Abstract
Complex molecular mechanisms control B-cell fate to become a memory or a plasma cell. Interleukin-24 (IL-24) is a class II family cytokine of poorly understood immune function that regulates the cell cycle. We previously observed that IL-24 is strongly expressed in leukemic memory-type B cells. Here we show that IL-24 is also expressed in human follicular B cells; it is more abundant in CD27+ memory B cells and CD5-expressing B cells, whereas it is low to undetectable in centroblasts and plasma cells. Addition of IL-24 to B cells, cultured in conditions shown to promote plasma cell differentiation, strongly inhibited plasma cell generation and immunoglobulin G (IgG) production. By contrast, IL-24 siRNA increased terminal differentiation of B cells into plasma cells. IL-24 is optimally induced by BCR triggering and CD40 engagement; IL-24 increased CD40-induced B-cell proliferation and modulated the transcription of key factors involved in plasma cell differentiation. It also inhibited activation-induced tyrosine phosphorylation of signal transducer and activator of transcription-3 (STAT-3), and inhibited the transcription of IL-10. Taken together, our results indicate that IL-24 is a novel cytokine involved in T-dependent antigen (Ag)–driven B-cell differentiation and suggest its physiologic role in favoring germinal center B-cell maturation in memory B cells at the expense of plasma cells.
Introduction
T cell–dependent (TD) antibody responses are initiated by activation of naive B cells in the T cell–rich extrafollicular areas of secondary lymphoid organs. This initial B-cell activation generates short-lived plasma cells and recruits germinal center cell (GC) precursors into B-cell follicles.1-3 Actively cycling centroblasts in the dark zone of GCs are the cells in which ongoing immunoglobulin (Ig) hypermutation takes place. Centroblasts then differentiate into small nondividing centrocytes in the GC light zone. Centrocytes presumably undergo either apoptosis or differentiate into plasma cells or memory cells with affinity-maturated B-cell receptors (BCRs) and cytokine-driven class switching toward immunoglobulin G (IgG)/A/E class antibodies. Apoptosis within the GC is thought to play a key role in the selection of Ig V-region mutated high-affinity variants.4-6
Human GC B cells express CD38 and CD20 surface markers, whereas plasma cells are CD38hiCD20− and memory cells are CD38−CD20hi.7,8 The balance between plasma cells and memory cells is tightly regulated by complex and still not fully understood molecular mechanisms.9
The initiation of the GC response requires the interaction of costimulatory B-cell surface receptors with ligands present at the surface of T cells and antigen presenting cells (APCs). A major B-cell molecule in this respect is CD40, which was first identified in B cells10 and is needed for isotype switching and affinity maturation.11,12 CD40 activation is involved in proliferation,13 rescue from apoptosis,14 and maturation into memory B cells15,16 as well as prevention of maturation of B cells into plasma cells.17,18 GC B cells can therefore be rescued ex vivo from cell death by CD40 stimulation, a property used to generate memory B cells in vitro.15 The choice for GC B cells to become memory cells or plasma cells is tightly controlled by a sequential expression and repression of transcription factors. Several transcription factors crucial for plasma cell commitment have been identified. They include B lymphocyte–induced maturation protein 1 (BLIMP1), interferon-regulatory factor 4 (IRF4), X-box–binding protein 1 (XBP1), and signal transducer and activator of transcription 3 (STAT-3), which must be up-regulated. These events follow the functional inactivation of the transcriptional repressor paired box protein 5 (PAX5) and the CD40-mediated down-regulation of B-cell lymphoma 6 (BCL6).9 However the existence of other factors selectively controlling the developmental program of plasma and memory B cells remains an important and open issue.
We have previously observed that interleukin-24 (IL-24), a class II cytokine of the IL-10 family,19 is highly expressed in chronic lymphocytic leukemia (CLL) cells20 ; IL-24 protected these cells, which are mostly in the G0/G1 phase of the cell cycle, from apoptosis. CLL cells are antigen-experienced B cells and display surface antigen phenotype and transcription profile similar to those of memory B cells.21,22 However, the immune function of IL-24 remains poorly understood. IL-24 was observed by immunohistochemistry in some 20% CD19+ cells purified from phytohemagglutinin-stimulated peripheral blood mononuclear cells (PBMCs).23 In turn, IL-24 stimulated PBMCs to produce IL-6, tumor necrosis factor-α, and interferon-γ,23 whereas its own production was stimulated by a wide spectrum of cytokines and T-helper type 2 cytokines failed to induce substantial expression of IL-24.24 Although IL-24 was suggested to be a “pro TH1” cytokine, evidence for its physiologic role in the immune system is still lacking. Our results prompted us to investigate the effect of IL-24 and its expression in normal B-cell populations from human tonsils. Herein we report that IL-24 gene expression is tightly regulated during antigen-driven maturation of B-cell response. It is down-regulated in naive B cells upon transition to the centroblast stage with strongly polarized expression within the pool of memory versus plasma cells. Taken together, we identified IL-24 as a new important component of the terminal differentiation program in B cells acting in concert with other factors to inhibit the differentiation of GC B cells into mature plasma cells, thereby shifting their maturation toward the memory B-cell pathway.
Methods
B-cell isolation and culture
Human tonsils were obtained from children (age 2-6 years) undergoing tonsillectomy. Briefly, tonsils were minced and cell suspension was filtered through 70-μm cell strainer (Becton Dickinson), and mononuclear cells were separated on Ficoll-Hypaque (GE Healthcare) density gradient. The resultant mononuclear cells were submitted to B cell–negative isolation kit (Dynal Biotech) and stained for further fluorescence-activated cell sorting (FACS) analysis and/or cell sorting. B cells comprised between 50% and 80% of mononuclear cells and were 97% to 99% pure after purification as assessed by CD19 staining.
Sorted CD20+CD38+ GC B cells were cultured in RPMI medium supplemented with 10% fetal calf serum, penicillin (100 U/mL), streptomycin (100 mg/mL), 2mM l-glutamate, and 1mM sodium pyruvate (Invitrogen). The following reagents were used for stimulation experiments: recombinant IL-2 (rIL-2; 100 ng/mL), rIL-10 (50 ng/mL), rIL-24 (100 ng/mL), CD40 ligand trimer (400 ng/mL; all from R&D Systems), anti–human IgM, AffiniPure F(ab′)2 fragment of rabbit IgG fraction (10 μg/mL; from Jackson ImmunoResearch), CpG ODN2006-G5, (5μM) CpG ODN2006-G5 CONTROL (5μM; both from InvivoGen), and lipopolysaccharide (LPS) from Escherichia coli (10 μg/mL; VWR International SAS).
Monoclonal antibodies and flow cytometry
The following monoclonal antibodies (MAbs) were used: fluorescein isothiocyanate (FITC)–conjugated anti-CD5, phycoerythrin (PE)–conjugated anti-CD19, FITC-conjugated anti-CD20, PE-conjugated anti-CD27, FITC- or PE–conjugated anti-CD38, FITC-conjugated anti-CD44, PE-conjugated anti-IgD (all from BD Biosciences Pharmingen). For IL-24 receptor (IL-24R) chains, allophycocyanin (APC)–anti–IL-22R MAb and isotype control APC-IgG1k; PE-anti–IL-20Rα and isotype control PE-IgG1k; and biotinylated goat anti–IL-20Rβ and control biotinylated goat IgG were followed by streptavidin-FITC. Anti–IL-24R chains were from R&D Systems; isotype controls and streptavidin (SA)–FITC, from BD Pharmingen. Fluorescence was analyzed using a FACSCalibur and the analysis of the data was performed using CellQuest software (BD Biosciences). For intracellular staining, cells were fixed with 4% paraformaldehyde–phosphate-buffered saline (PBS) and permeabilized with methanol and labeled with control biotinylated goat IgG or biotinylated goat IgG anti–IL-24 (R&D Systems), followed by SA-FITC. Fluorescence was analyzed using a FACSCalibur, and data analysis was performed using CellQuest software.
Cell-cycle analysis
Nuclear DNA content was measured using propidium iodide staining and fluorescence-activated cell sorter analysis, as previously described.25 Briefly, B cells (106/mL) were washed with cold 1× PBS and resuspended in 1 mL of EtOH 70% and kept on ice for at least 10 minutes. Cells were then centrifuged and the pellet was suspended in 1 mL of 1× PBS plus RNase (40 μg) plus PI (18 μL; all from Sigma-Aldrich) at room temperature. The percentage of doublets was estimated by forward scatter (FSC) linear versus FSC area analysis.
Proliferation assays
Proliferation assays were performed in round-bottom 96-well plates. Cells (105/200 μL/well) were incubated at 37°C under various conditions. Each condition was performed in triplicate. After incubation, 0.5 μCi (0.0185 MBq) of [methyl3H]thymidine was added to each well for 15 hours at 37°C before harvesting and counting. Plates were then harvested in a Wallac Printed Filtermat A with a Harvester 96 (Tomtec) and were dried for at least 1 hour. Dried Printed Filtermat was put into a Wallac sample bag and 10 mL of Betaplate Scint was added. Incorporation of thymidine was measured with the Wallac 1205 Betaplate liquid scintillation counter (all components are from PerkinElmer).
ELISA
Cells (106/mL) were cultured in various conditions and supernatants were collected at days 7 and 11. Detection of IgG in supernatants was performed with a human IgG quantitative enzyme-linked immunosorbent assay (ELISA) kit (Bethyl) following the manufacturer's instructions.
RNA interference
Tonsillar B cells (10 × 106 cells/mL for each condition) were transfected as described previously20 with 100nM IL-24 siRNA (Dharmacon), using the Human B Cell Nucleofectort Kit (Amaxa) following the manufacturer's instructions (selected program U-15). Cells were processed as indicated after nucleofection. Control siRNA or human IL-24 siRNA was synthesized, annealed, desalted, deprotected, and purified by Dharmacon. To estimate transfection efficacy, cells were cotransfected with 20nM fluorescent control siRNA: siGLO RISC-free siRNA (Dharmacon), a fluorescently labeled siRNA with impaired ability for RNA-induced silencing complex (RISC) interaction and with X4 mismatches with known human genes, and 100nM IL-24 siRNA. After transfection, cells were cultured with plasma B-cell differentiation conditions for 6 days and immunostaining was performed using anti-CD38 and anti-CD20 MAbs.
Immunostaining on tissue sections
Immunohistochemistry (IHC) was performed on 6-μm sections of OCT-embedded (TissueTec), nitrogen-frozen human tonsils. Nonspecific antibody binding sites were blocked with 10% normal goat or rabbit serum for 30 minutes at room temperature.
Incubation with a polyclonal goat antibody to IL-24 (R&D Systems) was then performed overnight at 4°C. IL-24 was detected by a biotinylated rabbit anti–goat antibody (Vector Elite). Controls included omission of the primary antibody and substitution of IL-24 by a goat IgG (sc-2028; Santa Cruz Biotechnology). Double staining of IL-24 and CD20 was performed on sections fixed by 4% paraformaldehyde. Nonspecific antibody binding sites were blocked with 10% normal rabbit serum for 30 minutes at room temperature. IL-24 was detected by a biotinylated rabbit anti–goat antibody (Vector Elite) followed by Alexa Fluor 488–streptavidin (Molecular Probes). A second step of saturation with 10% normal goat serum was applied. Incubation with anti-CD20cy MAb (clone L26; Dako) was then performed overnight at 4°C and detected with Alexa Fluor 594-F(ab′)2 of goat anti–mouse IgG (Molecular Probes). DAPI (4,6 diamidino-2-phenylindole; Thermo Fischer Scientific) was used for counterstaining. Images of immunofluorescence stainings were acquired on a Nikon 80i microscope coupled to a CCD Nikon DS Camera.
Real-time PCR
Cells (106/mL) cultured under various conditions were washed in 1× PBS, resuspended in lysis buffer RLT, and stored at −80°C, and RNA was purified using the RNeasy Mini Kit (QIAGEN). Reverse transcription was performed with random hexamers using the SuperScript first-strand synthesis system for reverse-transcription–polymerase chain reaction (RT-PCR; Invitrogen Life Technologies). Transcript expression was analyzed by quantitative real-time PCR using either LightCycler Sybr Green technology (for BCL6, BLIMP1, IRF4, PAX5, and β-actin) or TaqMan technology (for IL-24, IL-10, and GAPDH). Thermocycling was performed on a LightCycler 480 (LC480) instrument (Roche Diagnostic). The SYBR Green Master Mix (Roche) was used to perform real-time PCR. After incubation at 95°C for 10 minutes, 40 cycles of amplification were performed. Each cycle consisted of 15 seconds at 95°C, 15 seconds at 60°C, and 30 seconds at 72°C. For each sample, serial dilutions were made to determine the PCR efficiency. The oligonucleotides used for light cycler were as follows: BCL6 forward (5′-CTGGCTTTTGTGACGGAAAT-3′) BCL6 reverse (5′-AACCTGAAAACCCACACTCG-3′); BLIMP1 forward (5′-GTGTCAGAACGGGATGAACA-3′); BLIMP1 reverse(5′-GCTCGGTTGCTTT AGACTGC-3′); IRF4 forward (5′-ACCGAAGCTGGAGGGACTAC-3′); IFR4-reverse (5′-GTGGGGCACAAGCATAAAAG-3′); PAX5 forward (5′-GGAGGAGTGA ATCAGCTTGG-3′); and PAX5 reverse (5′-GGCTTGATGCTT CCTGTCTC-3′).
Results were analyzed using the ABI Prism 7700 sequence detection system software (PE Applied Biosystems). Each reaction was performed in duplicate and results were normalized by the cycle threshold (Ct) of GAPDH or actin expression. Relative expression was calculated by measuring the difference of normalized Ct between condition A versus condition B and applying the following formula: difference of expression between A and B = 2CtA − CtB.
Western blot
Freshly isolated B cells (10 × 106) were stimulated with rIL-2, rIL-10, and CD40L for 3 days, washed, and incubated or not with rIL-24 for the indicated time. Cells were lysed at 4°C for 30 minutes in lysis buffer (20mM tris(hydroxymethyl)aminomethane-Hcl, pH 7.5, 140mM NaCl, 1mM ethylenediaminetetraacetic acid, 50 U/mL aprotinin, 1mM phenylmethylsulphonyl fluoride, 1mM sodium orthovanadate) containing 1% Nonidet P-40 detergent. Proteins were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels, electrotransferred onto polyvinylidene fluoride membranes (Amersham), and blotted sequentially with anti–pY705-STAT-3 (B-7) MAb (Tebu-bio), anti-STAT-3 MAb (BD Transduction Laboratories), anti–pY694-STAT-5 Ab (Cell Signaling Technology), and anti-STAT-5 MAb (BD Transduction Laboratories) after dehybridization. Blots were revealed using goat anti–mouse horseradish peroxidase conjugated or goat anti–rabbit horseradish peroxidase conjugated (Bio-Rad Laboratories) and enhanced chemiluminescence detection (Amersham). IL-24 was detected by an anti–IL-24 MAb (R&D Systems) in extracts from cells after 3 days of transfection with control or IL-24–specific siRNA.
Statistical analysis
All statistics were carried using the Mann-Whitney U test.
Results
IL-24 inhibits germinal center B-cell maturation into plasma cells
The generation of memory B cells and plasma cells from ex vivo–cultured GC B cells has been previously established.15 To this end, sorted CD20hiCD38hi GC B cells must be rescued from apoptosis and driven to proliferate; this occurs if cells are incubated with IL-2 plus IL-10 and cocultured on CD40L+ cells for 3 to 4 days. Under these stimulatory conditions, cells down-regulate CD20 and CD38. If these cells are cultured again for 3 to 4 days in the same condition they acquire a CD20+CD38− memory phenotype.15 On the contrary, if CD40L+ cells are removed from the secondary culture, the differentiation is shifted toward CD38hiCD20− plasmablasts. In our experiments, we used the described15 plasma cell differentiation model, however instead of CD40L+ transfectants, we used soluble trimeric CD40L to cross-link CD40 in the primary culture. At day 3, secondary cultures were performed in the presence of IL-2 plus IL-10 with or without the addition of IL-24 (100 ng/mL).
As expected, GC B cells down-regulated CD20 and CD38 surface molecules after 3 days in culture and up to 17% CD38hiCD20− plasma cells were generated after 7 days (Figure 1A). In contrast, if IL-24 was added to the secondary culture, the number of plasma cells was reduced by as much as 85% (mean, 2.8% vs 12.2%, P = .002; Figure 1B). In parallel the percentage of CD20+CD38− increased by nearly a half, in IL-24–cultured cells compared with secondary cultures without IL-24 (Figure 1A). When cultured cells were analyzed by size, it appeared that CD38hiFSChi plasmablasts almost disappeared from IL-24–cultured cells (0.3% vs 10.3%, R3 region in a representative dot plot). Progression from more resting (FSClo) cells in the R1 region to more activated (FSCint) cells in the R2 region was strongly inhibited in the presence of IL-24 (5.6% vs 21.9% in R2, Figure 1A representative data).
rIL-24 inhibits plasma cell maturation. (A) GC B cells from tonsils were stained and CD20hiCD38hi cell sorted and cultured for 3 days with IL-2, IL-10, and soluble CD40L. A secondary culture was performed with IL-2 + IL-10 with or without IL-24 for 4 additional days and cells analyzed at day 7 by immunostaining using anti-CD38 and anti-CD20 MAbs. CD38+ cells were also analyzed based on their size (CD38 vs FSC) showing CD38hiFSChi, CD38+FSC+, and CD38+FSClo R1 to R3 population, respectively. (Right) For each condition, cells were stained with propidium iodide for cell-cycle analysis. Percentages on histograms from left to right show the respective amounts of hypodiploid cells, and cells in G0/G1 and S+G2/M phases of the cell cycle. (B) The respective percentages of CD20−CD38+ and CD20+CD38− in secondary culture at day 7 with or without IL-24 were calculated (n = 6). (C) Secondary culture with IL-24 inhibits IgG production. GC B cells were cultured as above in IL-2 + IL-10 + CD40-L for 3 days and thereafter in IL-2 + IL-10 with or in the absence of IL-24. Supernatants from cells were collected at days 7 and 10 and the concentration of IgG was measured by ELISA (n = 5). (D) Cells cultured with or without the addition of IL-24 in the secondary culture were restimulated at day 7 or day 11 with IL-2 + IL-10 + CD40-L for 3 more days and tritiated thymidine (0.5 μCi [0.0185 MBq]/well per 100 000 cells) was incorporated overnight after 3 days (days 10 and 14).
rIL-24 inhibits plasma cell maturation. (A) GC B cells from tonsils were stained and CD20hiCD38hi cell sorted and cultured for 3 days with IL-2, IL-10, and soluble CD40L. A secondary culture was performed with IL-2 + IL-10 with or without IL-24 for 4 additional days and cells analyzed at day 7 by immunostaining using anti-CD38 and anti-CD20 MAbs. CD38+ cells were also analyzed based on their size (CD38 vs FSC) showing CD38hiFSChi, CD38+FSC+, and CD38+FSClo R1 to R3 population, respectively. (Right) For each condition, cells were stained with propidium iodide for cell-cycle analysis. Percentages on histograms from left to right show the respective amounts of hypodiploid cells, and cells in G0/G1 and S+G2/M phases of the cell cycle. (B) The respective percentages of CD20−CD38+ and CD20+CD38− in secondary culture at day 7 with or without IL-24 were calculated (n = 6). (C) Secondary culture with IL-24 inhibits IgG production. GC B cells were cultured as above in IL-2 + IL-10 + CD40-L for 3 days and thereafter in IL-2 + IL-10 with or in the absence of IL-24. Supernatants from cells were collected at days 7 and 10 and the concentration of IgG was measured by ELISA (n = 5). (D) Cells cultured with or without the addition of IL-24 in the secondary culture were restimulated at day 7 or day 11 with IL-2 + IL-10 + CD40-L for 3 more days and tritiated thymidine (0.5 μCi [0.0185 MBq]/well per 100 000 cells) was incorporated overnight after 3 days (days 10 and 14).
As IL-24 regulates progression of B cells into the cell cycle,25 propidium iodide staining was performed in secondary cultures with or without IL-24 and DNA content analyzed. As expected from FSC parameter analyzing data, we confirmed that IL-24 inhibited entry/progression into the cell cycle in our experimental system as shown by a shift to the left of the G2/M peak and a broadened G0/G1 peak. The percentage of dead cells (hypodiploid) was also increased under IL-24 (Figure 1A right panel). To determine whether apoptosis affected only plasma cell precursors or more mature cells, cultures were performed until day 10 or 14 to generate plasma cells and IL-24 was added thereafter to the culture. Cell-cycle analysis and staining with annexin-V or Apo 2.7-PE were performed. Results show that IL-24 failed to kill mature cells (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
To further demonstrate that IL-24 inhibited plasma cell maturation, supernatants from cells cultured with or in the absence of IL-24 in the secondary culture were collected at days 7 and 11 and IgG concentration was measured by ELISA (Figure 1C). The abundance of IgG was significantly decreased in supernatants of cultures treated with IL-24 in comparison with cultures without IL-24, both at day 7 (P = .015) and at day 11 (P = .001). In fact, we found that IL-24 completely abrogated IL-10–dependent stimulation of IgG production in this system.
We next analyzed cell proliferation. After 7 days, cells cultured with or without IL-24 in the secondary culture (days 4 to 7) were restimulated for either 3 (day 10) or 7 (day 14) additional days with IL-2 plus IL-10 plus soluble CD40L, and thymidine incorporation was measured thereafter (Figure 1D). We found that cells cultured with IL-24 proliferated much better than cells cultured without IL-24 (P = .029 and P = .01 at days 10 and 14, respectively). Under these conditions, plasma cells could not be restored and most cells at days 10 and 14 were CD20+CD38− (supplemental Figure 2); in addition, the cell-surface expression of CD40 was not enhanced by IL-24 (data not shown). Sensitivity to CD40 may therefore reflect the predominance of memory cells in the culture.
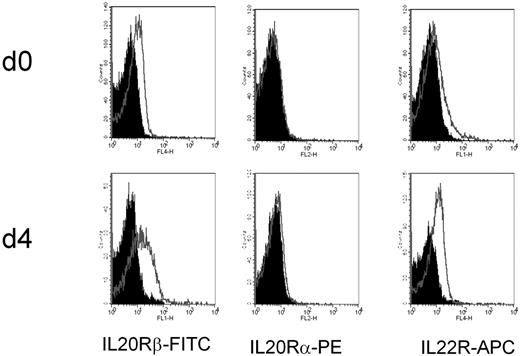
The expression of the 3 IL-24R chains was monitored by FACS in purified B cells (Figure 2). The IL-24R is made of a common IL-20Rβ chain, associated to either the IL-20Rα chain (generating type 1 IL-20R heterodimer) or to the IL-22R (type 2 IL-20R). IL-24 is a ligand of both IL-20Rs.19 As shown, the IL-20Rα chain is absent, thus only the type 2 heterodimer is present in B cells. This receptor is inducible as its expression increased after 3 days in the primary culture condition on the CD20+CD38+ cells (Figure 2). The expression of the 3 chains was monitored in CD20+, CD27+, and CD38+ populations and confirmed the lack of expression of IL-20Rα in tonsil B cells (supplemental Figure 3).
Expression of IL-20Rβ/IL-22R (IL-24R heterodimer) in tonsil B cells by flow cytometry. Tonsil B cells were purified and stained immediately (day 0) or after 3 to 4 days in culture with IL-2 + IL-10 + CD40L. Isotype APC-IgG1k, isotype PE-IgG1k, and biotinylated goat IgG followed by streptavidin, SA-FITC were used as controls.
Expression of IL-20Rβ/IL-22R (IL-24R heterodimer) in tonsil B cells by flow cytometry. Tonsil B cells were purified and stained immediately (day 0) or after 3 to 4 days in culture with IL-2 + IL-10 + CD40L. Isotype APC-IgG1k, isotype PE-IgG1k, and biotinylated goat IgG followed by streptavidin, SA-FITC were used as controls.
Taken together, these results demonstrated that rIL-24 inhibited the differentiation of plasma cells from GC B cells and suggested that, as it enhanced the response to CD40L, it may favor the differentiation of memory B cells.17,26
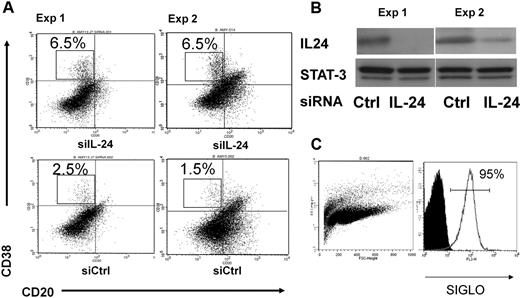
To investigate the effect of endogenous IL-24 in this process, we sorted GC B cells and transfected them with either control or IL-24–specific small interfering RNAs as described previously.20 Primary and secondary cultures were performed as in Figure 1. As shown in Figure 3A, cells transfected with IL-24 siRNAs generated 3 times more plasma cells (6.5% vs 1.5%-2.5%) than cells transfected with control siRNAs. In parallel, we failed to detect IL-24 by ELISA in these B-cell cultures (data not shown). The efficacy of transfection was monitored by cotransfecting the cells with IL-24 siRNA and random fluorescent siRNA siGLO at a 5:1 ratio. The efficacy ranged from 85% to 95% (Figure 3C), although 1 experiment of 6 on average led to a viable culture. Figure 3B shows that IL-24 siRNA significantly and specifically diminished the production of the IL-24 protein23 but not that of Stat3. Thus the blockade of intracellular IL-24 expression had an opposite effect on plasma cell maturation than the addition of rIL-24 to the culture of GC B cells. Both results support the notion that IL-24 is a physiologic repressor of plasma cell differentiation.
RNA interference. (A) The inhibition of endogenous IL-24 expression resulted in increased plasma cell numbers in GC B-cell cultures. Cell-sorted GC B cells (107) from 2 tonsils were transfected with 100nM control or IL-24–specific siRNAs, cultured the day after with IL-2 + IL-10 + CD40L for 3 days and with IL-2 + IL-10 for 4 more days, and cells were stained with CD38 and CD20 MAbs. (B) Western blot was performed in parallel at day 3 and shows the inhibition of IL-24 but not of STAT-3 protein expression in cells treated with IL-24–specific siRNA; control (random) RNA had no effect. (C) The efficacy of transfer was monitored by cotransfection of PE-random siRNA siGLO and IL-24–specific siRNA at a 1:5 ratio.
RNA interference. (A) The inhibition of endogenous IL-24 expression resulted in increased plasma cell numbers in GC B-cell cultures. Cell-sorted GC B cells (107) from 2 tonsils were transfected with 100nM control or IL-24–specific siRNAs, cultured the day after with IL-2 + IL-10 + CD40L for 3 days and with IL-2 + IL-10 for 4 more days, and cells were stained with CD38 and CD20 MAbs. (B) Western blot was performed in parallel at day 3 and shows the inhibition of IL-24 but not of STAT-3 protein expression in cells treated with IL-24–specific siRNA; control (random) RNA had no effect. (C) The efficacy of transfer was monitored by cotransfection of PE-random siRNA siGLO and IL-24–specific siRNA at a 1:5 ratio.
The expression of IL-24 in human B cells
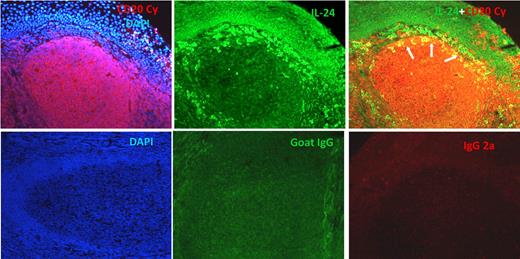
To visualize the expression of IL-24 within B cells in secondary lymphoid tissues, immunofluorescence staining was performed on frozen sections from human tonsils with anti-CD20 and anti–IL-24 Ab (Figure 4). Double-stained CD20+IL-24+ B cells are located within follicles and predominate at the periphery. Interestingly IL-24–expressing B cells were typically located at the periphery of secondary follicles immediately adjacent to the perifollicular T-cell area where more abundant IL-24–expressing T cells were also present (Figure 4 and data not shown). Of note, very few, if any, IL-24–expressing B cells were found within GC's dark zone, deep in the center of the light zone, T cell–rich paracortex area, and medullary cords (data not shown).
The detection of IL-24–positive cells in follicular B cells on OCT-embedded frozen sections. (Top left) Section was stained with anti-CD20cy MAb followed by a secondary Alexa 594 F(ab)′2-goat anti–mouse Ab, and DAPI-containing mounting medium. (Middle) Goat anti–human IL-24 Ab was followed by biotinylated rabbit anti–goat Ab and streptavidin–Alexa 488. (Right) Section was stained sequentially with anti–IL-24 then with anti-CD20 Abs. (Bottom panels from left to right) Controls are in order: DAPI, goat IgG + biotin–anti-goat + SA–Alexa 488, mouse IgG2a + Alexa 594 goat anti–mouse Ab.
The detection of IL-24–positive cells in follicular B cells on OCT-embedded frozen sections. (Top left) Section was stained with anti-CD20cy MAb followed by a secondary Alexa 594 F(ab)′2-goat anti–mouse Ab, and DAPI-containing mounting medium. (Middle) Goat anti–human IL-24 Ab was followed by biotinylated rabbit anti–goat Ab and streptavidin–Alexa 488. (Right) Section was stained sequentially with anti–IL-24 then with anti-CD20 Abs. (Bottom panels from left to right) Controls are in order: DAPI, goat IgG + biotin–anti-goat + SA–Alexa 488, mouse IgG2a + Alexa 594 goat anti–mouse Ab.
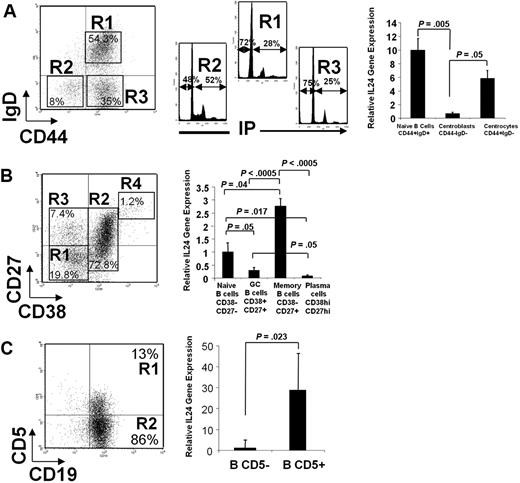
To quantitatively assess the expression of IL-24 in different B-cell populations, freshly isolated tonsil B cells were cell sorted and relative IL-24 mRNA expression was analyzed by quantitative RT-PCR (Figure 5). Cells stained with anti-CD44 and anti-IgD segregated into 3 populations26,27 : IgD+ naive cells (R1), IgD−CD44+ centrocytes (R3), and IgD−CD44− centroblasts (R2). As expected, the latter population is characterized by a high percentage, 52%, of cells in the S+G2/M phase of the cell cycle in comparison with R1 and R3 cells (Figure 5A). IL-24 mRNAs are abundant in naive cells. Within the population of GC B cells, IL-24 sharply decrease in centroblasts and is re-expressed at the centrocyte stage at a level close to that observed among naive cells. We also sorted cells based on their CD38 and CD27 expression (Figure 5B). The most abundant IL-24 mRNA expression was detected in memory CD38−CD27+ B-cell pool, intermediate in CD38−CD27− naive B cells, and low to undetectable in CD38+CD27+ GC B cells and in CD38hiCD27hi plasma cells. As we first described IL-24 in CD5+ malignant B cells, its expression was investigated within normal CD5-expressing B-cell pool from human tonsils. As shown in Figure 5C, CD19+CD5+ cells contained up to 30 times more IL-24 mRNA than CD19+CD5− cells.
Cells were cell sorted and mRNA was extracted, reverse transcribed, and measured by real-time PCR. Left is a representative dot plot of each experiment. (A) Tonsil B cells were stained and cell sorted according to their respective IgD+, IgD−CD44−, and IgD−CD44+ phenotype. Cell-cycle analysis was performed on all 3 sorted populations stained with propidium iodide (PI; note that 52% of IgD−CD44− centroblasts are in the S+G2/M phases of the cell cycle). (B) Cells were stained with CD27 and CD38 MAbs. (C) Cells were stained with CD19 and CD5 MAbs. (A-C) Histograms on the right show the relative expression of IL-24 mRNAs in sorted populations by quantitative polymerase chain reaction (qPCR). Mean ± SD from 3 to 6 experiments/cell population, each in duplicate, normalized for control GAPDH.
Cells were cell sorted and mRNA was extracted, reverse transcribed, and measured by real-time PCR. Left is a representative dot plot of each experiment. (A) Tonsil B cells were stained and cell sorted according to their respective IgD+, IgD−CD44−, and IgD−CD44+ phenotype. Cell-cycle analysis was performed on all 3 sorted populations stained with propidium iodide (PI; note that 52% of IgD−CD44− centroblasts are in the S+G2/M phases of the cell cycle). (B) Cells were stained with CD27 and CD38 MAbs. (C) Cells were stained with CD19 and CD5 MAbs. (A-C) Histograms on the right show the relative expression of IL-24 mRNAs in sorted populations by quantitative polymerase chain reaction (qPCR). Mean ± SD from 3 to 6 experiments/cell population, each in duplicate, normalized for control GAPDH.
CD40 engagement stimulates IL-24 expression in B cells
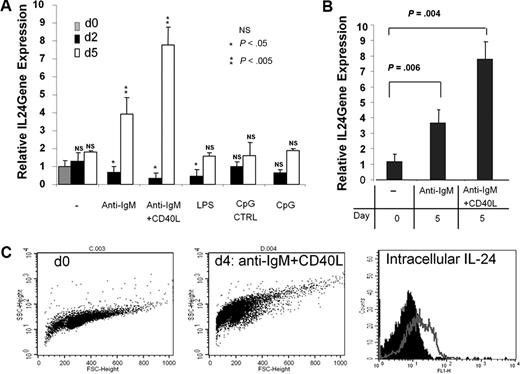
To investigate the nature of the physiologic inducers of IL-24 expression in B cells, we sorted GC B cells and stimulated them in various conditions and mRNAs were extracted after 2 and 5 days. The highest stimulation of IL-24 transcription was observed in cells cultured with anti-IgM plus CD40-L and to a lesser extent anti-IgM only (Figure 6A), whereas Toll-like receptor 4 and Toll-like receptor 9 triggering through LPS and CpG ODN stimulation, respectively, had no significant effects after 5 days of culture. Of note, a biphasic response of IL-24 expression after BCR triggering was observed. BCR engagement resulted in a transient decrease of IL-24 transcription within the first 24 to 48 hours followed by a sharp increase in its transcription at day 5. This time-dependent pattern was strongly increased by CD40 coengagement. In another set of experiments, IgD−CD44+ centrocytes were sorted and stimulated with anti-IgM alone or with anti-IgM plus CD40L. We found that both conditions enhanced the transcription of IL-24 after 5 days of culture (Figure 6B). Interestingly, there was no effect of CD40L addition on IL-24 expression at day 2 of culture. Intracellular expression of the IL-24 protein was monitored by FACS and paralleled that of mRNAs. As shown (Figure 6C), intracellular IL-24 in tonsil B cells was more abundant in cells cultured with anti-IgM plus CD40L for 5 days. Taken together, our data suggest that IL-24 is strongly up-regulated in human B cells during antigen (Ag)–specific T-dependent B-cell response.
BCR and CD40 stimulation are the optimal inducers of IL-24 mRNA expression. (A) GC B cells were cultured with medium only, F(ab)′2 anti-IgM, anti-IgM + CD40L, LPS, control CpG/ODN, or CpG/ODN for 2 and 5 days before RNA was extracted, reverse-transcribed, and analyzed by qPCR (n = 5). (B) CD44+IgD− centrocytes were sorted and restimulated or not with anti-IgM or anti-IgM + CD40L for 5 days and qPCR was performed (n = 3). (C) Intracellular expression of IL-24 was analyzed by FACS on fresh (day 0) or stimulated with anti-IgM + CD40-L for 4 days (day 4), purified tonsil B cells. Cells were fixed, permeabilized, and stained with biotinylated goat control IgG or goat anti–IL-24 Abs followed by SA-FITC as shown by black (day 0) or gray (day 4) histograms.
BCR and CD40 stimulation are the optimal inducers of IL-24 mRNA expression. (A) GC B cells were cultured with medium only, F(ab)′2 anti-IgM, anti-IgM + CD40L, LPS, control CpG/ODN, or CpG/ODN for 2 and 5 days before RNA was extracted, reverse-transcribed, and analyzed by qPCR (n = 5). (B) CD44+IgD− centrocytes were sorted and restimulated or not with anti-IgM or anti-IgM + CD40L for 5 days and qPCR was performed (n = 3). (C) Intracellular expression of IL-24 was analyzed by FACS on fresh (day 0) or stimulated with anti-IgM + CD40-L for 4 days (day 4), purified tonsil B cells. Cells were fixed, permeabilized, and stained with biotinylated goat control IgG or goat anti–IL-24 Abs followed by SA-FITC as shown by black (day 0) or gray (day 4) histograms.
IL-24 inhibits molecular mechanisms associated with plasma cell differentiation program
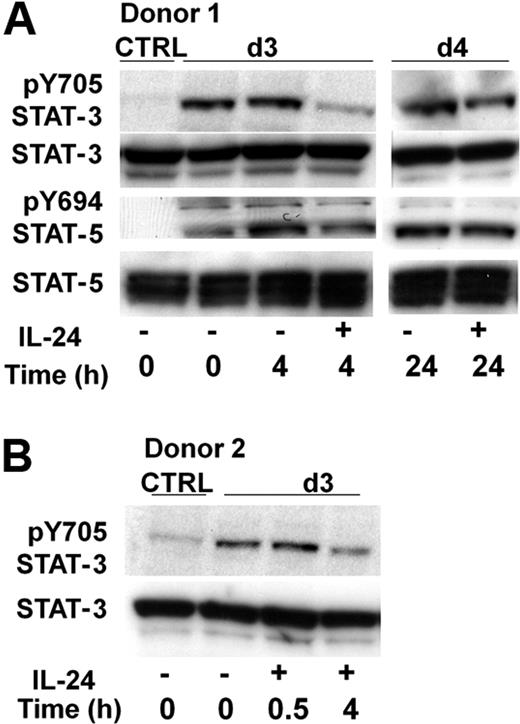
Several factors are thought to play a role in the development of memory or plasma cells from the GC B cells through a dynamic and multistep process. IL-2 induces B-cell proliferation, whereas IL-10 was shown to drive their differentiation into plasma cells. IL-2 activates STAT-3 and STAT-5 in lymphocytes through phosphorylation on Y705 and Y694 residues, respectively,28,29 whereas IL-10 activates STAT-1 and STAT-330 ; the latter was shown to be necessary for plasma cell differentiation.31 We cultured GC B cells with IL-2 plus IL-10 plus CD40-L for 3 days, then IL-24 was added or not to the culture. Both STAT-3 and STAT-5 were phosphorylated on tyrosine after 3 days of culture, however we observed a reduced phosphorylation of pY-STAT-3 but not of pY-STAT-5 after the addition of IL-24. STAT-3 phosphorylation was normal 30 minutes after the addition of IL-24 but was consistently and almost fully inhibited after 4 hours (Figure 7A-B), whereas it was almost restored after 24 hours (Figure 7A). This indicated a rapid but transient effect of IL-24 on pSTAT-3 in B cells.
IL-24 specifically inhibits the phosphorylation of STAT-3 on tyrosine in stimulated GC B cells. (A) Cells (10 × 106 cells/condition) were cultured for 3 days in IL-2 + IL-10 + CD40L and rIL-24 was added thereafter for the indicated time; cells were lysed and submitted to Western blot sequentially revealed after dehybridization, with anti–pY705-STAT-3, anti-STAT-3, anti–pY694-STAT-5, or anti-STAT-5 antibodies. Control was performed in parallel on unstimulated cells. (B) IL-24 transiently reduced STAT-3 phosphorylation. GC B cells from a second tonsil were cultured as in panel A and stimulated with IL-24 for 0.5 and 4 hours before Western blot.
IL-24 specifically inhibits the phosphorylation of STAT-3 on tyrosine in stimulated GC B cells. (A) Cells (10 × 106 cells/condition) were cultured for 3 days in IL-2 + IL-10 + CD40L and rIL-24 was added thereafter for the indicated time; cells were lysed and submitted to Western blot sequentially revealed after dehybridization, with anti–pY705-STAT-3, anti-STAT-3, anti–pY694-STAT-5, or anti-STAT-5 antibodies. Control was performed in parallel on unstimulated cells. (B) IL-24 transiently reduced STAT-3 phosphorylation. GC B cells from a second tonsil were cultured as in panel A and stimulated with IL-24 for 0.5 and 4 hours before Western blot.
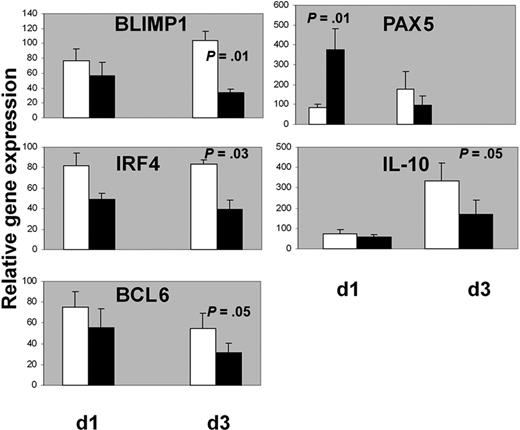
Activated STAT-3 has been shown to up-regulate BLIMP1 gene expression and promote plasma cell differentiation in murine and human B cells.32,33 In turn, BLIMP1 inhibits PAX5 and BCL6 expression.9,34,35 BCL6 is also inhibited by another factor necessary for plasma cell commitment, IRF4.36,37 We analyzed the expression of the above transcription factors in GC B cells for 3 days as described in the previous experiments and stimulated or not at day 3 with IL-24 (100 ng/mL). mRNAs were extracted after 1 and 3 days following the addition of IL-24 in the cultures, that is, at days 4 and 6 of the beginning of the culture, respectively (Figure 8). The addition of IL-24 decreased mRNA levels of IRF4 (83 ± 4 vs 38 ± 9, P = .03), BLIMP1 (104 ± 11 vs 34 ± 4, P = .01) and BCL6 after 3 days (54 ± 14 vs 31 ± 9 P = .05). In contrast, IL-24 rapidly stimulated PAX5 expression within 24 hours (85 ± 15 vs 376 ± 104, P = .01), albeit transiently as this augmentation was not observed after 3 days (175 ± 90 vs 98 ± 44, not significant). As IL-10 plays a role in plasma cell differentiation38 and activates STAT-3,39 its transcription was quantified in our experiments. The addition of IL-24 resulted in a delayed inhibition of the transcription of IL-10 after 3 days (334 ± 85 vs 168 ± 71, P = .05). These results prompted us to check whether inhibition of endogenous IL-24 by RNA interference resulted in opposite effects than that of rIL-24. RNAs from experiments 1 and 2 (Figure 3) were extracted at days 3 and 7. Some of the data mirror the results observed after the incubation with rIL-24: IL-10, IRF4, and to a lesser extent Blimp-1 transcripts were augmented. Three other experiments showed similar results (supplemental Figure 4).
IL-24 decreased mRNA levels of BLIMP1, IRF4, BCL6, and IL-10 and stimulated mRNA levels of PAX5 in GC B cells. Cells were cultured for 3 days with IL-2 + IL-10 + CD40L and IL-24 was added (black histograms) or not (white histograms) at day 3 then RNA was extracted after 1 or 3 more days for qPCR analysis. One hundred is the arbitrary value for unstimulated GC B cells at day 0 (n = 7 for IL-10 and n = 5 for the other transcripts).
IL-24 decreased mRNA levels of BLIMP1, IRF4, BCL6, and IL-10 and stimulated mRNA levels of PAX5 in GC B cells. Cells were cultured for 3 days with IL-2 + IL-10 + CD40L and IL-24 was added (black histograms) or not (white histograms) at day 3 then RNA was extracted after 1 or 3 more days for qPCR analysis. One hundred is the arbitrary value for unstimulated GC B cells at day 0 (n = 7 for IL-10 and n = 5 for the other transcripts).
Altogether, these results suggested that IL-24 coordinates multiple molecular events leading to the inhibition of plasma cell differentiation.
Discussion
This article describes for the first time the expression and function of IL-24 in human B cells and its inhibitory role on plasma cell differentiation. This cytokine was first described in terminally differentiated melanocytes40 and because its expression was gradually lost during advanced progression of melanoma and inversely correlated with proliferation,41 it gained interest as a potential anticancer agent,42 although there is no direct evidence that endogenous IL-24 is a tumor suppressor. IL-24 delivered by various routes killed several cancer cell lines, although the cytokine itself is not sufficient to kill primary melanoma cells. We reached the same conclusion by working on CLL and demonstrated that IL-24 was not detrimental and even protected nonstimulated cells from apoptosis in culture.20 However, IL-24 did induce a cell cycle block and apoptosis in cells preactivated with IL-2.25 As for normal B cells in our present work, the mechanism involved STAT-3 dephosphorylation by IL-24, although this mechanism was much faster in leukemic cells as it occurred in less than 5 minutes, whereas it took more than 30 minutes in GC B cells. IL-24 is strongly expressed in CLL cells,25 which are now believed to share many features with memory B cells, and this incited us to investigate the expression of this cytokine in normal B cells.
Here we present evidence that IL-24 strongly inhibits the plasma cell differentiation program in GC B cells. This cytokine worked on a previously published method in which GC B cells were driven into the cell cycle and rescued by apoptosis by coculture on a CD40 ligand–expressing cell line and a cocktail of IL-2 and IL-10 in the primary culture followed by a secondary culture without the CD40L+ cell line.15 The only modification here was the use of a soluble CD40L trimer. We also performed some experiments with the addition of IL-4 with similar results (data not shown). We consistently found that the addition of IL-24 to secondary cultures inhibited plasma cell differentiation. One possible mechanism behind this inhibition may be the direct killing of plasma cell precursors as we have observed a nearly 50% increase of death in cells cultured in the presence of IL-24 (Figure 1). Another mechanism may be a blockade of entry of these cells into the cell cycle. Cell-cycle control of B-cell development and differentiation in GC is a complex and poorly understood process, in which BCL6 plays a pivotal role.43-45 Our results imply that, at least in the culture system used here, a sustained proliferation drives plasma cell differentiation at the expense of memory B cells; this process is blocked by IL-24. The addition of IL-24 to the secondary culture not only inhibited the generation of CD38hiCD20− plasmablasts but also inhibited the production of IgG. As it was inactive on mature cells, it is therefore likely that IL-24 acted at the early steps of commitment toward plasma cells by inhibiting the cell cycle. In parallel IL-24 “sensitized” the cells in the secondary culture to CD40L-induced proliferation. As memory but not plasma cells respond to CD40 stimulation, this suggested that IL-24 redirected the differentiation of GC B cells toward the memory B-cell pathway. The latter may be a passive choice as IL-24 closes down the plasma cell differentiation pathway, thereby leaving only the memory pathway available. Alternatively, it may also be active by enhancing the responsiveness of B-cell to CD40-induced maturation. Although not demonstrated in vivo, it is possible that continued stimulation through CD40 may direct centrocytes toward the memory B-cell differentiation pathway.46 In fact, CD40 signaling is not absolutely needed for GC expansion and may act only at the initial and final stages of commitment toward memory B-cell maturation.47 We also demonstrate that IL-24 is differentially expressed by normal B-cell functional subpopulations and during Ag-driven B-cell response. The transcription kinetics of IL-24 indicates that this molecule is rapidly down-regulated after BCR and CD40 engagement, the stimulatory conditions mimicking the in vivo response to TD antigen stimulation. The inverse correlation between proliferation and IL-24 expression is also observed in vivo as IL-24 is clearly shut down at the centroblast stage. Therefore a possible sequence of events is that during a TD response, B cells are driven to proliferate and that this mechanism is coordinated with the down-regulation of IL-24 expression. Once centroblasts have differentiated in centrocytes, IL-24 is up-regulated and maintained all the way through the differentiation of centrocytes toward memory B cells. This is suggested by the up-regulation of IL-24, 5 days after stimulation with anti-IgM plus CD40-L (Figure 6). In contrast, in the case of commitment of centrocytes to differentiate into plasma cells, they down-regulate IL-24. This later observation may be contradictory with the notion that IL-24 expression is associated with terminal differentiation in B and in non-B cells.20,40 One possibility is that plasma cells from tonsils or those generated in vitro are immature plasmablasts prone to proliferate extensively. It would be worth analyzing whether IL-24 is re-expressed in long-lived/terminally differentiated plasma cells such as those present in the bone marrow.48,49
The notion that IL-24 is a physiologic inhibitor of plasma cell differentiation is supported by its ability to inhibit the transcription and activation of key factors known to be involved in plasma cell versus memory B-cell commitment.33,37-40 Terminal differentiation of a B cell into a plasma cell requires the inactivation of the transcriptional repressor PAX5.34,35 The up-regulation of PAX5 expression by IL-24 is therefore meaningful. The activation of STAT-3 in B cells is required for plasma cell maturation as it induces the expression of BLIMP1, IRF4, and XBP1 transcription factors.9 As IL-10 induced pSTAT-3 and plasma cell differentiation,38,40 its activity must be inhibited by IL-24 in GCs. As on the other hand IL-10 antagonized the action of IL-24 on human PBMCs,23 both cytokines therefore appear to have antagonistic functions.
The sources of the cytokines that induce or repress STAT-3 in GCs are therefore of interest. Our data using siRNA suggest that IL-24 is an intrinsic/autocrine regulator of GC B-cell differentiation. IL-24 may also act in a paracrine manner to block the generation of plasma cells within GC microenvironment. IL-24 may be secreted for instance by memory cells or by CD5+ B cells, which are abundant in the tonsils. How IL-24 interplays with other cytokines especially of the IL-10 family remains unclear. It should be mentioned in this respect that IL-19 is a ligand of the type 1 IL-20R, whereas IL-20 is a ligand for both (IL-20R1 and IL-20R2) IL-24 receptors.19,50 In addition, IL-22, a TH17 cytokine, is a ligand for the IL-22R chain, a component of the IL-20R1.50 IL-19, IL-20, and IL-22 may therefore compete with IL-24 for binding to its receptor. Taken together, our data indicate that, within the IL-10 family of cytokines, IL-24 is a novel important player controlling TD Ag-driven B-cell differentiation process by blocking plasma cell differentiation process and favoring memory B-cell development pathway.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
We thank Dr Thierry Rousselet for providing tonsils.
Authorship
Contribution: G.M. performed cell purifications, cultures, sorting, functional assays, RNA extractions and reverse transcriptions, and FACS analysis; L.B.-D. performed the cell sorting and qPCR; H.G.-G. performed the siRNA and Western blots; I.D.-G. performed the sections and immunostainings; R.K. participated in the design and discussion; and A.D. designed the experiments, participated in the cultures and functional assays, and wrote the article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ali Dalloul, Laboratoire de Thérapie Cellulaire Bâtiment Hématologie Transfusion, CHU de Nancy, Nancy Université, EA 4369, 54000 Nancy, France; e-mail: a.dalloul@chu-nancy.fr.
References
Author notes
*L.B.-D. and H.G.-G. contributed equally to this work.

![Figure 1. rIL-24 inhibits plasma cell maturation. (A) GC B cells from tonsils were stained and CD20hiCD38hi cell sorted and cultured for 3 days with IL-2, IL-10, and soluble CD40L. A secondary culture was performed with IL-2 + IL-10 with or without IL-24 for 4 additional days and cells analyzed at day 7 by immunostaining using anti-CD38 and anti-CD20 MAbs. CD38+ cells were also analyzed based on their size (CD38 vs FSC) showing CD38hiFSChi, CD38+FSC+, and CD38+FSClo R1 to R3 population, respectively. (Right) For each condition, cells were stained with propidium iodide for cell-cycle analysis. Percentages on histograms from left to right show the respective amounts of hypodiploid cells, and cells in G0/G1 and S+G2/M phases of the cell cycle. (B) The respective percentages of CD20−CD38+ and CD20+CD38− in secondary culture at day 7 with or without IL-24 were calculated (n = 6). (C) Secondary culture with IL-24 inhibits IgG production. GC B cells were cultured as above in IL-2 + IL-10 + CD40-L for 3 days and thereafter in IL-2 + IL-10 with or in the absence of IL-24. Supernatants from cells were collected at days 7 and 10 and the concentration of IgG was measured by ELISA (n = 5). (D) Cells cultured with or without the addition of IL-24 in the secondary culture were restimulated at day 7 or day 11 with IL-2 + IL-10 + CD40-L for 3 more days and tritiated thymidine (0.5 μCi [0.0185 MBq]/well per 100 000 cells) was incorporated overnight after 3 days (days 10 and 14).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/9/10.1182_blood-2009-05-220251/4/m_zh89990947740001.jpeg?Expires=1769141810&Signature=FGv1Jc7Kuzk4c5L6dvucp5zlpHFfUi-E3Pgvzqd9-1jAAOE5p4EF16hW4K~yuymD5fxivRKqREVux0Idch87bHbH8Mo7A9yuAKdNTr35W4YXgVxm8eYENzNhqcp2lgWiqrZc0~Nr56GeYfkgwfPXYjyMhYlygT84aNeQb0YgJ7GRmOibe5W-cYdnPL~l~nsOPHWoRPoL7PeBgDI3gMKvFXDOtxV6pP1j1pY9PSEsTqvxVx79G5ifdtii1U7PYD0gd0dXGwtlkWSKVJ1T6OG45w0X7pQSTubDMVl1HJ~nH58D0O512xu~pDbKq-sWlwtTkSdmVW96n-MLe9WXAbBgow__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal