
Tissues were paraffin-embedded and stained with hematoxylin and eosin or with Prussian blue for detection of iron by the use of standard techniques.26
Fine-linkage mapping
Because homozygous sph3J and sph4J mice breed poorly, F2 intercrosses were established with the use of ovarian transplantation.27 Mice carrying homozygous mutant ovaries were mated to Mus musculus castaneous (CAST/Ei) mice to generate F1 hybrids (obligate heterozygotes), which were intercrossed to produce F2 progeny. Tail DNA was obtained from F2 mice and the segregation of anonymous DNA markers followed by the use of standard protocols.28 The sphIhj mutation was linkage mapped with the use of affected and unaffected mice from the breeding colony and selective genotyping for markers on chromosomes known to contain membrane skeleton genes.
Sequencing
Oligonucleotides (Integrated DNA Technologies Inc) amplifying all 52 exons of the Spna1 gene were designed with the use of public sequence data (Ensembl, http://www.ensembl.org; National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov). Polymerase chain reaction (PCR) amplification products were purified (AMPure; Agencourt Biosciences Corp) and sequenced with the use of the automated dye termination technique (ABI Prism Model 3700 genetic analyzer; Applied Biosystems) as described.29 Sequences were analyzed by the use of Sequencher 4.1 licensed software.
Genotyping
The sph3J mutation creates, and the sph4J destroys, an RsaI polymorphism in Spna1 exons 43 and 52, respectively, allowing genotyping of progeny by RsaI digestion of PCR products visualized on ethidium bromide-stained 2% agarose gels. For sph3J the oligonucleotides (5′→3′) CTCAGGAAATGGTGCCATTCAAAGTGGTTG and CAGTGAGATGTGCTTTTATGATAATCTGTC were used as forward and reverse primers, respectively. For sph4J, forward and reverse primers TTCTGATGGCTCTCCATCTGTGAG and AATCTTGGCACCCTAGGTAGAGAC were used. SphIhj was genotyped by sequencing a 483-bp Spna1 exon 38 PCR product amplified with forward and reverse primers TCTTCAGTGTCACGGAAGATCAGC and CACTTCTAGTGCTATGACTGGGTC. The 483-bp PCR product was sequenced by use of the reverse primer to establish the presence of the cytosine to thymine sphIhj mutation.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting
Hemoglobin-depleted RBC ghosts were prepared as described.30 MgCl2 (2mM) was included in the lysis buffer to prevent selective depletion of tropomyosin31 when blotting with tropomyosin-specific antibody. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed with the use of 4% Steck32 or 10% Laemmli33 gels. Gels were loaded semiquantitatively (equal volume of packed ghosts/lane), as described.30,34 Ghost preparations from multiple animals of each genotype and multiple blots per antibody were performed to minimize loading variation. Gels were stained with Coomassie blue or transferred to Hybond-ECL nitrocellulose membranes (Amersham Biosciences) for Western blotting as described.30 Primary antibodies to spectrin, ankyrin, protein 4.1, protein 4.2, band 3, p55, glycophorin C, protein 4.9, and β- and γ-adducin have been previously described.30,34,35 β-actin antibody was obtained from Abcam Inc; α-adducin from Santa Cruz Biotechnology Inc; tropomyosin from the Developmental Studies Hybridoma Bank; and CapZα, CapZβ, and tropomodulin from Lifespan Biosciences Inc. Bound antibody was detected with horseradish peroxidase-conjugated immunoglobulin G (Bio-Rad Laboratories) and visualized with the use of ECL reagents from PerkinElmer.
Northern blotting and reverse transcriptase PCR
Total RNA was extracted from E14.5 fetal livers, adult spleen, and whole blood as previously described.36 Wild-type NOD and B6 reticulocyte RNA were isolated after phenylhydrazine treatment.37 Northern blotting of total RNA transferred to Hybond-N+ membrane (Amersham Biosciences) and reverse transcriptase (RT)–PCR of DNAse I-treated RNA were performed with the use of standard protocols.38 Hybridization probes were a 736-bp Spna1 PCR fragment of exon 52 amplified by the use of forward primer GCTCTCACCCCGGAACAAGTGTCATT and reverse primer GCCAAAGTCATTCACTTTAATGGT, a 493-bp Spna1 PCR fragment of exons 1 to 3 amplified with forward primer GAGCTGAGCAGAGATGGAAACTCCA and reverse primer GTGGCTTCGTGGGCAAAATGGTCTT, band3 cDNA,34 and Gapdh (Seegene Inc).
Quantitative RT-PCR
To determine the percentage of the sph3J mutant allele that is alternatively spliced, quantitative real-time RT-PCR was performed with the use of the Applied Biosystems 7900HT platform and associated reagents per the manufacturer's instructions. The quantity of all mutant Spna1 cDNA present was assayed by generating a 228-bp fragment of Spna1 with a forward exon 41 primer (GAATGAAAAGACAGCCTCTTTGGCC) and a reverse exon 43 primer (TGCTGGAAGCTCTGAAGACTGGCAT), which is upstream of the alternative splice site. The quantity of mutant cDNA that does not undergo alternative splicing was determined by generating a 278-bp fragment with an exon 41/42 forward primer (GGTGGAATCCTGGATAGGTGAGAA) and an exon 43 reverse primer (GCAGTTTCTGCCTGTGAACTCTAGA) that recognizes the deleted region. The difference between the relative quantities of each product was calculated to determine the amount of mutant Spna1 that undergoes alternative splicing. Amplification of β-glucuronidase (Gusb) was performed as a control. All reactions were performed in triplicate.
Flow cytometry
Band 3 was quantitated with the use of eosin-5-maleimide (Invitrogen)39 or a band 3 antibody recognizing extracellular epitopes (provided by Dr Xiuli An, New York Blood Center), as described.39,40 Cells were labeled with CD71 and Ter-119 antibodies to differentiate reticulocytes from mature RBCs.41 Band 3 was detected with Alexa Fluor 647–conjugated secondary antibody (Invitrogen). Median fluorescent intensities (at least 4 mice/strain) were measured with a FACSVantageSE/DiVA (BD BioSciences) flow cytometer.
Image acquisition and assembly
SDS-PAGE gels and Western autoradiograms were scanned with the use of a HP Scanjet 4070 Photosmart Scanner and HP Precisionscan Pro software (Hewlett-Packard) at 600 dpi. Peripheral blood and tissue section images were acquired with a Leica DMRXE microscope equipped with a Leica DFC 300 FX cooled-CCD color camera and LeicaCam software (Leica Microsystems Inc). Smears were examined with a 100×/1.4 oil immersion objective, tissue sections with a 20×/0.60 objective. Scanning electron photomicrographs were acquired with PCI Quartz Imaging software built into the Hitachi S3000N VP Scanning electron Microscope. Final images were assembled with Adobe Illustrator 14.0.0 (Adobe).
Statistical analysis
Tukey honestly significant difference or the 2-tailed Student t test was used to identify significant differences between mean values with JMP version 7.0 software.
Results
Identification of mutant mice
Two severely anemic, autosomal-recessive mouse mutations were recovered from breeding colonies at The Jackson Laboratory, one from the Mouse Mutant Resource (http://mousemutant.jax.org/index.html) and the other from the Neuroscience Mutagenesis Facility (http://nmf.jax.org). The mutations were provisionally designated nm3286 (the 3286th new mutant identified at The Jackson Laboratory) and nmf4 (the 4th mutant identified in the Neuroscience Mutagenesis Facility), respectively. Ihj (Iasi hereditary jaundice) is a mutant mouse strain with features of severe HS that arose spontaneously in the mouse colony of Petre Andrei University. Its phenotypic characteristics have been presented elsewhere.22,23 All mutations are maintained by crossing heterozygotes because homozygotes breed poorly. Heterozygotes are phenotypically normal in all traits examined to date.
Genetic mapping, sequencing, and genotyping confirm that nm3286 and nmf4 are defects in the α-spectrin gene Spna1
We established F2 intercrosses to map nm3286 and nmf4. Both mapped to a 2.5-cM interval between D1Mit407 and D1Mit105 on distal mouse chromosome 1. Spna1 encoding α-spectrin, an obvious candidate gene, maps to this interval. We established matings between heterozygous nm3286 and sph, nmf4 and sph, and nm3286 and nmf4 mice to test for allelism. Each cross produced approximately 25% severely anemic progeny, indicating that the new mutant strains are alleles of each other and of sph. In accordance with the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/mgihome/nomen/strains.shtml), the official genetic designation for nm3286 is Spna1sph-3J and for nmf4 is Spna1sph-4J (abbreviated sph3J and sph4J, respectively).
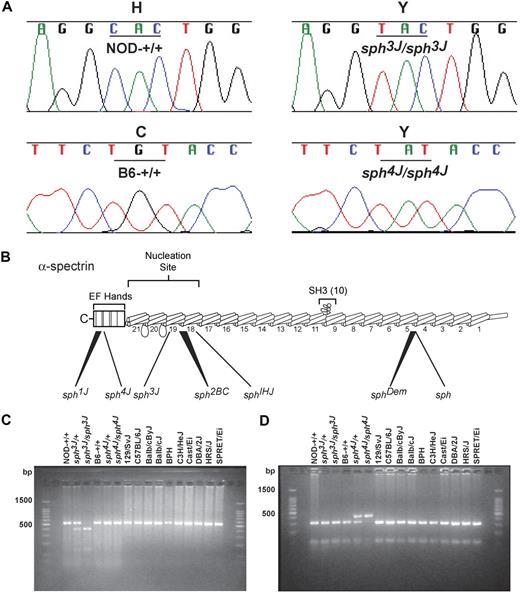
To define the molecular defect, we sequenced all 52 Spna1 exons (Ensembl transcript ENSMUST00000027817) in sph3J and sph4J and their respective wild-type control strains. In sph3J, a cytosine to thymine transition in Spna1 exon 43 was identified that changes a histidine to tyrosine (H2012Y; Figure 1A) in repeat 19, which forms part of the αβ-spectrin dimer nucleation site (Figure 1B). In sph4J, a guanine to adenine transversion in exon 52 resulted in a cysteine to tyrosine substitution (C2384Y: Figure 1A) in the C-terminal EF Hand domain (EF hand 3; Figure 1B).
Identification of sph3J and sph4J defects in Spna1. (A) A cytosine to thymine change in the sph3J allele causes a histidine to tyrosine substitution at residue 2012 of α-spectrin, and a guanine to adenine change in the sph4J allele causes a cysteine to tyrosine substitution at residue 2384 of α-spectrin. (B) Schematic representation of the α-spectrin monomer is depicted with repeat 10 shown as a Src homology domain (SH3; redrawn from Walensky et al6 ). The approximate locations of 4 previously identified sph alleles (sph, sphDem, sph1J, sph2BC), sph3J and sph4J, and the sphIhj allele are indicated. sph has a single nucleotide deletion in repeat 5 that results in a premature stop codon43 ; sphDem has an in-frame deletion of exon 11 that deletes 46 amino acids from repeat 521 ; sph2BC has a guanine to thymine substitution in intron 41 that results in a frame shift and premature stop codon in repeat 1942 ; sph1J is a deletion of the C-terminal 13 amino acids42 ; sph3J is a missense mutation in repeat 19 of the αβ-spectrin dimer nucleation site; sph4J is a missense mutation in the C-terminus within the EF-hand domain just upstream of sph1J deletion; and sphIhj is a premature stop codon in repeat 18. (C) The sph3J mutation creates an RsaI recognition site that was used to genotype mice after PCR amplification. The wild-type fragment is 536 bp. RsaI digestion produces fragments of 401 and 135 bp in mutant mice. (D) The sph4J mutation destroys an RsaI site and was used to genotype mice after PCR amplification. RsaI digestion does not produce the wild-type fragments of 315 and 105 bp but instead a 420-bp fragment. Note that the smallest PCR fragments, 135 bp and 105 bp, are not visible. All inbred strains of mice tested for each allele retain the wild-type genotypes.
Identification of sph3J and sph4J defects in Spna1. (A) A cytosine to thymine change in the sph3J allele causes a histidine to tyrosine substitution at residue 2012 of α-spectrin, and a guanine to adenine change in the sph4J allele causes a cysteine to tyrosine substitution at residue 2384 of α-spectrin. (B) Schematic representation of the α-spectrin monomer is depicted with repeat 10 shown as a Src homology domain (SH3; redrawn from Walensky et al6 ). The approximate locations of 4 previously identified sph alleles (sph, sphDem, sph1J, sph2BC), sph3J and sph4J, and the sphIhj allele are indicated. sph has a single nucleotide deletion in repeat 5 that results in a premature stop codon43 ; sphDem has an in-frame deletion of exon 11 that deletes 46 amino acids from repeat 521 ; sph2BC has a guanine to thymine substitution in intron 41 that results in a frame shift and premature stop codon in repeat 1942 ; sph1J is a deletion of the C-terminal 13 amino acids42 ; sph3J is a missense mutation in repeat 19 of the αβ-spectrin dimer nucleation site; sph4J is a missense mutation in the C-terminus within the EF-hand domain just upstream of sph1J deletion; and sphIhj is a premature stop codon in repeat 18. (C) The sph3J mutation creates an RsaI recognition site that was used to genotype mice after PCR amplification. The wild-type fragment is 536 bp. RsaI digestion produces fragments of 401 and 135 bp in mutant mice. (D) The sph4J mutation destroys an RsaI site and was used to genotype mice after PCR amplification. RsaI digestion does not produce the wild-type fragments of 315 and 105 bp but instead a 420-bp fragment. Note that the smallest PCR fragments, 135 bp and 105 bp, are not visible. All inbred strains of mice tested for each allele retain the wild-type genotypes.
The sph3J mutation created a RsaI digestion site in exon 43, whereas sph4J destroyed a RsaI digestion site in exon 52. PCR amplification with forward and reverse primers flanking the sph3J mutation produced a wild-type fragment of 536 bp that was RsaI digested to produce 401- and 135-bp fragments in the mutant (Figure 1C). Amplification with forward and reverse primers flanking the sph4J mutation produced a mutant fragment of 420 bp that could not be RsaI digested into wild-type allele fragments of 315 and 105 bp (Figure 1D). These polymorphisms allow genotyping of sph3J and sph4J mice and were exploited to demonstrate that the respective mutations segregate with the mutant phenotypes and are not present in other wild-type inbred strains of mice (Figure 1C-D). Thus, sph3J and sph4J mutations located to novel positions within Spna1 compared with previously described sph alleles (Figure 1B).2,6,21,42,43
Severe hemolytic anemia in sph3J and sph4J
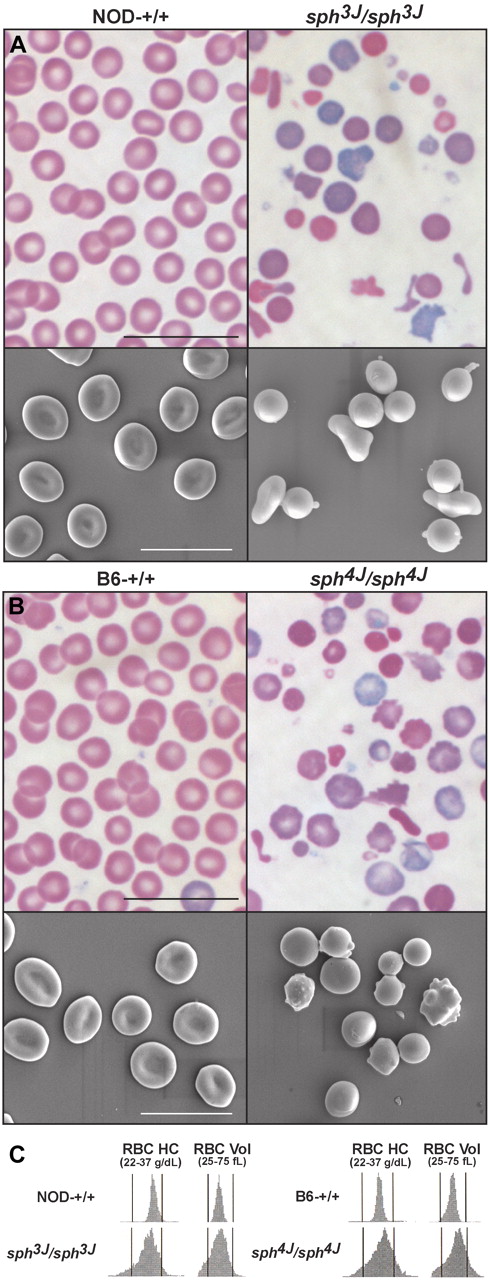
sph3J and sph4J homozygotes displayed characteristics of severe hemolytic anemia. The RBC count, hemoglobin, and hematocrit values were significantly decreased (Table 1), whereas the percentage reticulocytes and spleen weights were dramatically increased, indicating a compensatory acceleration of erythropoiesis (Table 2). Blood smears prepared from sph3J and sph4J mice revealed similar morphologic characteristics (Figure 2A-B). Marked anisocytosis was present, with many microcytes and spherocytes. Significant polychromatophilia was observed, reflecting the high degree of reticulocytosis. Poikilocytosis was particularly dramatic, with many bizarrely shaped cells and fragments. These morphologic changes were reflected in the markedly increased RBC distribution width in both mutants (Table 1). Scanning electron microscopy revealed spherocytic RBCs in both mutants with evidence of membrane loss (blebbing and vesicle shedding; Figure 2A-B). In sph3J homozygotes, frequent irregular sphero-elliptocytes also were seen.
Hematologic values by genotype
| Genotype . | RBC, × 106/μL . | Hgb, g/dL . | Hct, % . | MCV, fL . | MCH, pg . | MCHC, g/dL . | CHCM, g/dL . | RDW, % . | HDW, g/dL . |
|---|---|---|---|---|---|---|---|---|---|
| Male mice | |||||||||
| NOD-+/+ (5) | 9.1 ± 1.1 | 14.8 ± 2.2 | 44.6 ± 4.9 | 49.2 ± 1.8 | 16.5 ± 0.9 | 33.5 ± 1.3 | 32.8 ± 1.7 | 13.7 ± 0.6 | 2.1 ± 0.2 |
| sph3J/sph3J (5) | 3.3 ± 0.2* | 5.5 ± 0.2* | 16.6 ± 1.7* | 50.4 ± 5.5 | 16.8 ± 1.1 | 33.4 ± 2.2 | 29.2 ± 1.6† | 35.6 ± 1.5* | 4.6 ± 0.2* |
| B6-+/+ (8) | 9.8 ± 1.2 | 15.1 ± 1.7 | 45.6 ± 4.7 | 46.6 ± 1.2 | 15.5 ± 0.3 | 33.2 ± 0.9 | 31.0 ± 0.3 | 14.4 ± 2.0 | 1.9 ± 0.2 |
| sph4J/sph4J (20) | 3.2 ± 0.3* | 5.3 ± 0.4* | 18.7 ± 1.7* | 59.2 ± 5.3* | 16.6 ± 0.9† | 28.1 ± 1.4* | 26.5 ± 2.3* | 33.2 ± 2.7* | 4.6 ± 0.6* |
| Female mice | |||||||||
| NOD-+/+ (5) | 9.1 ± 0.2 | 15.3 ± 0.1 | 43.6 ± 0.7 | 46.1 ± 4.2 | 16.9 ± 0.2 | 35.1 ± 0.4 | 36.0 ± 0.5 | 14.5 ± 0.4 | 2.4 ± 0.1 |
| sph3J/sph3J (5) | 2.8 ± 0.7* | 4.5 ± 1.1* | 14.2 ± 2.6* | 52.4 ± 7.6 | 16.2 ± 0.8‡ | 31.1 ± 3.0† | 29.4 ± 1.8* | 32.9 ± 2.5* | 4.9 ± 0.3* |
| B6-+/+ (7) | 9.0 ± 1.3 | 13.8 ± 1.7 | 41.1 ± 3.4 | 46.0 ± 1.6 | 15.4 ± 0.4 | 33.6 ± 1.6 | 31.9 ± 1.3 | 15.5 ± 2.4 | 1.8 ± 0.1 |
| sph4J/sph4J (10) | 3.2 ± 0.2* | 5.4 ± 0.4* | 19.6 ± 1.9* | 60.3 ± 1.3* | 16.6 ± 0.7† | 27.6 ± 1.6* | 25.9 ± 2.9* | 33.5 ± 2.6* | 4.4 ± 0.7* |
| Genotype . | RBC, × 106/μL . | Hgb, g/dL . | Hct, % . | MCV, fL . | MCH, pg . | MCHC, g/dL . | CHCM, g/dL . | RDW, % . | HDW, g/dL . |
|---|---|---|---|---|---|---|---|---|---|
| Male mice | |||||||||
| NOD-+/+ (5) | 9.1 ± 1.1 | 14.8 ± 2.2 | 44.6 ± 4.9 | 49.2 ± 1.8 | 16.5 ± 0.9 | 33.5 ± 1.3 | 32.8 ± 1.7 | 13.7 ± 0.6 | 2.1 ± 0.2 |
| sph3J/sph3J (5) | 3.3 ± 0.2* | 5.5 ± 0.2* | 16.6 ± 1.7* | 50.4 ± 5.5 | 16.8 ± 1.1 | 33.4 ± 2.2 | 29.2 ± 1.6† | 35.6 ± 1.5* | 4.6 ± 0.2* |
| B6-+/+ (8) | 9.8 ± 1.2 | 15.1 ± 1.7 | 45.6 ± 4.7 | 46.6 ± 1.2 | 15.5 ± 0.3 | 33.2 ± 0.9 | 31.0 ± 0.3 | 14.4 ± 2.0 | 1.9 ± 0.2 |
| sph4J/sph4J (20) | 3.2 ± 0.3* | 5.3 ± 0.4* | 18.7 ± 1.7* | 59.2 ± 5.3* | 16.6 ± 0.9† | 28.1 ± 1.4* | 26.5 ± 2.3* | 33.2 ± 2.7* | 4.6 ± 0.6* |
| Female mice | |||||||||
| NOD-+/+ (5) | 9.1 ± 0.2 | 15.3 ± 0.1 | 43.6 ± 0.7 | 46.1 ± 4.2 | 16.9 ± 0.2 | 35.1 ± 0.4 | 36.0 ± 0.5 | 14.5 ± 0.4 | 2.4 ± 0.1 |
| sph3J/sph3J (5) | 2.8 ± 0.7* | 4.5 ± 1.1* | 14.2 ± 2.6* | 52.4 ± 7.6 | 16.2 ± 0.8‡ | 31.1 ± 3.0† | 29.4 ± 1.8* | 32.9 ± 2.5* | 4.9 ± 0.3* |
| B6-+/+ (7) | 9.0 ± 1.3 | 13.8 ± 1.7 | 41.1 ± 3.4 | 46.0 ± 1.6 | 15.4 ± 0.4 | 33.6 ± 1.6 | 31.9 ± 1.3 | 15.5 ± 2.4 | 1.8 ± 0.1 |
| sph4J/sph4J (10) | 3.2 ± 0.2* | 5.4 ± 0.4* | 19.6 ± 1.9* | 60.3 ± 1.3* | 16.6 ± 0.7† | 27.6 ± 1.6* | 25.9 ± 2.9* | 33.5 ± 2.6* | 4.4 ± 0.7* |
Number in parentheses indicates n (sample size). All values are stated as mean ± SD. All mice were ≥6 weeks of age.
CHCM indicates cell hemoglobin concentration mean; Hct, hematocrit; HDW, hemoglobin distribution width Hgb, hemoglobin; MCH, mean cell hemoglobin; MCHC, mean cell hemoglobin concentration; MCV, mean cell volume; RBC, red blood cell count; and RDW, red cell distribution width.
P < .001 vs sex-matched control (+/+) mice.
P < .01 vs sex-matched control (+/+) mice.
P < .05 vs sex-matched control (+/+) mice.
Percent reticulocytes and spleen weights, total bilirubin, and plasma iron levels by genotype
| Genotype . | Spleen weight, % body weight . | Reticulocyte, % . | Total bilirubin, mg/dL . | Plasma iron, μg/dL . |
|---|---|---|---|---|
| NOD-+/+ | 0.35 ± 0.06 (8) | 2.8 ± 0.8 (3) | 0.63 ± 0.11 (7) | 278.4 ± 59.5 (7) |
| sph3J/sph3J | 4.68 ± 0.96* (9) | 88.4 ± 2.5* (4) | 1.23 ± 0.18* (7) | 485.7 ± 42.4* (7) |
| B6-+/+ | 0.27 ± 0.08 (4) | 3.3 ± 1.5 (3) | 0.51 ± 0.25 (6) | 148.5 ± 32.8 (6) |
| sph4J/sph4J | 4.27 ± 1.07*; (17) | 91.5 ± 1.1* (3) | 1.87 ± 0.81* (15) | 213.6 ± 65.5† (15) |
| Genotype . | Spleen weight, % body weight . | Reticulocyte, % . | Total bilirubin, mg/dL . | Plasma iron, μg/dL . |
|---|---|---|---|---|
| NOD-+/+ | 0.35 ± 0.06 (8) | 2.8 ± 0.8 (3) | 0.63 ± 0.11 (7) | 278.4 ± 59.5 (7) |
| sph3J/sph3J | 4.68 ± 0.96* (9) | 88.4 ± 2.5* (4) | 1.23 ± 0.18* (7) | 485.7 ± 42.4* (7) |
| B6-+/+ | 0.27 ± 0.08 (4) | 3.3 ± 1.5 (3) | 0.51 ± 0.25 (6) | 148.5 ± 32.8 (6) |
| sph4J/sph4J | 4.27 ± 1.07*; (17) | 91.5 ± 1.1* (3) | 1.87 ± 0.81* (15) | 213.6 ± 65.5† (15) |
Number in parentheses indicates sample number. All values are reported as mean ± SD.
P < .001 vs control (+/+) mice.
P < .05 vs control (+/+) mice.
Abnormal erythrocyte morphology. (A) Peripheral blood smears and scanning electron photomicrographs from control (NOD-+/+) and homozygous mutant (sph3J/sph3J) mice, and (B) control (B6-+/+) and homozygous mutant (sph4J/sph4J) mice. Bars, 10 μm. (C) RBC hemoglobin concentration (RBC HC) and volume distribution (RBC Vol) histograms.
Abnormal erythrocyte morphology. (A) Peripheral blood smears and scanning electron photomicrographs from control (NOD-+/+) and homozygous mutant (sph3J/sph3J) mice, and (B) control (B6-+/+) and homozygous mutant (sph4J/sph4J) mice. Bars, 10 μm. (C) RBC hemoglobin concentration (RBC HC) and volume distribution (RBC Vol) histograms.
The RBC mean cell volume was significantly increased in sph4J mutants but not in sph3J mutants (Table 1). However, the combination of high numbers of large reticulocytes (Table 2), many small spherocytes (some extremely so), and many cell fragments precluded our obtaining accurate and meaningful mean cell volume measurements via automated cell counting. The mean cell hemoglobin and mean cell hemoglobin concentration demonstrated statistically significant differences in all groups except sph3J male mice. The cell hemoglobin concentration mean, which is directly measured on a cell-by-cell basis on the Advia 120 analyzer and is analogous to the calculated MCH concentration value, was decreased in all mutant mice (Table 1). Examination of RBC volume histograms confirmed populations of both small and large cells in each mutant (Figure 2C). Moreover, significant populations of small cells (left-shifted portion of RBC volume histograms; Figure 2C) and dehydrated cells (right shifted portion of RBC hemoglobin concentration histograms; Figure 2C) indicated the presence of dehydrated cells in both mutants. Total bilirubin levels and plasma iron levels were significantly increased (Table 2), reflecting hemolysis. All hematologic observations were consistent with severe hemolytic anemia in sph3J and sph4J homozygotes. Sph4J closely mimicked HS, whereas sph3J demonstrated features of both HS and HE, as in rare human patients with spherocytic hereditary elliptocytosis (SphHE).6
Histopathology confirms extreme hemolysis in mutant mice
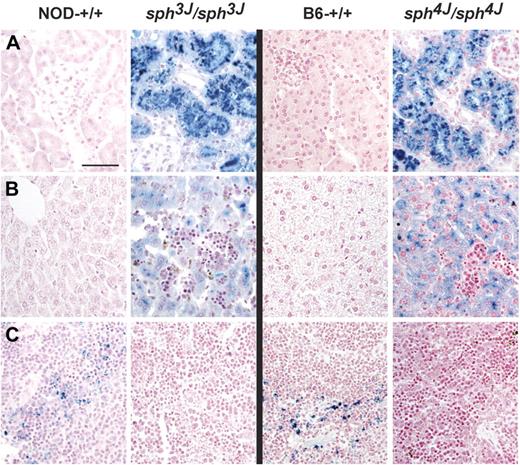
Histologic analyses demonstrated changes consistent with severe hemolysis in both mutants. Prussian blue staining revealed extensive iron accumulation in the kidney (Figure 3A) and liver (Figure 3B). Numerous extramedullary hematopoietic foci were observed in the liver (Figure 3B). There was massive splenomegaly (Table 2) attributable to the expansion of red pulp areas. Unlike normal spleens, mutant spleens were devoid of iron (Figure 3C), likely reflecting extensive use as the result of markedly accelerated erythropoiesis, consistent with other mouse models with severe anemia.2
Tissue pathology. Prussian blue staining of kidney (A), liver (B), and spleen (C) tissue sections from (left to right) NOD control (NOD-+/+), sph3J/sph3J mutant, B6 control (B6-+/+), and sph4J/sph4J mutant mice. Note the extensive iron accumulation in kidney tubules and liver parenchyma of mutant mice. Mutant liver sections also contain numerous foci of extramedullary hematopoiesis. Bars, 100 μm.
Tissue pathology. Prussian blue staining of kidney (A), liver (B), and spleen (C) tissue sections from (left to right) NOD control (NOD-+/+), sph3J/sph3J mutant, B6 control (B6-+/+), and sph4J/sph4J mutant mice. Note the extensive iron accumulation in kidney tubules and liver parenchyma of mutant mice. Mutant liver sections also contain numerous foci of extramedullary hematopoiesis. Bars, 100 μm.
Differential alterations in membrane skeletal proteins in sph3J and sph4J mice
Quantitative SDS-PAGE and Western blot analysis of RBC membrane skeletal proteins in severely anemic mice is complicated by the fact that significantly more hemoglobin is retained on mutant membranes than on normal membranes. Whereas normal “white” ghosts can be obtained from control RBCs, mutant ghosts remain extremely red.34 Consequently, loading equal protein concentrations per well results in an underrepresentation of the membrane skeleton fraction in mutant samples. To overcome this difficulty, we loaded equal volumes of packed ghosts per well and repeated each analysis multiple times to control for loading variation.
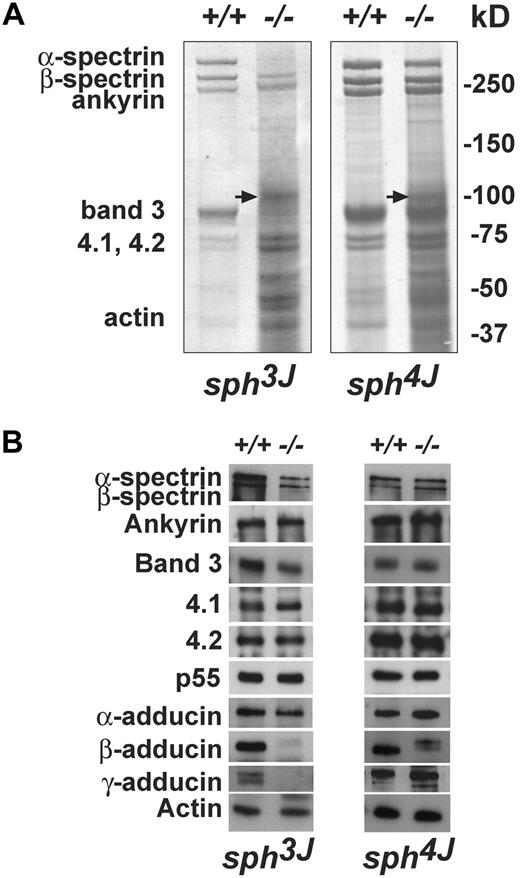
SDS-PAGE analysis and Western blotting revealed unique membrane skeleton alterations in sph3J and sph4J RBC ghosts. In sph3J, α-spectrin was markedly reduced with an accompanying significant decrease in β-spectrin levels (Figure 4A-B left).34 Surprisingly, band 3 also was significantly reduced in sph3J. In comparison, in sph4J, the decrement in α- and β-spectrin was minimal, with no apparent decrement in band 3 (Figure 4A-B right). In both mutants, ankyrin, protein 4.1, protein 4.2, and p55 all appeared normal by Western blotting (Figure 4B). Loss of band 3 has not previously been described in α-spectrin–deficient mouse RBCs.
Erythrocyte membrane skeletal defects. (A) Coomassie blue-stained SDS-PAGE gels. Equal volumes of normal and mutant packed RBC ghosts/lane were loaded. The high percentage of circulating reticulocytes in mutants results in numerous additional bands and an overall smeared appearance, especially in the lower molecular weight ranges, on SDS-PAGE gels. The species indicated by the arrow is unknown; it is not band 3 because it is also present in band 3-null RBC ghosts.34 (B) Western blots of RBC ghost proteins probed with the indicated antibodies. Each panel is representative of results from multiple blots. +/+, wild-type; −/−, homozygous mutant.
Erythrocyte membrane skeletal defects. (A) Coomassie blue-stained SDS-PAGE gels. Equal volumes of normal and mutant packed RBC ghosts/lane were loaded. The high percentage of circulating reticulocytes in mutants results in numerous additional bands and an overall smeared appearance, especially in the lower molecular weight ranges, on SDS-PAGE gels. The species indicated by the arrow is unknown; it is not band 3 because it is also present in band 3-null RBC ghosts.34 (B) Western blots of RBC ghost proteins probed with the indicated antibodies. Each panel is representative of results from multiple blots. +/+, wild-type; −/−, homozygous mutant.
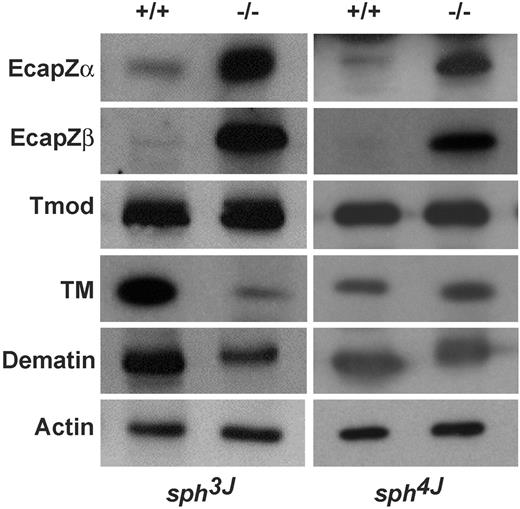
Further analyses of proteins at the RBC junctional complex in sph3J RBCs revealed decreased α-adducin and absent β- and γ-adducin (Figure 4B left). EcapZα and EcapZβ were markedly increased, tropomyosin and dematin (protein 4.9) were decreased, whereas tropomodulin and actin appeared normal (Figure 5 left). It is surprising that γ-adducin was lacking in sph3J RBCs because we and others35,44 have shown up-regulation of γ-adducin in β-adducin–null RBCs. In contrast, sph4J RBCs, which have normal-to-slightly increased levels of α-adducin, do up-regulate γ-adducin in the presence of significantly decreased levels of the β-adducin subunit (Figure 4B right). EcapZα and EcapZβ are also markedly increased in sph4J. Tropomodulin, tropomyosin, and dematin appear normal (Figure 5 right).
Other erythrocyte actin-associated proteins. Western blots of RBC ghost proteins probed with the indicated antibodies. Each panel is representative of results from multiple blots. TM indicates tropomyosin; Tmod, tropomodulin; +/+, wild-type; −/−, homozygous mutant.
Other erythrocyte actin-associated proteins. Western blots of RBC ghost proteins probed with the indicated antibodies. Each panel is representative of results from multiple blots. TM indicates tropomyosin; Tmod, tropomodulin; +/+, wild-type; −/−, homozygous mutant.
Clearly, the sph3J and sph4J mutations result in unique RBC membrane skeleton protein profiles. As with band 3 deficiency, adducin and dematin deficiency have not previously been noted in any of the sph alleles.
Reevaluation of other sph mutant strains
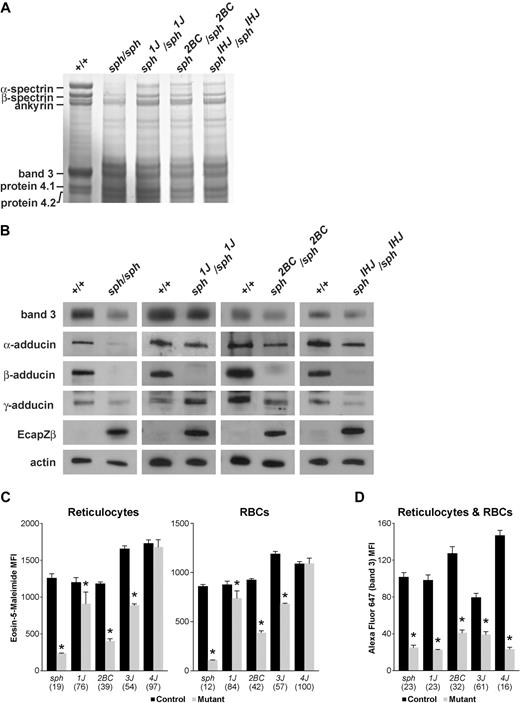
SDS-PAGE gels and Western blots demonstrate that band 3 also was reduced in sph, sph1J, and sph2BC (Figure 6A-B). β-adducin was absent and EcapZβ markedly increased (Figure 6B) in these strains. As observed in sph3J, α- and γ-adducin were reduced in sph and sph2BC (Figure 6B). Similar to sph4J, α-adducin appeared relatively normal, whereas there was increased γ-adducin in sph1J membranes (Figure 6B).
Band 3 and adducin deficiency across sph alleles. (A) Coomassie blue-stained SDS-PAGE gel assessment of RBC ghosts proteins from several sph alleles. Equal volumes of normal and mutant packed RBC ghosts/lane were loaded. (B) Western blots of RBC ghosts proteins probed with the indicated antibodies. Each panel is representative of results from multiple blots. (C) Eosin-5-malemide staining of erythroid band 3 was measured for mature RBCs and reticulocyte gated populations and is presented as MFI. Eosin-5-malemide MFI is significantly (*) reduced (P < .05) in RBCs and reticulocytes of all sph alleles examined except sph4J. (D) Alexa Flour 647 fluorescent intensity staining of erythroid band 3 was measured for combined RBC and reticulocyte-gated populations because no difference was observed between the cell types (data not shown) and is presented as MFI. Alexa Flour 647 MFI is significantly (*) reduced (P < .05) in all sph alleles examined. All plotted values in panels C and D are expressed as mean ± standard error. Values in parentheses below the x-axis represent the percentage of respective control.
Band 3 and adducin deficiency across sph alleles. (A) Coomassie blue-stained SDS-PAGE gel assessment of RBC ghosts proteins from several sph alleles. Equal volumes of normal and mutant packed RBC ghosts/lane were loaded. (B) Western blots of RBC ghosts proteins probed with the indicated antibodies. Each panel is representative of results from multiple blots. (C) Eosin-5-malemide staining of erythroid band 3 was measured for mature RBCs and reticulocyte gated populations and is presented as MFI. Eosin-5-malemide MFI is significantly (*) reduced (P < .05) in RBCs and reticulocytes of all sph alleles examined except sph4J. (D) Alexa Flour 647 fluorescent intensity staining of erythroid band 3 was measured for combined RBC and reticulocyte-gated populations because no difference was observed between the cell types (data not shown) and is presented as MFI. Alexa Flour 647 MFI is significantly (*) reduced (P < .05) in all sph alleles examined. All plotted values in panels C and D are expressed as mean ± standard error. Values in parentheses below the x-axis represent the percentage of respective control.
To quantitate the decrement in band 3, RBCs were stochiometrically labeled with eosin-5-maleimide and analyzed by flow cytometry.39 In accordance with the Western blot data, the median fluorescent intensity (MFI) was significantly decreased in all mutant alleles except sph4J, which did not differ significantly from its wild-type control (Figure 6C). Sph1J showed a relatively minor, but significant, decrement (84% of normal). To establish that decreased staining did not simply reflect loss of Rh or Rh-associated glycoprotein protein, which also are recognized by eosin-5-maleimide,45 the experiment was repeated with the use of a mouse-specific antibody recognizing extracellular band 3 epitopes.40 In this case, the MFI was significantly decreased in all mutant RBCs (Figure 6D). However, when expressed as percentage of wild-type, band 3 labeling was substantially less than eosin-5-maleimide only in sph1J and sph4J. This finding is in direct contrast to Western blot data for these mutants (Figures 4,6). Furthermore, Western blotting of sph1J and sph4J RBC ghosts with the extracellular band 3 antibody resulted in blots similar to those shown in Figures 4 and 6, which were probed with band 3 antibodies raised to intracellular epitopes. We conclude that reduction of band 3 antibody MFI compared with eosin-5-maleimide MFI likely results from a conformational change that prevents antibody binding to sph1J and sph4J RBCs.
Ihj (Iasi hereditary jaundice) is the eighth sph allele, Spna1sph-Ihj
While this study was in progress, we imported Ihj to The Jackson Laboratory. We mapped Ihj to mouse chromosome 1 and confirmed that Ihj is allelic with sph3J. Sequencing identified a cytosine to thymine transition in Spna1 exon 38 that changes a glutamine to a stop codon (Q1853Stop) in repeat 18. The official genetic designation for Ihj is Spna1sph-Ihj.
SDS-PAGE and Western blotting confirmed a pattern very similar to that observed in sph, sph2BC, and sph3J. There was α-spectrin deficiency with significant loss of band 3, absent β-adducin, decreased α- and γ-adducin, and up-regulated EcapZ (Figure 6A-B). A generalized gain of EcapZβ was seen in all sph alleles and likely reflects loss of β-adducin, as described previously in β-adducin–null RBCs.30,44,46 Up-regulated EcapZ is thought to compensate for the lack of adducin in actin protofilament capping. Notably, sph1J and sph4J, both of which carry mutations in the C-terminal EF hand domain (Figure 1B legend),2 closely resemble each other and contain the most membrane band 3 relative to the other sph alleles.
The sph3J mutation produces a cryptic splice site resulting in alternative splicing and production of an unstable RNA
By Northern blotting, erythroid α-spectrin transcripts were barely detectable in reticulocytes and spleens of adult sph3J mice but appeared at normal levels in sph4J tissues (Figure 7A). This finding was unexpected, given that the sph3J mutation, like sph4J, produces a single amino acid substitution and would not be predicted to activate RNA decay mechanisms.
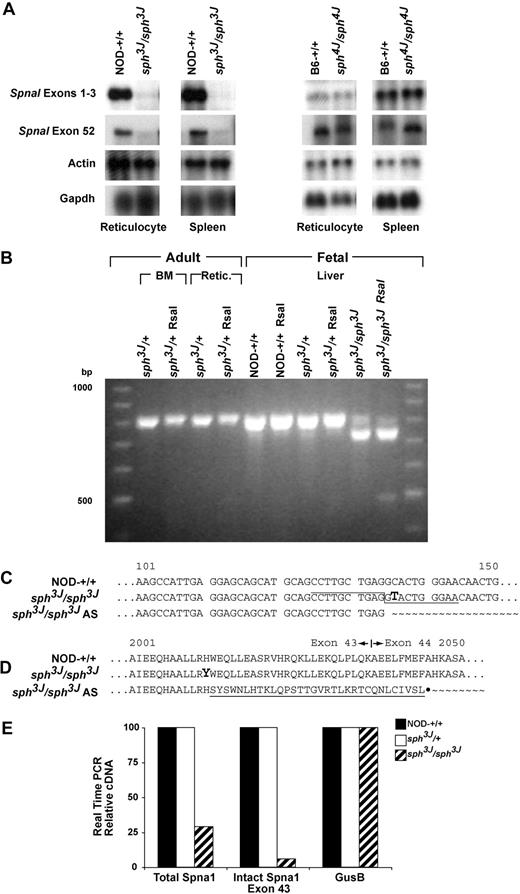
Erythroid molecular defects. (A) Northern blotting of total RNA extracted from adult reticulocytes and spleen. Note the near absence of Spna1 message in homozygous sph3J mice when blotted with either a probe upstream or downstream of the mutation. Spna1 RNA is normal in sph4J homozygotes. Actin and/or Gapdh expression levels were used to normalize RNA loading. (B) RT-PCR amplification of Spna1 exons 41 to 45 on total RNA extracted from adult sph3J/+ bone marrow cells (BM) and reticulocytes (Retic.); and from E14.5 fetal liver. Amplification was followed by RsaI digestion and is shown along with undigested products. Note the production of 2 distinct products in homozygous sph3J fetal liver. (C) Sequencing of the distinct products revealed that the C to T (bold) mutation creates a CCTTGCTGAG/gtactgggaa (over-, underlined) alternative splice (AS) site that eliminates the last 76 bp of Spna1 exon 43. (D) Sequence analysis of the H2012Y (bold) and alternative splicing predicted that AS produces 32 mutant amino acids followed by a stop codon (underlined). (E) Real time RT-PCR demonstrated that total Spnal message was 27% of NOD control in sph3J and was only 6% of control when alternatively spliced mutant transcripts are discounted. Spna1 message levels were at control levels for heterozygous sph3J samples. Gusb amplification was used as a housekeeping gene and was equal for all samples.
Erythroid molecular defects. (A) Northern blotting of total RNA extracted from adult reticulocytes and spleen. Note the near absence of Spna1 message in homozygous sph3J mice when blotted with either a probe upstream or downstream of the mutation. Spna1 RNA is normal in sph4J homozygotes. Actin and/or Gapdh expression levels were used to normalize RNA loading. (B) RT-PCR amplification of Spna1 exons 41 to 45 on total RNA extracted from adult sph3J/+ bone marrow cells (BM) and reticulocytes (Retic.); and from E14.5 fetal liver. Amplification was followed by RsaI digestion and is shown along with undigested products. Note the production of 2 distinct products in homozygous sph3J fetal liver. (C) Sequencing of the distinct products revealed that the C to T (bold) mutation creates a CCTTGCTGAG/gtactgggaa (over-, underlined) alternative splice (AS) site that eliminates the last 76 bp of Spna1 exon 43. (D) Sequence analysis of the H2012Y (bold) and alternative splicing predicted that AS produces 32 mutant amino acids followed by a stop codon (underlined). (E) Real time RT-PCR demonstrated that total Spnal message was 27% of NOD control in sph3J and was only 6% of control when alternatively spliced mutant transcripts are discounted. Spna1 message levels were at control levels for heterozygous sph3J samples. Gusb amplification was used as a housekeeping gene and was equal for all samples.
To determine whether the sph3J mutation creates unstable Spna1 transcripts detectable in immature erythrocyte precursors, we performed RT-PCR on bone marrow, reticulocyte, and fetal liver RNA and digested the products with RsaI to distinguish wild-type from mutant transcripts. No sph3J mutant RsaI-digestible fragments (483 and 329 bp) were detected by agarose gel electrophoresis in any of the heterozygous cDNA samples (Figure 7B). Instead, only the expected 812-bp wild-type allele was detected. In homozygous sph3J fetal liver, however, 2 cDNA products were observed. The expected 812-bp product was present in small amounts relative to a smaller cDNA product that greatly predominated. Moreover, most of the smaller cDNA appeared to be RsaI resistant (Figure 7B). Sequencing revealed that this smaller product was missing the last 76 bp of Spna1 exon 43 and, as a result, the RsaI restriction site (Figure 7C). The cytosine to thymine transition in the sph3J allele introduces a CCTTGCTGAG/gtactgggaa alternate isoform/cryptic donor site, as predicted by the web-accessible Splice Site Predictor (ASSP; http://es.embnet.org/%7emwang/assp.html) application.47 Sequence analysis predicted that the alternatively spliced mutant sph3J cDNA properly encodes the first 2012 residues of α-spectrin followed by an additional 32 mutant amino acids and a stop codon (Figure 7D). Because the alternatively spliced Spna1 transcript contains a downstream exon junction complex (exon 44), it is presumably degraded in heterozygous and homozygous sph3J mice by nonsense-mediated mRNA decay.48,49
We used real-time PCR to confirm that the alternatively spliced mutant Spna1 transcript is highly unstable. Total Spna1 cDNA was assayed by generating a fragment of Spna1 with an exon 41 forward primer and an exon 43 reverse primer that is upstream of the alternative, mutant splice site. The quantity of cDNA not undergoing alternative splicing was assayed by generating a similar fragment with a reverse primer in the deleted region (see “Quantitative RT-PCR”). These results were compared to deduce the relative quantities of sph3J transcription and occurrence of alternative splicing. Total Spna1 transcription was reduced 3.45-fold in sph3J mice and the amount of message not being alternatively spliced was further reduced 16.67-fold, compared with the wild-type (Figure 7E). Interestingly, Spna1 transcription in heterozygous mice remained at wild-type levels and was absent of any alternative splicing. Transcription of Gusb was assayed as an internal control and was unchanged in heterozygous and homozygous samples. When these results were calculated relative to the wild-type control, homozygous sph3J mice expressed 29% of control total Spna1 message levels. Of this, only 6% of cDNA was derived from the full-length mutant allele after alternative splicing (Figure 7E). Thus, approximately 80% of homozygous sph3J transcripts contain the alternative splice site and are presumably destroyed by nonsense-mediated mRNA decay. It is noteworthy that the alternatively spliced transcripts were not detected in heterozygous transcripts. This finding is probably the result of the highly unstable nature of the alternative transcript in the presence of the wild-type allele transcript. Furthermore, no truncated α-spectrin protein was observed for sph3J or any other sph mice by Western blotting.
Discussion
In this report we describe 3 new sph alleles, sph3J, sph4J, and sphIhj. sph3J carries a missense mutation (H2012Y) in repeat 19 of α-spectrin, sph4J a missense mutation (C2384Y) in the EF hand domain, and sphIhj a premature stop codon in repeat 18 (Q1853Stop). The sph3J missense mutation introduces a cryptic splice site that produces a highly unstable Spna1 RNA transcript and, as a result, markedly diminished stable incorporation of α-spectrin onto the RBC membrane. Expression in the sph4J allele, however, is normal, and the RBC membrane α-spectrin content appears normal by Western blotting.
As with other sph alleles, sph3J and sph4J homozygotes show a phenotype of severe hemolytic anemia characterized by dramatically decreased RBC count, hemoglobin, and hematocrit with evidence of extensive hemolysis (iron accumulation in the kidney proximal tubules, elevated bilirubin levels) and a compensatory increase in erythropoietic rate (massive splenomegaly, extreme reticulocytosis). The RBC morphology in both mutations shows a high degree of anisocytosis and poikilocytosis, evident on peripheral blood smears and by scanning electron microscopy. Spherocytes predominate in sph4J; hence, sph4J mice show all the major features of HS. Extensive spherocytosis is also seen in sph3J; however, frequent sphero-elliptocytes also are seen. In this respect, sph3J shows features of SphHE, a rare disorder in humans that is poorly understood at the molecular level,6 although humans with SphHE typically show greater numbers of ellipsoidal cells than are found in sph3J.
By SDS-PAGE gels and Western blotting of major membrane skeleton proteins we identified differential alterations in sph3J versus sph4J RBC ghosts. In fact, we identified changes not previously detected in any of the allelic sph mutant strains. Evaluation of available sph mutant mice revealed absent β-adducin and significant decrements in membrane band 3 content in all except sph4J. Band 3 is normal in sph4J, whereas β-adducin is present in the membrane, although at reduced levels. While these studies were in progress, we identified sphIhj as another member of the sph allelic series and confirmed loss of RBC membrane β-adducin and decreased band 3 in this allele as well. Thus, with the notable exception of sph4J, absent β-adducin and reduced band 3 is found in all α-spectrin–deficient sph alleles examined. The authors of a recent study50 revealed a direct interaction of the tail domain of both α- and β-adducin with the cytoplasmic domain of band 3 (CDB3) in vitro. Moreover, in vitro studies predict 3 populations of band 3—ankyrin bound, adducin bound, and freely diffusing.50,51 It seems likely that loss of band 3 in vivo involves the population interacting with adducin. Normal ankyrin levels in all sph mice suggest tetrameric band 3 content is normal.39
We can only speculate as to why band 3 deficiency was not detected in previous studies of the sph mutants upon SDS-PAGE.2,20,21,42 A likely contributing factor is improved SDS-PAGE techniques and, in the past, reliance on discontinuous Laemmli gels for mouse studies, which often fail to clearly delineate band 3 protein upon Coomassie blue staining.21 In many cases, band 3 was the standard to which everything else was compared. Furthermore, to our knowledge, neither Western blotting of sph membranes for band 3 content nor eosin labeling to quantitate band 3 in sph RBCs by flow cytometry has been performed previously.
In addition to absent β-adducin, α-adducin is diminished, γ-adducin absent, and dematin is decreased in sph3J RBCs. α- and γ-adducin also are diminished in sph, sph2BC, and sphIhj, whereas α-adducin remains in the presence of elevated γ-adducin in sph1J and sph4J. This finding correlates with the sph, sph2BC, sph3J, and sphIhj mutations all producing premature stop codons and the sph1J and sph4J mutations resulting in stable α-spectrin translation with an altered extreme C-terminus.
In both sph1J and sph4J, the anemia is very severe, as severe as in any of the sph allelic series of mutants. Previous studies revealed that the C-terminus of α-spectrin binds protein 4.2 in vitro but not if the terminal 13 amino acids that are deleted in sph1J are excluded.16 However, this cannot explain the severity of the anemia in sph1J because complete 4.2 deficiency causes very mild HS.52 It is unlikely that loss of band 3 accounts for the severity of the anemia; band 3 is normal in sph4J, and there is a relatively minor decrement in sph1J (although significant compared with its strain-specific control) as measured by Western blotting and eosin-5-maleimide staining. Likewise, loss of adducin does not account for the severity of anemia. β-adducin knockout mice have relatively mild anemia in comparison with sph alleles, as do α-adducin null mice lacking all 3 adducins.30,35,46 However, flow cytometric analysis of the band 3 indicates that both sph1J and sph4J may have conformational changes in the extracellular domain that, possibly, are detrimental to RBC stability.
Our results clearly indicate that the C-terminal domain of α-spectrin, specifically the last 13 amino acids and the highly conserved cysteine, participates in interactions that are critical to membrane stability. Two nonmutually exclusive possibilities likely account for the extremely unstable RBCs in sph1J and sph4J. First, the EF hand domain of α-spectrin, which aligns with the actin-4.1 binding domain of β-spectrin, is required to stabilize the spectrin-actin-4.1 complex, as suggested many years ago by Cohen and Langley53 and more recently by Korsgren et al.16 Second, our results are in keeping with the notion that a membrane bridge in addition to the classic protein 4.1-p55-glycophorin C linkage exists at the RBC junctional complex that involves interactions between spectrin, adducin, band 3, and, most likely, additional proteins that remain to be identified.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs Thomas Chase and David Harrison of The Jackson Laboratory for critical review of the manuscript.
This work was supported by National Institutes of Health grants HL088468 and HL075714 (to L.L.P.), HL081608 (to S.E.L.); RR01183 (The Jackson Laboratory Mouse Mutant Resource); and The National Cancer Institute (CA34196, The Jackson Laboratory).
National Institutes of Health
Authorship
Contribution: R.F.R. and L.L.P. designed experiments and analyzed data; A.J.L. maintained the sph4J allele; C.S.B. maintained and provided the sph, sph1J, and sph2BC alleles; M.V.C. and A.F.M.C. identified and performed initial characterization of the sphIhj allele in their facility in Iasi, Romania; D.R.C. mapped sphIhj; R.F.R. and A.J.L. performed experiments; S.E.L. provided valuable reagents and insights and assisted in data analysis; R.F.R. maintained the sph3J allele and wrote the manuscript; and S.E.L. and L.L.P. edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luanne L. Peters, The Jackson Laboratory, 600 Main St, Bar Harbor, ME 04609; e-mail: luanne.peters@jax.org.