Abstract
Chromosomal translocations involving the immunoglobulin heavy chain (IGH) locus define common subgroups of B-cell lymphoma but are rare in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Recent fluorescent in situ hybridization and molecular cloning studies have identified several novel IGH translocations involving genes that play important roles in normal hemopoiesis, including the cytokine receptor genes CRLF2 and EPOR, all members of the CCAAT enhancer-binding protein gene family, as well as genes not normally expressed in hemopoietic cells including inhibitor of DNA binding 4. IGH translocation results in deregulated target gene expression because of juxtaposition with IGH transcriptional enhancers. However, many genes targeted by IGH translocations are also more commonly deregulated in BCP-ALL as a consequence of other genetic or epigenetic mechanisms. For example, interstitial genomic deletions also result in deregulated CRLF2 expression, whereas EPOR expression is deregulated as a consequence of the ETV6-RUNX1 fusion. The possible clinical importance of many of the various IGH translocations in BCP-ALL remains to be determined from prospective studies, but CRLF2 expression is associated with a poor prognosis. Despite their rarity, IGH chromosomal translocations in BCP-ALL therefore define not only new mechanisms of B-cell transformation but also clinically important subgroups of disease and suggest new targeted therapeutic approaches.
Introduction
It is nearly 50 years since the cytogenetic description of the first recurrent chromosomal translocation in malignancy, the Philadelphia chromosome in chronic myeloid leukemia (CML).1 Since then, a plethora of different chromosomal translocations has been identified in nearly all forms of malignancy.2 Many recurrent translocations are the primary genetic cause of disease; that is, the production of tumor-specific gene products derived from the fusion of genes normally on 2 different chromosomes, or alternatively, deregulated gene expression, arising as a consequence of chromosomal translocation, play a pivotal role in the emergence of the neoplastic phenotype. Molecular analysis of these translocations was initially conducted via positional cloning strategies, led by cytogenetic observations3 but now increasingly from whole genome and high-throughput transcriptional studies.4 These studies, supplemented by sophisticated animal models,5 define mechanisms of transformation and suggest new targeted therapeutic approaches. Successful inhibition of the B-cell precursor acute lymphoblastic leukemia (BCP-ALL) fusion protein in CML by imatinib6 is a paradigm that urgently needs to be emulated, although whether other malignancies will have such a vulnerable “Achilles heel” as CML remains to be determined.
However, as more is understood about the genetic complexities of cancer, problems emerge concerning the relative importance of the multiple and different abnormalities that exist within different subpopulations of the neoplastic clone, raising questions concerning which to target therapeutically. Clues may be derived either from the biologic functions of the involved genes or from clinical associations with specific subtypes of disease; but in many instances, these data may not be available. For example, are all chromosomal translocations or deletions and gene mutations of equal importance in the pathogenesis of disease, or are some secondary events, or perhaps even epiphenomena, of little or no biologic significance, reflecting the relative “genetic instability” of cancer cells? In short, which are “drivers” of disease, and which merely “passengers?”7 This problem becomes more acute with the availability of whole genome sequences. The genome sequence of a case of acute myeloid leukemia (AML) with minimal myeloid differentiation, lacking any cytogenetic abnormality, compared with that of normal skin fibroblasts derived from the same person, revealed an estimated 750 somatically acquired mutations.8,9 Most mutations occurred in nonconserved and non–protein-coding regions of the genome and therefore are presumed to have no role in disease pathogenesis. Of the 64 mutations within conserved regions, only 12 occurred in protein-coding regions, with the remaining 52 in noncoding but highly conserved DNA sequences that presumably have regulatory functions. Of the protein-coding mutations, 7 were predicted to alter protein function and therefore might be considered “drivers.” Four mutations, involving NPM1, NRAS, IDH1, and a conserved region of chromosome 10, were found to be recurrent in a screen of 180 other cases of AML. IDH1 (isocitrate dehydrogenase) has also been found to be mutated in primary brain tumors and results in production of the onco-metabolite, 2-hydroxyglutarate, which may contribute to neoplastic transformation by the production of reactive oxygen species.10
The oncogenic potential of NPM1, NRAS, and IDH1 is therefore well established from both their known biologic activities and the presence of mutations in several tumor types. However, whether any of the other apparently sporadic mutations were of pathogenetic significance remains unclear and must await more detailed functional analysis, a formidable undertaking if all tumors have similar frequencies of sporadic mutations. The picture emerging from unbiased whole genome approaches to malignancy therefore is one of increasingly genetic complexity, with a few of commonly mutated gene “mountains” and a much larger number of gene “hills” mutated at low frequency.11
In this article, we review recent results on the nature and consequences of immunoglobulin heavy chain (IGH) chromosomal translocations in BCP-ALL. Several previously unidentified IGH translocations have been reported in BCP-ALL over the past 3 years.12-17 All are relatively rare events, occurring individually in less than 1% of all cases and many are probably sporadic. This is in marked contrast to the situation in mature B-cell malignancies, where specific IGH translocations are commonly found in most subtypes of disease and underlie disease pathogenesis.18 One response to the finding of many different and perhaps sporadic IGH translocations in BCP-ALL might be to discount these events as rare curios or perhaps epiphenomena, of little biologic or clinical significance to the pathogenesis of most cases of BCP-ALL. In contrast, despite their rarity, we argue that the genes identified from their direct involvement in IGH translocations nevertheless define important new mechanisms of B-cell precursor transformation, define new prognostic subgroups of disease, and indicate new therapeutic targets. Moreover, we demonstrate that several of the genes targeted by IGH translocations in BCP-ALL are also deregulated as a consequence of other mechanisms in significant subgroups of patients.
General considerations of IGH translocations in B-cell malignancies
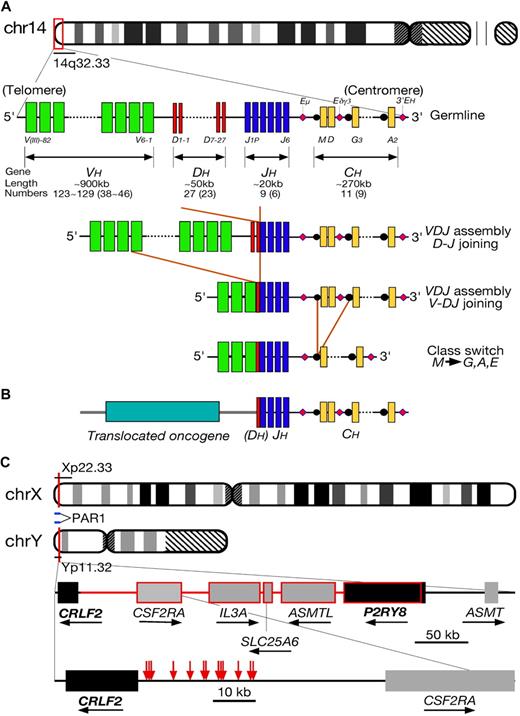
Chromosomal translocations involving the immunoglobulin loci have long been recognized to play a central role in the pathogenesis of several subtypes of B-cell malignancy.18 Burkitt lymphoma is characterized by translocations involving MYC on chromosome 8q24, follicular lymphoma by involvement of BCL2 on 18q21, and mantle cell lymphoma by involvement of CCND1 on 11q13. Each translocation is present in the great majority of cases and, thus, like the Philadelphia chromosome in CML, plays an important role in disease pathogenesis. How the specificity of each type of chromosomal translocation might arise is beyond the scope of this article but presumably represents differences in the higher-order chromatin structures in B cells at different stages of differentiation.19 IGH translocations directly involve either the joining (J) or switch (S) regions of the IGH locus, regions physiologically involved in VDJ and class-switching recombination, respectively (Figure 1A). The principal consequence of IGH translocations is deregulated transcription of the translocated gene because of the physical juxtaposition of transcriptional enhancers within the IGH locus (Figure 1B). In mature B-cell malignancies, the enhancers are composed of the Eμ enhancer lying between the IGHJ and IGHMS segments and the 2 centromeric enhancers lying distal to both IGH constant region Cα (IGHCA) gene segments. In normal B-cell precursors, another transcriptional enhancer, located between IGHCD and IGHG3, is also functional and contains many Ikaros transcription factor recognition motifs.20 This may be pathologically significant because approximately 5% of BCP-ALL demonstrate an interstitial deletion within the IGH locus on chromosome 14q32 with biallelic deletion of the IGHJ/IGHCM and IGHCD sequences extending to the IGHCD-IGHCG3 interval.21,22 Interestingly, Ikaros itself is also recurrently deleted in BCP-ALL, but the possible biologic and clinical consequences of these IGH deletions remain to be determined.23 Importantly, none of the IG translocations alone is sufficient for the emergence of the neoplastic phenotype; concurrent activation of synergistic oncogenes and loss of tumor suppressor gene functions are necessary.
Overview of IGH translocations. (A) Schematic of the IGH locus. The human IGH locus is located adjacent to the telomere of chromosome 14q. The locus is composed of dispersed variable (VH, denoted by green bars), diversity (DH, red), and joining (JH, blue) region segments that undergo somatic recombination in B-cell precursors to produce a functional VDJ unit. These are expressed with constant region gene segments (CH, yellow) with transcription being driven by enhancers (denoted by pink diamonds). (B) A diagram of IGH translocation in BCP-ALL. As a result of an error of D-J or V-DJ recombination, a partner gene from another chromosomal region becomes adjacent to J or DJ region. Expression of the translocated gene is driven by the transcriptional enhancers within the IGH locus. Usually, the entire open reading frame of the translocated gene is maintained without mutation. (C) Schema showing breakpoints of CRLF2/IGH translocation and PAR1 interstitial deletion. Red arrows indicate the positions of break points of the cloned IGH translocations (falling in the region immediately 5′ of CRLF2), whereas the portion consisting of lines and frames in red represents the deletion between P2RY8 and CRLF2 resulting in fusion of the 2 genes.
Overview of IGH translocations. (A) Schematic of the IGH locus. The human IGH locus is located adjacent to the telomere of chromosome 14q. The locus is composed of dispersed variable (VH, denoted by green bars), diversity (DH, red), and joining (JH, blue) region segments that undergo somatic recombination in B-cell precursors to produce a functional VDJ unit. These are expressed with constant region gene segments (CH, yellow) with transcription being driven by enhancers (denoted by pink diamonds). (B) A diagram of IGH translocation in BCP-ALL. As a result of an error of D-J or V-DJ recombination, a partner gene from another chromosomal region becomes adjacent to J or DJ region. Expression of the translocated gene is driven by the transcriptional enhancers within the IGH locus. Usually, the entire open reading frame of the translocated gene is maintained without mutation. (C) Schema showing breakpoints of CRLF2/IGH translocation and PAR1 interstitial deletion. Red arrows indicate the positions of break points of the cloned IGH translocations (falling in the region immediately 5′ of CRLF2), whereas the portion consisting of lines and frames in red represents the deletion between P2RY8 and CRLF2 resulting in fusion of the 2 genes.
IGH translocations in BCP-ALL
In contrast to the lymphomas, BCP-ALL lack common recurrent IG translocations. Why this should be is unclear as normal B-cell precursors express high levels of recombinase activating genes (RAGs) necessary to induce double-strand DNA breaks associated with VDJ recombination, whereas levels are much lower in peripheral B cells; concurrent expression of RAG and activation-induced deaminase (not normally expressed in B-cell precursors) may be necessary for optimal generation of IGH translocations.24 However, it is now apparent that there is a plethora of rare IGH translocations in this disease. Their identification has been facilitated by the use of IGH“breakapart” probes located centromeric of the IGHC and within the (telomeric parts of the) IGHV regions on interphase fluorescence in situ hybridization (FISH); these probes should normally be physically adjacent, whereas the IGHV signal is moved from the der(14) to the derivative partner chromosome by an IGH translocation, which results in a “split” signal in interphase cells by most IGH translocations. A list of all cloned IGH translocations in BCP-ALL is shown in Table 1. These cases have been garnered from cytogenetic samples collected in many different laboratories worldwide, over many years, from patients who have been treated with different chemotherapeutic protocols. Thus, the precise frequency and possible prognostic significance of each individual IGH translocation in BCP-ALL remain to be determined from uniformly treated series of patients. Overall, IGH translocations are seen in approximately 2% to 3% of all cases of BCP-ALL.14,35,36 The CRLF2/IGH translocation is the most frequent, followed by inhibitor of DNA binding 4 (ID4) and the collective CCAAT enhancer-binding protein (CEBP) translocations; the remaining IGH translocations appear to be sporadic. For reasons that are unclear, most cases of IGH translocation present in adolescents or young adults, outside the peak incidence of childhood BCP-ALL. Whether this reflects a postnatal, rather than a fetal, origin of these malignancies is not known. The spectrum of partner genes is specific to BCP-ALL; with the exception of translocations involving hTERT and BCL2, these translocations are not seen in malignancies of mature B cells. Importantly, although there are some similarities, there is no obvious common motif to unify all the various translocations functionally.
Summary of IGH chromosomal translocations in BCP-ALL
| Target gene . | Location . | Functions in normal B cells . | Presumed mechanism of transformation . | Deregulated expression in BCP-ALL without IGH translocation? . | Comments . | References . |
|---|---|---|---|---|---|---|
| CRLF2 | Xp22Yp11 | Heterodimerization with IL7RA to mediate TSLP signaling | Homodimerization mediating constitutive STAT5 phosphorylation | Also deregulated by interstitial deletion on X/Y chromosome. Juxtamembrane CRLF2 mutations also seen | Associated with DS, JAK2 mutation, and poor prognosis | 17, 25, 26 |
| EPOR | 19p133 cases | Not expressed in normal B-cell precursors | Constitutive STAT5 phosphorylation | EPOR also a documented target gene of ETV6-RUNX1 fusion | EPOR expression also seen in many other malignancies | 16 |
| ID4 | 6p2114 cases | Not expressed in normal B-cell precursors | Unknown | High-level expression also seen BCP-ALL without IGH translocation | 12, 15 | |
| CEBPD | 8q1144 cases | Not expressed in normal B-cell precursors | Unknown | Association with DS and t(9;22) | 14, 27, 28 | |
| CEBPA | 19q13 | Not expressed in normal B-cell precursors | Unknown | Mutated in AML subject to acquired uniparental disomy in AML | 13, 14 | |
| CEBPE | 14q11 | Not expressed in normal B-cell precursors | Unknown | Predisposition to BCP-ALL within CEBPE | 14 | |
| CEBPB | 20q133 cases | Unknown | 14 | |||
| CEBPG | 19q13Single case | Unknown | 14 | |||
| IL3 | 5q31 | Cytokine | — | Associated with hypereosinophilia | 29 | |
| IGF2BP1 | 17q21.33 cases | RNA binding protein | Enhanced translation of oncogenic proteins | 32 | ||
| MIR125B | 11q24.12 cases | Intergenic microRNA | Down-regulation of p53 | Deregulated by interstitial insertion into IGH locus | 30; AUPO (2008) | |
| BCL9 | 1q212 cases/1 cell line | Involved in WNT signaling; binds to β-catenin | Deregulated WNT signaling | CEMO-1 cell line harbors this translocation | 31; AUPO (2008) | |
| hTERT | 5p15.3Single case | Maintenance of telomeres | Identical translocations also seen in mature B-cell malignancies | Nagel et al (2009), MS under revision | ||
| LHX4 | 1q21Single case | LIM homeobox domain protein: ? ectopic expression in BCP-ALL | Unknown | Normally involved in control of pituitary development | 33 | |
| DUX4 | 10q26Single case (cell line) | Unknown | Unknown | Cell line only; nature of deregulated transcript uncertain | AUPO (2008) | |
| BCL2 | 18q21 Single case (cell line) | Suppression of apoptosis | Cell line only; derived from 15-year-old with BCP-ALL with no obvious antecedent lymphoma | 34 |
| Target gene . | Location . | Functions in normal B cells . | Presumed mechanism of transformation . | Deregulated expression in BCP-ALL without IGH translocation? . | Comments . | References . |
|---|---|---|---|---|---|---|
| CRLF2 | Xp22Yp11 | Heterodimerization with IL7RA to mediate TSLP signaling | Homodimerization mediating constitutive STAT5 phosphorylation | Also deregulated by interstitial deletion on X/Y chromosome. Juxtamembrane CRLF2 mutations also seen | Associated with DS, JAK2 mutation, and poor prognosis | 17, 25, 26 |
| EPOR | 19p133 cases | Not expressed in normal B-cell precursors | Constitutive STAT5 phosphorylation | EPOR also a documented target gene of ETV6-RUNX1 fusion | EPOR expression also seen in many other malignancies | 16 |
| ID4 | 6p2114 cases | Not expressed in normal B-cell precursors | Unknown | High-level expression also seen BCP-ALL without IGH translocation | 12, 15 | |
| CEBPD | 8q1144 cases | Not expressed in normal B-cell precursors | Unknown | Association with DS and t(9;22) | 14, 27, 28 | |
| CEBPA | 19q13 | Not expressed in normal B-cell precursors | Unknown | Mutated in AML subject to acquired uniparental disomy in AML | 13, 14 | |
| CEBPE | 14q11 | Not expressed in normal B-cell precursors | Unknown | Predisposition to BCP-ALL within CEBPE | 14 | |
| CEBPB | 20q133 cases | Unknown | 14 | |||
| CEBPG | 19q13Single case | Unknown | 14 | |||
| IL3 | 5q31 | Cytokine | — | Associated with hypereosinophilia | 29 | |
| IGF2BP1 | 17q21.33 cases | RNA binding protein | Enhanced translation of oncogenic proteins | 32 | ||
| MIR125B | 11q24.12 cases | Intergenic microRNA | Down-regulation of p53 | Deregulated by interstitial insertion into IGH locus | 30; AUPO (2008) | |
| BCL9 | 1q212 cases/1 cell line | Involved in WNT signaling; binds to β-catenin | Deregulated WNT signaling | CEMO-1 cell line harbors this translocation | 31; AUPO (2008) | |
| hTERT | 5p15.3Single case | Maintenance of telomeres | Identical translocations also seen in mature B-cell malignancies | Nagel et al (2009), MS under revision | ||
| LHX4 | 1q21Single case | LIM homeobox domain protein: ? ectopic expression in BCP-ALL | Unknown | Normally involved in control of pituitary development | 33 | |
| DUX4 | 10q26Single case (cell line) | Unknown | Unknown | Cell line only; nature of deregulated transcript uncertain | AUPO (2008) | |
| BCL2 | 18q21 Single case (cell line) | Suppression of apoptosis | Cell line only; derived from 15-year-old with BCP-ALL with no obvious antecedent lymphoma | 34 |
Some IGH translocations, including those involving CRLF2, hTERT, and DUX4 involve genes immediately adjacent to the telomeres and cannot therefore be detected by conventional cytogenetics alone.
AUPO indicates authors' unpublished observations.
Involvement of the cytokine receptor CRLF2 in both IGH translocations and interstitial deletion of the PAR1
The cytokines that drive proliferation of B-cell precursor subpopulations appear to be at least partially species-specific and may vary at different stages of embryonic and adult development.37 In adult mice, B-cell lymphopoiesis is strictly interleukin-7 (IL-7) dependent. However, the precise role of this cytokine in human B-cell lymphopoiesis is more controversial.38 Another cytokine, which may have similar species-specific roles, is thymic stromal lymphopoietin (TSLP).39 TSLP was originally identified as a cytokine produced from a derived murine thymic stromal cell line able to support growth of a pre-B-cell line and the development of IgM+ B cells from normal mouse fetal liver cells.40 Murine TSLP shows little sequence homology (43%) to its human counterpart (Figure 2A). Similarly, the specific receptor for TSLP (cytokine receptor-like factor 2 [CRLF2]) differs significantly with only 39% amino acid sequence homology (Figure 2B). CRLF2 interacts with IL-7RA to mediate high-affinity TSLP binding and signaling.39 As might be anticipated from the resulting lack of both IL-7 and TSLP signaling, the murine IL-7RA mutant mouse has a more severe immunodeficient phenotype than the IL-7 mutant. However, surprisingly, given the in vitro data, inactivation of murine CRLF2 resulted in no obvious B-cell phenotype in adult mice, although in one model TSLP signaling was necessary for full lymphoid recovery after sublethal irradiation.41,42 Human, but not murine, TSLP induces dendritic cell activation and proliferation of some derived myeloid cell lines but has no obvious proliferative effects on lymphocytes; thus far, its possible effects on T- and B-cell lymphopoiesis remain unclear. One study has shown that TSLP may induce proliferation of BCP-ALL cell lines.43 There are no reported mutations of either TSLP or CRLF2 in humans or mice associated with immunodeficiency. Therefore, despite its discovery as a cytokine signaling pathway able to support B-cell growth and differentiation in vitro, there has been little evidence for either TSLP or CRLF2 playing a pivotal role in human or murine B-cell development.
Sequence alignments of TSLP, CRLF2, and EPOR. (A) Lack of homology of mouse and human CRLF2 amino acid sequences. Extracellular domain (green), transmembrane region (pink), and intracellular domain (blue). Blue box represents box 1 necessary for JAK activation; red box, cytokine receptor motif WSXWS sequence. (*) denotes identity; (:) indicates strong homology; and (.) indicates weak homology. (B) Lack of homology of mouse and human TSLP amino acid sequences. (C) Comparison of human CRLF2 and EPOR to show limited homology within box 1 of both proteins.
Sequence alignments of TSLP, CRLF2, and EPOR. (A) Lack of homology of mouse and human CRLF2 amino acid sequences. Extracellular domain (green), transmembrane region (pink), and intracellular domain (blue). Blue box represents box 1 necessary for JAK activation; red box, cytokine receptor motif WSXWS sequence. (*) denotes identity; (:) indicates strong homology; and (.) indicates weak homology. (B) Lack of homology of mouse and human TSLP amino acid sequences. (C) Comparison of human CRLF2 and EPOR to show limited homology within box 1 of both proteins.
However, we and others have shown direct involvement of CRLF2 in BCP-ALL as a consequence, not only of IGH chromosomal translocation but also more commonly as a result of an interstitial genomic deletion.17,25,26 CRLF2 maps to the pseudoautosomal regions (pseudoautosomal region 1 [PAR1]) of the short arms of both sex chromosomes; the gene maps just more than 1 Mb from the telomere. This, and the fact that the IGH locus is also at the telomere of chromosome 14q, renders the translocations cytogenetically cryptic; FISH studies are essential for detection. Both sex chromosomes are involved in translocation with IGH. The precise frequency of the IGH translocation in adolescent and adult patients remains unknown. However, in an unselected series of pediatric cases, the frequency of the translocation was 0.8%. The consequences of CRLF2/IGH translocation are deregulated expression of CRLF2 at the cell surface.17,26
Fascinatingly, deregulated expression of CRLF2 may also be seen after a small interstitial deletion within PAR1. Again, this event is cytogenetically cryptic and has to be detected by FISH, quantitative PCR approaches, or array-based imbalance mapping (ie, array comparative genomic hybridization or single nucleotide polymorphism arrays). Interstitial deletion of PAR1 was more common than the CRLF2/IGH translocation (occurring overall in 4.2% of pediatric cases in our series and in 7% of cases in the St Jude's series) and occurred in younger patients than with the CRLF2/IGH translocation. In all series reported to date, there was a strong association with Down syndrome (DS), with approximately 50% to 60% of DS BCP-ALL exhibiting the deletion. Molecular mapping revealed a recurrent rearrangement involving CRLF2 with a centromeric gene P2RY8. RT-PCR experiments showed expression of a chimeric fusion mRNA composing the first noncoding exon of P2RY8 with the entire coding region of CRLF2 (Figure 1C).25,26 DNA sequence analysis of the genomic breakpoints showed high conservation of the deletion breakpoints with flanking recombination signal sequences, indicating that the deletions arise because of aberrant RAG activity, as has been observed with other recurrent deletions in BCP-ALL.23 As with the CRLF2/IGH translocation, the principal consequence of the deletion was expression of CRLF2 at the cell surface. One copy of the IL-3 receptor gene (IL3RA) is also lost as a consequence; whether this contributes to the pathogenesis of disease is not known.
In experimental models, deregulated expression of CRLF2 alone does not appear to result in B-cell progenitor transformation. Expression of human CRLF2 in the murine IL-3–dependent pro-B-cell line Ba/F3 did not result in cytokine-independent cell growth, and retroviral transfection of normal mouse fetal liver cells resulted in short-term proliferation.17,25 Similarly, knockdown of CRLF2 mRNA in a derived human BCP-ALL cell line harboring the CRLF2/IGH translocation resulted in slowed proliferation but not cell death, emphasizing the role for secondary, cooperating oncogenetic events.17 In 3 cases with PAR1 deletion in DS BCP-ALL, an activating mutation of CRLF2 (F232C) occurring within the juxtamembrane region of CRLF2 has been observed.26 However, the most common associated mutation reported to date is JAK2, with approximately one-third of DS BCP-ALL cases with PAR1 deletion exhibiting concurrent JAK2 mutations. Like CRLF2, JAK2 mutations do not transform Ba/F3 cells. In contrast, cotransfection of both CRLF2 and mutated JAK2 into either Ba/F3 cells or Ba/F3 cells transduced with human IL-7RA resulted in marked synergy with cytokine-independent cell growth.25,26 Whether JAK2 is the physiologic and only CRLF2 kinase remains controversial, and it is possible that mutations or alterations in other kinases might synergize with CRLF2.44 The necessity of IL-7RA for the proliferative effect of deregulated CRLF2 expression is also unclear; it is possible that both IGH translocation and PAR1 deletion result in CRLF2 homodimerization, leading directly to JAK2 and STAT5 phosphorylation and thus proliferation.
JAK2 mutations are associated with a poor prognosis in BCP-ALL.45 However, preliminary data also indicate that CRLF2 expression is also associated with poor outcome, defining a new poor prognostic subgroup in otherwise good-risk prognosis BCP-ALL26,46 (G.C., M. Zimmerman, R. Romey, S. Gesk, J. Harbott, A. Schrauder, A. Moericke, S. Igraeli, T.A., M.J.S.D., R.S., M. Schrappe, and M. Stanulla, manuscript under review). Given that CRLF2 expression can be readily detected by flow cytometry on BCP-ALL blasts, we would recommend the addition of CRLF2 antibodies to the routine diagnostic immunophenotypic panels. The restricted expression of CRLF2 in normal human tissues and the association with poor prognostic disease therefore make this an excellent target for immunotherapy, comparable with HER2 amplification in breast cancer. Whether inhibition of CRLF2 signaling might have a role in suppressing normal lymphopoiesis, and therefore have a possible role in maintenance therapy for BCP-ALL, remains to be determined.
EPOR/IGH translocations and EPOR expression in BCP-ALL
Like CRLF2, the erythropoietin receptor (EPOR) is recurrently involved in BCP-ALL by 2 distinct mechanisms. First, rare cases may exhibit chromosomal translocation t(14;19)(q32;p13), sometimes as the only detectable cytogenetic abnormality.16 The translocation results in high-level EPOR expression. However, EPOR is also a transcriptional target gene for the ETV6-RUNX1 fusion protein resulting from t(12;21)(p13;q22) that occurs in approximately 25% of pediatric BCP-ALL.47 From analysis of the available electronic RNA expression data, EPOR mRNA does not appear to be expressed at appreciable levels in normal human B-cell precursors, indicating that EPOR expression in BCP-ALL is ectopic.48 Sequence analysis of EPOR and CRLF2 showed that both have the box 1 motif required for JAK interaction (Figure 2C), and it is likely that deregulated EPOR, like CRLF2, contributes to B-cell precursor transformation by constitutive STAT5 phosphorylation. As ETV6-RUNX1 induces resistance to transforming growth factor-β–mediated growth suppression rather than proliferation, it is possible that EPOR expression confers a survival advantage in this context.49 Ectopic EPOR expression is also seen in other malignancies, including breast carcinoma.50
Other components of the cytokine signaling pathways do not appear to be commonly implicated in the pathogenesis of lymphoid malignancies. The cytokine IL-3 is occasionally involved in IGH translocation, results in deregulated IL-3 expression, and is seen specifically in BCP-ALL with associated eosinophilia.51 This has now been recognized as a specific disease entity by the World Health Organization.29
ID4/IGH translocations in BCP-ALL
Originally reported in a single case, we have subsequently shown chromosomal translocation t(6;14)(p22;q32) to be a recurrent event in a series of 13 cases; all involve the ID4 gene on chromosome 6p22.3.12,15 The precise overall frequency and the possible prognostic and clinical significance of the ID4 translocation remain unknown, but from our initial series of cases, it may be associated with relatively good prognosis disease. All patients presented with low peripheral blood blast counts (median, 3 × 109/L; range, 1-11 × 109/L) and given the older age of most patients, most appeared to respond well to chemotherapy. However, this needs to be confirmed from prospective studies of uniformly treated cohorts of patients. Unlike both the CRLF2 and CEBP translocation cases, most ID4/IGH cases had a complex karyotype with several other structural and/or numerical chromosomal changes. A common karyotypic/FISH feature was deletion of the CDKN2A locus on chromosome 9p21 in all cases, suggesting possible synergy with ID4 dysregulation.
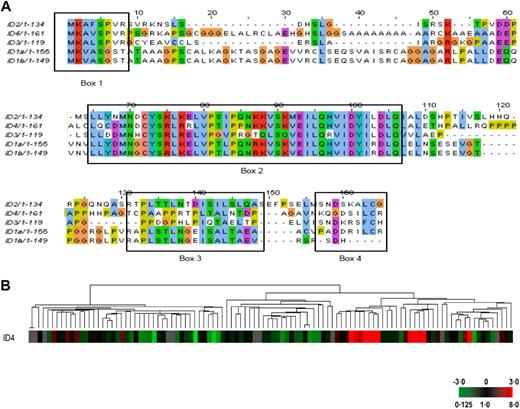
In contrast to the CEBP/IGH translocations, only ID4 of the ID gene family is involved in IGH translocation in BCP-ALL, suggesting that ID4 has specific transforming properties. How ID4 contributes to neoplastic transformation of normal B-cell precursors remains unknown. ID4 was first identified through its sequence homology to 3 related genes (ID1-ID3).52 All 4 family members share a highly homologous and conserved helix-loop-helix motif found in many developmentally regulated transcription factors but lack the basic DNA binding domain found in basic-helix-loop-helix (bHLH) family of transcription factors (Figure 3A). ID proteins cannot act directly as transcription factors but bind to bHLHs and affect their ability to bind DNA and thus inhibit their functions. In hemopoietic cells, ID proteins bind to E-box protein family members, including E2A, but also other transcription factors, including PAX5, pRb, and ETS.53,54 ID proteins therefore act as dominant negative inhibitors of transcription factor activity. However, several other roles, for example, in cell-cycle control independent of transcription factor binding and nuclear localization, have also been described.55 Of all 4 ID family members, ID4 has been least studied. In terms of oncogenesis, ID4 can behave either as an oncogene or as a tumor suppressor gene depending on cellular context. Overexpression of ID4 in mouse p16Ink4a/Arf−/− astrocytes, and glioblastoma cells drives both cyclin E expression to induce proliferation and Notch activation to induce a neural “stem cell–like” state.56 ID4 is not expressed in human hemopoietic cells.48,57 In addition, B-cell precursors from mouse ID4 knockout mice do not show any obvious proliferative defects but differentiate more rapidly than wild-type counterparts (M.C., M.J.S.D., F. Sablitsky, unpublished observations, 2008). However, in contrast to neural cell malignancies, ID4 appears to behave as a tumor suppressor gene in mature lymphoid cells, with heavy methylation of the ID4 promoter seen in all cases of chronic lymphocytic leukemia.58 Loss of ID4 accelerates disease progression in one mouse model of chronic lymphocytic leukemia.59 Overexpression of ID4 in transformed lymphoid cell lines results in rapid G1 cell-cycle arrest58 (P.D., T.A., Y.F.L., E.L.K., and M.J.S.D., unpublished observations, September 2009).
Comparison of ID4 amino acid sequences and RNA expression in BCP-ALL. (A) Protein sequence comparison of the human ID proteins. The region of highest homology between ID family proteins is within the HLH region (box 2), although there are other small areas of homology (boxes 1, 3, and 4). An amino-terminal CDK2-dependent phosphorylation site has been identified (SPVR) in all IDs except ID1 (box 1).86 All members except ID3 contain a D-box region (RXXLXXXN in box 3) that targets the protein for proteosomal degradation.48 Phosphorylation status may have implications for nuclear localization and cell-cycle effect of ID proteins.86,87 (B) Dendrogram from unsupervised clustering of gene expression data obtained from 106 cases of pediatric BCP-ALL lacking ETV6-RUNX1, BCR-ABL, or MLL-AF4 chromosomal translocations showing clustering of cases with high-level ID4 expression. (G.C., T.A., R.S., and M.J.S.D., unpublished observations, August 2008). Hierarchical clustering analysis was carried out considering only those clones that had a 2-fold difference in expression from the mean on at least 2 arrays (4421 clones). Each column represents one sample. The expression of ID4 is displayed as a variation in color as indicated in the lower right corner. Gray represents data that were omitted because they were not well measured.
Comparison of ID4 amino acid sequences and RNA expression in BCP-ALL. (A) Protein sequence comparison of the human ID proteins. The region of highest homology between ID family proteins is within the HLH region (box 2), although there are other small areas of homology (boxes 1, 3, and 4). An amino-terminal CDK2-dependent phosphorylation site has been identified (SPVR) in all IDs except ID1 (box 1).86 All members except ID3 contain a D-box region (RXXLXXXN in box 3) that targets the protein for proteosomal degradation.48 Phosphorylation status may have implications for nuclear localization and cell-cycle effect of ID proteins.86,87 (B) Dendrogram from unsupervised clustering of gene expression data obtained from 106 cases of pediatric BCP-ALL lacking ETV6-RUNX1, BCR-ABL, or MLL-AF4 chromosomal translocations showing clustering of cases with high-level ID4 expression. (G.C., T.A., R.S., and M.J.S.D., unpublished observations, August 2008). Hierarchical clustering analysis was carried out considering only those clones that had a 2-fold difference in expression from the mean on at least 2 arrays (4421 clones). Each column represents one sample. The expression of ID4 is displayed as a variation in color as indicated in the lower right corner. Gray represents data that were omitted because they were not well measured.
Deregulated ID4 expression in BCP-ALL may also arise from mechanisms other than chromosomal translocation; 11.5% of 297 unselected cases of pediatric BCP-ALL showed a high ID4 expression as defined by an expression of at least 100-fold higher than the median expression of all samples using quantitative RT-PCR. Moreover, analyzing genome-wide unselected gene expression data using unsupervised clustering algorithm, those samples with a high ID4 expression clustered together, suggesting distinct biologic subgroup(s) of disease (G.C., T.A., R.S., and M.J.S.D., unpublished observations, August 2008; Figure 3B). What mechanism(s) underlies the expression of ID4 in these cases is not known. It is also unknown in cases lacking the ID4/IGH translocation whether ID4 is a “driver” or a “passenger.” If ID4 expression genuinely acts a “driver,” this might make ID4 a therapeutic target in BCP-ALL, as there is only very low-level or absent expression of ID4 in most normal adult tissues.60,61
CEBP family/IGH translocations in BCP-ALL
CEBPs compose a family of 5 multifunctional basic leucine zipper (bZIP) transcription factors. The protein family is defined by a conserved carboxy-terminal bZIP domain, composing a leucine zipper dimerization and basic DNA-binding domains. All but CEBPG exhibit an amino-terminal transcriptional transactivation domain; CEBPG therefore, like ID4, is thought to act as a natural dominant negative inhibitor of CEBP functions. All 5 CEBP genes are involved in IGH translocation at various frequencies, implicating the conserved bZIP domain in B-cell precursor transformation.14 The IGH translocation involving CEBPD is the most common and has been best characterized clinically.27 Data from 44 cases have recently been collated.28 The median age at presentation was 13 years, and the median white blood cell count at presentation was low (9 × 109/L). The t(8;14)(q11;q32) was the sole acquired cytogenetic abnormality in 13 of 44 cases, occurring in conjunction with t(9;22)(q34;q11) in 7 cases. The translocation occurred in 12 patients with DS, none of whom had a t(9;22)(q34;11). The linkage of the CEBPD/IGH translocation with DS appears to be specific among the CEBP/IGH translocations; to our knowledge, only one other CEBP translocation (involving CEBPE) has been reported in DS BCP-ALL.14 Why CEBPD/IGH translocations and the PAR1 interstitial deletion should preferentially occur in persons with DS is not yet clear. CEBPA translocations are the next most frequent, whereas CEBPE, CEBPB, and CEBPG translocations are probably sporadic. Given the variable age of the patients and the different chemotherapy protocols used, the prognostic significance of the CEBP/IGH translocations is not known. However, overall, the rate of early relapse appears to be low; whether this translates into a good survival is not yet clear.
Although rare, both individually and collectively and therefore potentially not of great clinical significance, the CEBP/IGH translocations are of great interest biologically. First, it is remarkable from a genetic perspective that all 5 CEBP family members are involved in IGH chromosomal translocations in the one disease. The closest parallel is the involvement of the cyclin D gene family in various mature B-cell malignancies.18,62,63 Second, it is all the more remarkable given that the CEBP genes suppress cell growth and induce differentiation under many conditions both in vitro and in vivo. For example, transduction of CEBPD in BCR-ABL+ CML cell lines resulted in prompt cell-cycle arrest, apoptosis, and differentiation into neutrophils.64 This result is of some interest because, in BCP-ALL, the t(8;14)(q11;q32) involving CEBPD is often seen specifically in conjunction with the t(9;22)(q34;q11).28 However, the apparent discrepancies are most marked in the case of CEBPA. CEBPA is normally expressed exclusively in cells of the myeloid and monocytic lineages with no detectable expression in normal B-cell precursors.65 Loss of CEBPA expression results in enhanced cell proliferation and self-renewal of normal hemopoietic stem cells.66 Expression of CEBPA in committed lymphoid cells suppresses the lymphoid differentiation program and, moreover, may drive the reprogramming of mature sIg+ B cells to macrophages, albeit only at low frequency and under strong selective pressures.67 CEBPA expression in uncommitted hemopoietic stem cells usually leads to up-regulation of PU.1 and down-regulation of PAX5, with consequent suppression of B-cell differentiation and commitment to the myeloid lineage.68 However, BCP-ALL expressing any of the CEBP gene family appear to have typical BCP-ALL immunophenotypes with no overt evidence for commitment to other lineages. Third, in myeloid cells, CEBPA is unequivocally a tumor suppressor gene, with loss of CEBPA function observed in AML by several different mechanisms.69 Approximately 7% of all cases of AML exhibit either monoallelic or biallelic acquired mutations/deletions of CEBPA, and the region is also subject to acquired uniparental disomy. Some familial cases of AML exhibit germline CEBPA mutations.70 Furthermore, both transcription and translation of CEBPA may be suppressed directly or indirectly by various AML-associated chromosomal translocations.71 Consequently, the deregulated expression of all 5 CEBP genes in BCP-ALL is unexpected and constitutes something of “a spanner in the works!”65
How deregulated CEBP expression contributes to the neoplastic transformation B-cell precursors remains unknown. The involvement of all family members suggests that transformation is mediated via the common dimerization and DNA binding regions with concurrent loss of the growth-suppressive activities of the amino-terminal transactivational domains. There are at least 3 possible mechanisms, all of which require the presence of prior genetic alterations before the CEBP/IGH translocation occurs. First, CEBP/IGH translocation may occur in a committed B-cell precursor where the myeloid differentiation pathway has already been “blocked.” There is some evidence for this in a case of BCP-ALL with t(14;14)(q11;q32) involving CEBPE where the same IGH DJ rearrangement was seen in both a VDJ rearrangement and at the chromosomal translocation breakpoint sequence, suggesting that the translocation occurred in a clone that had already undergone transformation.14 Second, there may be selective translation of short CEBP protein isoforms or alternatively posttranslational modifications that allow proliferation.65,72 Alternatively, despite the lack of B-cell phenotype in most CEBP mutant mice, the CEBP genes may have unsuspected functions in normal human B-cell precursors; recent genome-wide association studies have shown that single nucleotide polymorphisms within CEBPE may predispose to BCP-ALL.73
The involvement of the CEBP gene family and ID4 in BCP-ALL was unanticipated from previous biologic studies; if ID4 and CEBP deregulation contribute to the neoplastic transformation of normal B-cell precursors, it seems probable they must all act in a strictly cell context–dependent fashion.
MIR125B1/IGH translocation and insertion
MicroRNA genes play pivotal roles in normal and malignant hemopoiesis. One gene, MIR125B1 on chromosome 11q23.3, has been implicated in the pathogenesis of various forms of myelodysplasia evolving to AML exhibiting t(2;11)(p21;q24) and more rarely in BCP-ALL with t(11;14)(q24;q32)30,74 (T.A., R.S., and M.J.S.D., unpublished observations, August 2008); both translocations result in deregulated expression of MIR125B1. In human myeloid cells, overexpression of this microRNA resulted in blocked myeloid differentiation74 ; the effects of deregulated expression in B-cell precursors are so far unknown. Recent studies, however, have shown that MIR125B1 binds to the 3′ untranslated region of p53 mRNA and suppresses p53 protein levels, at least in neuroblastoma and human lung fibroblasts.75 Whether there are other more cell context–dependent effects of MIR125B1 remains to be explored. MIR125B1 is also highly expressed in germinal center B cells where it down-regulates both IRF4 and PRDM1.76
MicroRNAs are often found as multiple copies in the human genome. Given the direct involvement of MIR125B1 in both myeloid and lymphoid malignancies, it is therefore of interest that a homolog of this gene, MIR125B2, is located on chromosome 21q21 (http://www.mirbase.org/); whether the extra copy of this microRNA in persons with DS contributes to their increased susceptibility to both BCP-ALL and acute megakaryoblastic leukemias is currently unknown.77 High levels of MIRNA125B2 have been reported in DS AML.78 In BCP-ALL, however, expression appears to be restricted to ETV6-RUNX1 leukemias.79
Conclusions
In conclusion, BCP-ALL exhibit a limited number of common recurrent chromosomal translocations, including EVT6-RUNX1, BCR-ABL, and MLL-AF4. Each of these cytogenetically defined subgroups is associated with a specific gene expression signature and a specific response to chemotherapy.80,81 BCP-ALL, however, also exhibit a plethora of much rarer IGH translocations. Is it important clinically to identify and dissect out these rare genetic events or can they be regarded as curios? Detection of IGH translocations in clinical material is relatively simple using the IGH“breakapart” probe set in interphase FISH experiments, but determining precisely which partner gene is involved may be difficult because of the large number of involved genes.
The expression of CRLF2 arising as a consequence of either IGH translocation or interstitial deletion probably becomes mandatory in clinical samples because of its association with a poor prognosis in otherwise “good-risk” patients. Expression of CRLF2 can be readily detected in clinical material using flow cytometry with commercially available antibodies. However, all IGH translocations, even those that are apparently sporadic, merit further scientific and clinical study. First, unlike somatic mutations, which may be either “drivers” or “passengers,” IGH translocations invariably act as direct pointers to genes not previously known to be involved in oncogenesis. Molecular cloning of IGH translocations has allowed rapid, unambiguous identification of genes that play pivotal roles in disease pathogenesis,18 and despite their relative scarcity and diversity in BCP-ALL, this disease appears to be no exception. Second, deregulated target gene expression may be seen more commonly in BCP-ALL resulting from other mechanisms. Third, and perhaps more importantly in terms of clinical application, target genes defined by IGH translocations may also lead to new therapeutic approaches. For example, expression of CRLF2 in BCP-ALL may allow the development of therapeutic antibodies. BCL9 was originally identified from its involvement in t(1;14)(q21;q32) in BCP-ALL, and its expression indicates constitutive WNT signaling in a cell type–specific fashion.31,82 The interaction of BCL9 with β-catenin is potentially targetable therapeutically, not only for BCP-ALL with the BCL9 chromosomal translocation but in all forms of WNT-dependent malignancy.83 Similar remarks also apply to IGF2BP1 and MIR125B1 that have pathogenetic roles not only in BCP-ALL but also in several other types of malignancy.84,85
Collectively, these data indicate that IGH translocations in BCP-ALL may lead to important new mechanistic insights into cancer pathogenesis and may have possible diagnostic and therapeutic significance. The diversity of the targeted genes, including cytokine receptors, transcription factors, and signaling adapter molecules and miRNAs, reflects the heterogeneity of pathogenic mechanisms that underlie these diseases and malignancy as a whole, with no single obvious overarching mechanism to encompass all cases. Thus, both molecular cytogenetic and whole genome approaches in BCP-ALL indicate some common molecular abnormalities, but also a bewildering complexity of potentially “driving” somatic mutations and translocations. This perhaps should not be surprising given that B-cell precursors at different stages of differentiation have very different gene expression profiles.48 Practically, this diversity will have profound implications for the development and rational use of targeted therapies. Development of novel targeted therapeutics may not be possible if the population of affected persons is too small for this to be commercially viable. Even if effective targeted therapeutics are developed, the genetic composition of tumors will require comprehensive assessment from routine clinical material to maximize efficacy and minimize toxicities with such therapeutics.
Acknowledgments
This work was supported by the United Kingdom Medical Research Council, MRC Technology, the Hope Foundation for Cancer Research, Leicester, United Kingdom (Jemima Sellicks Fellowship; P.D.), and the Deutsche Krebshilfe and the KinderKrebsInitiative Buchholz/Holm Seppensen.
Authorship
Contribution: All authors contributed material (some previously unpublished) and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martin J. S. Dyer, MRC Toxicology Unit, Leicester University, Hodgkin Building Room 402, Lancaster Rd, Leicester, United Kingdom LE1 9HN; e-mail: mjsd1@le.ac.uk.