Abstract
The adapter protein Slp65 is a key component of the precursor-B (pre-B) cell receptor. Slp65-deficient mice spontaneously develop pre-B cell leukemia, but the mechanism by which Slp65−/− pre-B cells become malignant is unknown. Loss of Btk, a Tec-family kinase that cooperates with Slp65 as a tumor suppressor, synergizes with deregulation of the c-Myc oncogene during lymphoma formation. Here, we report that the presence of the immunoglobulin heavy chain transgene VH81X prevented tumor development in Btk−/−Slp65−/− mice. This finding paralleled the reported effect of a human immunoglobulin heavy chain transgene on lymphoma development in Eμ-myc mice, expressing transgenic c-Myc. Because activation of c-Myc strongly selects for spontaneous inactivation of the p19Arf-Mdm2-p53 tumor suppressor pathway, we investigated whether disruption of this pathway is a common alteration in Slp65−/− pre-B cell tumors. We found that combined loss of Slp65 and p53 in mice transformed pre-B cells very efficiently. Aberrations in p19Arf, Mdm2, or p53 expression were found in all Slp65−/− (n = 17) and Btk−/−Slp65−/− (n = 32) pre-B cell leukemias analyzed. In addition, 9 of 10 p53−/−Slp65−/− pre-B cell leukemias manifested significant Mdm2 protein expression. These data indicate that malignant transformation of Slp65−/− pre-B cells involves disruption of the p19Arf-Mdm2-p53 tumor suppressor pathway.
Introduction
B cells are produced in the bone marrow (BM) through a complex process of cellular differentiation, characterized by the ordered rearrangement of immunoglobulin (Ig) heavy (H) and light (L) chain gene segments encoding the B-cell antigen receptor (BCR) (reviewed in Meffre et al1 and Herzog et al2 ). In pro-B cells productive V(D)J recombination results in cell-surface deposition of the pre-BCR, comprising Ig μ H chain and the surrogate light chain (SLC) components λ5 and VpreB.2-4 The pre-BCR serves as an important checkpoint to monitor proper expression of a functional immunoglobulin heavy chain (IgH) chain and triggers clonal expansion, whereby pre-B cells acquire the capacity to respond to low concentrations of the proliferation factor interleukin-7 (IL-7).2-4 After a limited number of cell divisions, large pre-B cells stop cycling and differentiate into small, resting pre-B cells in which IgL chain rearrangement is initiated.
The adaptor protein Slp65 (also known as Blnk or Bash) is a key component in the signaling pathway downstream of the pre-BCR and the BCR. Slp65, when phosphorylated by Syk, provides docking sites for various molecules, including Bruton tyrosine kinase (Btk) and phospholipase C-γ (PLC-γ). Btk then phosphorylates PLC-γ, which leads to its full activation and the generation of second messengers.3,5
In humans, mutations in SLP65 or BTK result in defective pre-B cell proliferation and an almost complete arrest of B-cell development at the pro-B to pre-B cell transition, associated with the immunodeficiency disorder agammaglobulinemia.3 In contrast, mice deficient for Slp65 or Btk show only a partial arrest at the large cycling pre-B cell stage, whereas a nearly complete block is present in Btk/Slp65 double-deficient mice.6-8 Importantly, at the age of approximately 6 months 5% to 10% of Slp65-deficient mice develop pre-B cell leukemia, expressing high levels of pre-BCR on the cell surface.6,9 Although Btk-deficient mice do not develop pre-B cell tumors, we found that Slp65 and Btk cooperate as tumor suppressors, whereby Btk exerts its tumor suppressor function independently of its kinase activity.8,10 It has been reported that in a substantial fraction of human pre-B cell acute lymphoblastic leukemia (ALL), including cases expressing the oncogenic BCR-ABL1 tyrosine kinase fusion protein, SLP65 expression is defective as the result of aberrant splicing.11,12 Because subsequent analyses indicated that SLP65 deficiency may be an infrequent event in human pre-B lineage ALL,13,14 the importance of loss of SLP65 expression as one of the primary causes of pre-B ALL in human remains unclear.
The mechanism by which Slp65 exerts its tumor suppression function in mice has not yet been elucidated. During the transition of large cycling into small resting pre-B cells, Slp65−/− or Btk−/− cells fail to efficiently down-regulate SLC and IL-7 receptor (IL-7R) expression, resulting in an increased proliferative response to IL-7 in vitro.6,8 In a recent work,15 the authors show that Slp65 also down-regulates IL-7–mediated proliferation and survival through direct inhibition of Jak3, which is an essential IL-7R signaling component. The IL-7R pathway promotes cellular survival, proliferation, and maturation, involving induction of the antiapoptotic protein Bcl-2 and the proto-oncogene c-Myc.16,17 B-cell development can be partially restored in Jak-3–deficient mice when they are bred to mice coexpressing a rearranged IgH chain transgene and a c-Myc transgene.18
Similar to Slp65−/− mice, Eμ-myc transgenic mice, which express c-Myc under the control of the IgH intronic enhancer, develop rapid-onset pre-B cell malignancies.19 Interestingly, the loss of Btk or PLC-γ2 synergizes with the deregulation of c-Myc during lymphoma formation in Eμ-myc mice.20,21 The presence of the Eμ-myc transgene substantially increases the proliferative potential of B-cell progenitors in response to IL-7.20,21 Activation of c-Myc is required for progression of quiescent cells into the S-phase of the cell cycle, but c-Myc also can induce the p53 protein, which protects against oncogenic transformation of proliferating cells (see Sherr22 for review). c-Myc activates the p19Arf tumor suppressor that interferes with the E3 ubiquitin protein ligase Mdm2 and thereby stabilizes and activates p53, resulting in cell-cycle arrest or apoptosis. It has been shown that in Myc-induced lymphomagenesis the p19Arf-Mdm2-p53 circuitry often is disrupted, indicating that c-Myc activation strongly selects for spontaneous inactivation of this pathway.23
We hypothesized that malignant transformation of Slp65-deficient pre-B cells is not only dependent on constitutive proliferative signals provided by the pre-BCR and IL-7R but also requires loss of the protective checkpoint function of the p19Arf-Mdm2-p53 pathway. Therefore, we investigated whether disruption of the p19Arf-Mdm2-p53 pathway is a common alteration in Slp65−/− pre-B cell tumors. We found that loss of p53 enhanced lymphoma formation in Slp65-deficient mice and that Slp65-deficient pre-B cell tumors harbored aberrations in the p19Arf-Mdm2-p53 tumor suppressor pathway.
Methods
Mice and genotyping
Btk−/−,24 Eμ-2-22 Bcl-2 transgenic (Tg),25 p53−/−,26 Aicda−/−,27 and VH81X Tg28 mice were on the C57BL/6 background. Slp65−/− mice5 were on the Balb/c background. The different composite genotypes were on a mixed background, and in single experiments littermates were compared. For mouse genotyping, genomic DNA was analyzed by polymerase chain reaction (PCR) as previously described.8,27,28 Animals in tumor panels were killed after indication of tumor formation, or after a maximum period of 26 weeks. Mice were bred and maintained in the Erasmus MC animal care facility under pathogen-free conditions. Experimental procedures were reviewed and approved by the Erasmus MC committee of animal experiments.
Tumors were identified as described,8 and tumor load in lymphoid tissues was determined by flow cytometry on the basis of intracellular expression of SLC components and Ig μ heavy chain and surface expression of B220 and CD19 (supplemental Table 3A-C, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Quantitative reverse transcriptase PCR analysis
Total RNA was extracted by use of the GenElute Mammalian Total RNA Miniprep system (Sigma-Aldrich). One microgram of total RNA was DNAse digested (Invitrogen) and used as a template for cDNA synthesis by the use of Superscript II reverse transcriptase (Invitrogen) and random hexamer primers. For quantitative reverse-transcriptase (RT)–PCR, primers spanning at least 1 intron-exon junction were designed manually or with the use of ProbeFinder software (Roche Applied Science; supplemental Table 1). Quantitative RT-PCR was performed by use of the ABI Prism 7700 sequence detection system (Applied Biosystems). Ct-values were obtained by the use of SDS v1.9 software (Applied Biosystems) and normalized to glycereraldehyde-3-phosphate dehydrogenase (GAPDH).
Southern blotting
Genomic DNA (10 μg) was digested with various restriction endonucleases (New England Biolabs) and processed by Southern blotting by the use of nylon membrane (Hybond N+ [Amersham] or Nytran SPC [Whatman]). Fragments were visualized with a PhosphorImager and analyzed with ImageQuant (Molecular Dynamics). Probes specific for N-myc, a 990-bp PCR fragment from exon 3; for Gata-3, a 800-bp cDNA containing exons 1 to 3; for p53, a 1123-bp PCR fragment from intron 1 (probe 1) and a 614-bp fragment containing part of intron 10 and exon 11 (probe 2); and for Ink4a-Arf, a 815-bp PCR fragment from exon 1α were labeled by random priming according to standard procedures.
Flow cytometry, cell culture, and in vivo 5-bromo-2′-deoxyuridine labeling
Preparations of single-cell suspensions, flow cytometry procedures, and monoclonal antibodies have been described previously.8,24,29 BM-cell suspensions were depleted of erythrocytes by standard ammonium chloride lysis, and IL-7–driven cultures were performed as described previously.29 For intracellular staining of cytoplasmic proteins, cells were first stained for cell surface markers and subsequently fixed in 2% paraformaldehyde and permeabilized by the use of 0.5% saponin. Procedures for calcium flux measurements have been described previously.10 Events (1-5 × 105) were scored by the use of a FACSCalibur flow cytometer and analyzed with CellQuest software (BD Biosciences).
5-Bromo-2′-deoxyuridine (BrdU; BD Biosciences) was dissolved in phosphate-buffered saline at 2 mg/mL. Mice were injected intraperitoneally with 200 μL and killed at various time points. Total BM-cell suspensions were analyzed by flow cytometry for BrdU incorporation by the use of the BrdU flow kit (BD Biosciences) in conjunction with cell-surface marker expression.
Spectral karyotyping and fluorescence in situ hybridization
Tumor cells were cultured overnight in the presence of 100 U/mL IL-7 and 10 ng/mL colcemid (KaryoMAX Colcemid solution; Gibco-BRL) and subsequently with 30 ng/mL for 4 hours to arrest proliferating cells at metaphase. Cells were treated with 75mM KCl and fixed with methanol/acetic acid (3:1). Spectral karyotyping (SKY) was performed by the use of the Applied Spectral Imaging system (ASI) following manufacturer's protocols. Slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) containing DABCO/Vectashield (Vector Laboratories). Chromosomes were analyzed by Zeiss Axioplan 2 microscope equipped with the Spectra Cube system (ASI). At least 10 different metaphases from each sample were analyzed with the use of Skyview analysis software (ASI). Dual-colored fluorescence in situ hybridization (FISH) experiments were performed by the use of BAC probes RP23-246B9 for N-Myc and CT7-199M11 for IgH chain (Invitrogen) with the use of standard procedures.
Pre-B cell stimulation and Western blotting
Total BM cells were cultured in the presence of 100 U/mL IL-7 (Sigma-Aldrich) as described.29 Pre-B cells were stimulated with 20 mg/mL F(ab)2 fragment of polyclonal goat anti-mouse IgM (Jackson ImmunoResearch) at 37°C for 5 minutes. Cultured pre-B cells or pre-B leukemia cells were lysed in lysis buffer (20mM Tris; 137mM NaCl; 10mM EDTA (ethylenediaminetetraacetic acid); 100mM NaF; 1% Nonidet P-40; 10% glycerol; 1mM pefabloc/AEBSF; 4-[2-aminoethyl]-benzenesulfonyl fluoride hydrochloride, Roche Applied Science; 1mM Na3VO4) on ice for 20 minutes and centrifuged at 16 000g at 4°C for 20 minutes. Total cell lysates of were immunoprecipitated with antiphosphotyrosine (pTyr-100; Cell Signaling Technology). Samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electrotransferred to nitrocellulose membrane (Whatman) with the use of standard procedures. Blots were stained with antibodies to Mdm2 (C18), p53 (DO-1 and FL-393), or Syk (N-19), all purchased from Santa Cruz Biotechnology, or to p19ARF (ab80), from Abcam.
Results
Enhancement of cellular survival does not increase pre-B cell leukemia incidence in Slp65-deficient mice
The developmental arrest in Slp65 and Btk single- or double-deficient mice results in a dominating populating of pre-B cells with increased surface expression of SLC and IL-7R compared with wild-type mice.6,7 Although Slp65−/− pre-B cells display an increased proliferative response to IL-7 in vitro,6,9 the pre-B cell population in Slp65−/− mice contains significantly fewer cycling cells compared with wild-type littermates.9 In agreement with the absence of increased in vivo proliferation of Slp65−/− pre-B cells, we found that BM pre-B cell numbers were not increased in Slp65−/− mice and even significantly reduced in Slp65−/−Btk−/− mice (Figure 1A).
Enhancement of cellular survival does not increase pre-B cell leukemia incidence in Slp65-deficient mice. (A) Absolute numbers of pre-B cells (CD19+Ig L−cμ+) per hind leg in the indicated mice. Bars represent average values and the SEM of 9 to 14 animals per genotype. Asterisks indicate significant differences with P < .001 (t test). (B) Flow cytometric analysis of BM lymphoid cells from the indicated mouse groups. Total CD19+ B-lineage cells were gated and CD19/cκ profiles are displayed as dot plots. Percentages of cells within the indicated gates are given and data shown are representative of 3 to 4 animals per genotype. (C) Kaplan-Meier tumor-free survival estimates for Btk−/−Slp65−/− mice (n = 25) and Bcl-2 Tg Btk−/−Slp65−/− mice (n = 7).
Enhancement of cellular survival does not increase pre-B cell leukemia incidence in Slp65-deficient mice. (A) Absolute numbers of pre-B cells (CD19+Ig L−cμ+) per hind leg in the indicated mice. Bars represent average values and the SEM of 9 to 14 animals per genotype. Asterisks indicate significant differences with P < .001 (t test). (B) Flow cytometric analysis of BM lymphoid cells from the indicated mouse groups. Total CD19+ B-lineage cells were gated and CD19/cκ profiles are displayed as dot plots. Percentages of cells within the indicated gates are given and data shown are representative of 3 to 4 animals per genotype. (C) Kaplan-Meier tumor-free survival estimates for Btk−/−Slp65−/− mice (n = 25) and Bcl-2 Tg Btk−/−Slp65−/− mice (n = 7).
We previously reported that transgenic expression of the antiapoptotic protein Bcl-2 enhanced in vitro survival upon IL-7 withdrawal but did not rescue the developmental arrest of Slp65−/−Btk−/− pre-B cells.30 It is therefore conceivable that Bcl-2 overexpression, which is commonly associated with enhanced malignancy of hematologic tumors, may increase tumor incidence in Slp65−/− mice. To investigate this, we compared the incidence of pre-B cell leukemia in a panel of Slp65−/−Btk−/− mice and Eμ-2-22 Bcl-2 Tg Slp65−/−Btk−/− mice. We confirmed that in these crosses Bcl-2 overexpression did not rescue pre-B cell differentiation (Figure 1B) and found that the presence of the Eμ-2-22 Bcl-2 Tg had no effect on the incidence of pre-B cell leukemia in Slp65−/−Btk−/− mice (Figure 1C) or Slp65−/− mice (not shown).
These findings show that in Slp65−/− pre-B cells—in which pre-BCR/IL-7R–mediated proliferation and survival signals are increased—enhancement of cellular survival does not augment tumor formation. Therefore, acquisition of additional genetic changes is required for malignant transformation of Slp65−/− pre-B cells.
Expression of the VH81X IgH chain transgene prevents leukemia in Btk−/−Slp65−/− mice
Because (1) pre-BCR/IL-7R signaling induces c-Myc and its family member N-myc in large pre-B cells,16 and (2) loss of Btk or PLC-γ synergizes with deregulation of the c-Myc oncogene during lymphoma formation,20,21 we hypothesized that transformation of Slp65-deficient pre-B cells may involve enhanced Myc activation.
We used quantitative RT-PCR to establish that pre-B cell tumors from Slp65−/− (n = 16) and Btk−/−Slp65−/− (n = 27) mice expressed substantial levels of Myc transcripts. compared with cultured wild-type pre-B cells, c-Myc expression in Slp65−/− and Btk−/−Slp65−/− pre-B cell tumors was approximately 8-fold increased, whereas n-Myc was moderately increased only in Slp65−/− pre-B cell tumors (Figure 2A).
The VH81X Tg prevents tumor formation in Btk/Slp65 double-deficient mice. (A) Quantitative RT-PCR analyses of c-Myc and N-Myc expression in Slp65−/− (n = 16) and Btk−/−Slp65−/− (n = 27) pre-B cell tumors normalized with GAPDH. Values in wild-type pre-B cells, cultured for 5 days with 100 U/mL IL-7, were set to 1. For each of these groups, the horizontal line represents the mean of the relative expression level. (B) Effect of the VH81X Tg on pre-B cell proliferation in vivo. The absolute numbers of BrdU+ CD19+cμ+cκ− pre-B cells in the BM, 4 hours after intraperitoneal injection of a single dose of BrdU, as determined by flow cytometry. Average values and SEM of 3 animals per group are shown. (C) Effect of the VH81X Tg on pre-B cell proliferation in vitro. Results are displayed as fold expansion after 5 days of culture in the presence of IL-7 (100 U/mL), whereby the pre-B cell numbers at the start of the culture where set to 1. Bars represent average values and SEM of 7 to 8 animals per genotype. (D) Syk phosphorylation in pre-B cell cultures from non-VH81X Tg Slp65−/− and VH81X Tg Btk−/−Slp65−/− mice. Cells were either unstimulated (−) or stimulated for 5 minutes with polyclonal anti-IgM F(ab)2 fragments. The presence of phosphorylated Syk was detected in antiphosphotyrosine immunoprecipitates from total cell lysates, analyzed by Western blotting by the use of Syk-specific antibodies. (E) Kaplan-Meier tumor-free survival estimates for Btk−/−Slp65−/− mice (n = 20) and VH81X transgenic Btk−/−Slp65−/− mice (n = 25).
The VH81X Tg prevents tumor formation in Btk/Slp65 double-deficient mice. (A) Quantitative RT-PCR analyses of c-Myc and N-Myc expression in Slp65−/− (n = 16) and Btk−/−Slp65−/− (n = 27) pre-B cell tumors normalized with GAPDH. Values in wild-type pre-B cells, cultured for 5 days with 100 U/mL IL-7, were set to 1. For each of these groups, the horizontal line represents the mean of the relative expression level. (B) Effect of the VH81X Tg on pre-B cell proliferation in vivo. The absolute numbers of BrdU+ CD19+cμ+cκ− pre-B cells in the BM, 4 hours after intraperitoneal injection of a single dose of BrdU, as determined by flow cytometry. Average values and SEM of 3 animals per group are shown. (C) Effect of the VH81X Tg on pre-B cell proliferation in vitro. Results are displayed as fold expansion after 5 days of culture in the presence of IL-7 (100 U/mL), whereby the pre-B cell numbers at the start of the culture where set to 1. Bars represent average values and SEM of 7 to 8 animals per genotype. (D) Syk phosphorylation in pre-B cell cultures from non-VH81X Tg Slp65−/− and VH81X Tg Btk−/−Slp65−/− mice. Cells were either unstimulated (−) or stimulated for 5 minutes with polyclonal anti-IgM F(ab)2 fragments. The presence of phosphorylated Syk was detected in antiphosphotyrosine immunoprecipitates from total cell lysates, analyzed by Western blotting by the use of Syk-specific antibodies. (E) Kaplan-Meier tumor-free survival estimates for Btk−/−Slp65−/− mice (n = 20) and VH81X transgenic Btk−/−Slp65−/− mice (n = 25).
Interestingly, the incidence of lymphomas in Eμ-myc mice is greatly reduced by the introduction of a human IgH transgene.31 Such a transgene is thought to accelerate B-cell development, thus reducing the population size of cells most susceptible to oncogenic transformation by c-Myc. To investigate whether expression of an IgH chain transgene also reduces pre-B cell tumor incidence in Slp65-deficient mice, we used mice carrying the IgH chain transgene VH81X.28 First, we ascertained by flow cytometry that the VH81X transgene did not significantly affect the generation of large and small pre-B cell compartments in the BM of wild-type, Btk−/−, Slp65−/−, and Btk−/−Slp65−/− mice (supplemental Figure 1). Furthermore, the presence of the VH81X transgene did not affect in vivo pre-B cell proliferation: when mice were pulsed with a single dose of the thymidine analog BrdU, which is selectively incorporated into the DNA of large pre-B cells,29 comparable numbers of BrdU+ CD19+IgL−cμ+ pre-B cells were present in non-Tg and VH81X Tg Btk−/−Slp65−/− mice (Figure 2B). The high in vitro proliferative capacity of Btk−/−Slp65−/− pre-B cells in IL-7–driven cultures8 was also not significantly affected by the presence of the VH81X transgene (Figure 2C). We therefore conclude that the phenotypical abnormalities of Btk−/−, Slp65−/−, and Btk−/−Slp65−/− pre-B cells, which reflect their impaired cellular maturation from large cycling into small resting pre-B cells, were preserved in the presence of the prerearranged VH81X IgH chain transgene. When cultured VH81X transgenic and nontransgenic Btk−/−Slp65−/− pre-B cells were stimulated with Ig μ H chain-specific antibodies, we found similar Syk tyrosine kinase phosphorylation (Figure 2D), supporting the signaling competence of the VH81X IgH chain. Consistent with the essential role of Slp65 in calcium mobilization, we found that pre-BCR signaling-associated calcium fluxes in both VH81X transgenic and nontransgenic Btk−/−Slp65−/− pre-B cells were blunted (supplemental Figure 2).
To address the effect of the VH81X transgene on pre-B cell leukemia formation, we followed panels of Btk−/−Slp65−/− mice that did or did not carry the VH81X transgene. At 6 months of age, 16 of 20 Btk−/−Slp65−/− mice developed pre-B cell leukemia. In strong contrast, none of the 23 VH81X IgH chain transgenic Btk−/−Slp65−/− mice developed pre-B cell leukemia (Figure 2E). In addition, when we examined BM and spleen of these mice by flow cytometry at the age of 6 months we did not find evidence for lymphoproliferative disease.
Thus, expression of the prerearranged VH81X H chain transgene in early B-cell differentiation prevented oncogenic transformation of Btk−/−Slp65−/− pre-B cells, equivalent to findings in Eμ-myc Tg mice.31
Deficiency of Slp65 and p53 have cooperative effects in tumorigenesis
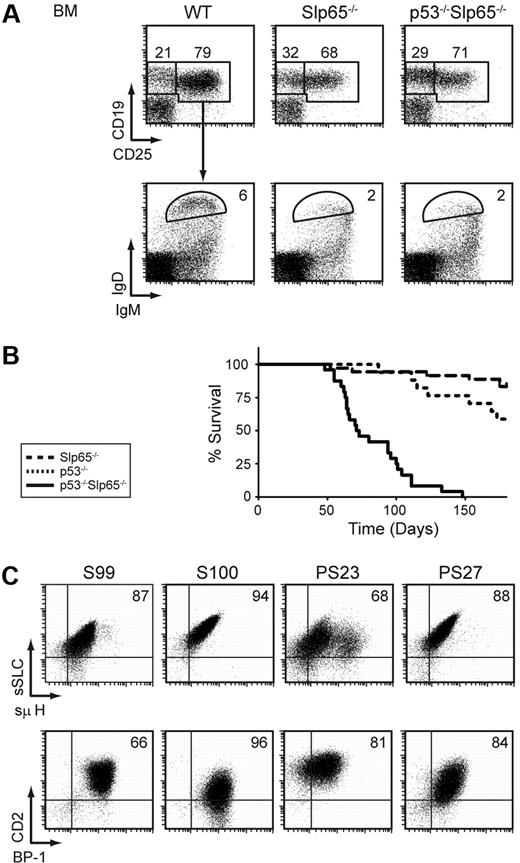
Next, we investigated the effect of p53-deficiency on pre-B cell tumor formation by crossing Slp65−/− mice onto a p53−/− background. In flow cytometric experiments, no significant differences were found between the sizes of the individual B-lineage subpopulations in Slp65−/− mice and p53−/−Slp65−/− mice: as shown in Figure 3A, Slp65−/− mice and p53−/−Slp65−/− mice have a similar defect in CD25 up-regulation upon pre-BCR signaling8 and have a similar reduction in the number of mature recirculating IgMlowIgD+ B cells. The proliferative capacity of Slp65−/− pre-B cells in IL-7–driven BM cultures in vitro was not affected by the concomitant absence of p53 (data not shown).
Slp65 and p53 cooperate as tumor suppressors in pre-B cells. (A) Flow cytometric analysis of total lymphoid fractions in BM (top). Total CD19+ fractions were gated and analyzed for IgM and IgD expression (bottom). Data are displayed as dot plots, and numbers indicate the percentages of cells within the gates and are representative for 4 5-week-old nontumor-bearing mice. (B) Kaplan-Meier tumor-free survival estimates for Slp65−/− mice (n = 36), p53−/− mice (n = 17), and p53−/−Slp65−/− mice (n = 24). (C) Characterization of pre-B cell tumors by flow cytometry. Dot plots for surface SLC and μ H chain expression (top) and dot plots for CD2 and BP-1 expression (bottom) in gated CD19+ cells from tumor samples of Slp65−/− (S99, S100) and p53−/−Slp65−/− (PS23, PS27) mice.
Slp65 and p53 cooperate as tumor suppressors in pre-B cells. (A) Flow cytometric analysis of total lymphoid fractions in BM (top). Total CD19+ fractions were gated and analyzed for IgM and IgD expression (bottom). Data are displayed as dot plots, and numbers indicate the percentages of cells within the gates and are representative for 4 5-week-old nontumor-bearing mice. (B) Kaplan-Meier tumor-free survival estimates for Slp65−/− mice (n = 36), p53−/− mice (n = 17), and p53−/−Slp65−/− mice (n = 24). (C) Characterization of pre-B cell tumors by flow cytometry. Dot plots for surface SLC and μ H chain expression (top) and dot plots for CD2 and BP-1 expression (bottom) in gated CD19+ cells from tumor samples of Slp65−/− (S99, S100) and p53−/−Slp65−/− (PS23, PS27) mice.
Whereas approximately 17% (6 of 36) of Slp65−/− mice developed pre-B cell leukemia at 6 months, all p53−/−Slp65−/− mice (n = 24) developed pre-B cell leukemia within 5 months (Figure 3B). All pre-B cell tumors in p53−/−Slp65−/− mice were positive for Ig μ H chain and SLC and showed variable surface expression of developmentally regulated markers, including CD2 and the metallopeptidase BP-1, similar to those normally found in Slp65−/− mice (Figure 3C). In contrast, 5 of 17 p53−/− single-deficient littermates developed T-cell lymphoma before the age of 5 months. Thus, loss of Slp65 on the p53−/− background resulted in rapid onset of pre-B cell tumors before the age at which T-cell tumors usually arise in p53−/− mice. We conclude that Slp65 and p53 cooperate to limit the oncogenic potential of DNA damage and/or sustained oncogenic signaling.
Activation-induced cytidine deaminase is not involved in malignant transformation of Slp65−/− pre-B cells
One of the malignancy-associated stress signals that activates p53 is DNA damage. We therefore investigated a possible role of DNA damage caused by the activation-induced cytidine deaminase (Aid) mutator protein or by aberrant V(D)J recombination, leading to chromosomal translocations.32
Human pre-B ALL that harbor the t(9;22)(q34;q11) Philadelphia translocation encoding the oncogenic BCR-ABL1 tyrosine kinase have defective pre-BCR signaling, express a truncated isoform of SLP65,12,33 along with AID,12,33 which acts as a BCR-ABL1–induced mutator. AID expression is normally restricted to germinal center B cells, where it initiates somatic hypermutation and class switch recombination. However, when expressed in nongerminal center cells, AID can induce mutation in various highly transcribed genes and thereby act as a genome-wide mutator.34
We analyzed Slp65-deficient pre-B cell tumors for Aid expression by quantitative RT-PCR. Remarkably, in most of the samples Aid transcripts were detected, albeit at variable levels. In a small fraction of leukemic samples Aid expression was in the same range as in Peyer patches (supplemental Figure 3A). It was thus conceivable that Aid-induced DNA damage could contribute to tumorigenesis in Slp65-deficient pre-B cells. However, when we sequenced putative Aid target regions, including the 5′ regions of the λ5 and p53 genes, as well as the JH4 intron of the IgH locus, from a panel of 15 Btk−/−Slp65−/− tumors, we did not find evidence for somatic hypermutation (data not shown). Next, we reasoned that if Aid would play an important role in malignant transformation of Slp65−/− pre-B cells, the frequency of pre-B cell leukemias would be significantly reduced in the absence of the Aicda gene encoding Aid. However, we found that the pre-B cell tumor incidences in Slp65−/− or Btk−/−Slp65−/− mice were similar in the presence (Aicda+/−) or absence (Aicda−/−) of Aid (supplemental Figure 3B). Therefore, we conclude that Aid-induced DNA damage is not involved in malignant transformation of Slp65−/− pre-B cells.
Chromosome 12 abnormalities in Slp65−/− pre-B cell tumors
When DNA breaks mediated by the V(D)J recombination machinery are not resolved properly, they can give rise to chromosomal alterations, in particular translocations, and lymphoid tumors.32 We examined metaphase spreads from Btk−/−Slp65−/− and p53−/−Slp65−/− tumor cells by SKY and found evidence for chromosomal instability mainly characterized by chromosome gains, in particular of chromosome 12 and 14 (supplemental Table 2). Interestingly, BCR-Abl transgenic mouse lymphomas often show gain of chromosome 12 and 14.35
Two leukemia samples, BS56 (Btk−/−Slp65−/−) and PS11 (p53−/−Slp65−/−), harbored an aberration involving intrachromosomal gain of chromosome 12 material (supplemental Figure 4A). Similar enlargements of chromosome 12 have been identified in pro-B cell leukemias derived from mice deficient for p53 and Artemis, a component of the nonhomologous end-joining DNA repair pathway.36 Because the observed chromosome 12 enlargement points to the possibility of N-myc amplification, FISH experiments were performed. We found a duplication (BS56) and dramatic amplification (PS11) of the N-myc locus (supplemental Figure 4B). Southern blotting of genomic EcoRI digests with an N-myc–specific probe confirmed amplification of N-myc in the BS56 and PS11 tumors and identified a third pre-B cell leukemia (PS08) with N-myc amplification (3 of 38 analyzed tumors) (supplemental Figure 4C). Quantitative RT-PCR experiments demonstrated that the 2 p53−/−Slp65−/− pre-B cell leukemias (PS08 in red and PS11 in green) that harbored substantial N-myc gene amplification expressed extremely high levels of N-myc and severely reduced levels of c-Myc transcripts (supplemental Figure 4D). Similar cross-regulation of Myc family members has been reported in the context of N-myc–related transformation.36 In contrast to chromosome 12 enlargements involving N-myc in Artemis/p53 double-deficient mice,36,37 we did not detect coamplification of IgH chain locus sequences in FISH and Southern blotting experiments (data not shown).
In summary, although part of the Slp65−/− pre-B cell leukemias exhibit chromosomal instability, we found no evidence for Rag-mediated translocations. Our SKY analyses revealed gain of chromosome 12 or N-myc amplification in 6 of 19 (∼ 32%) pre-B cell tumors.
Only a small subset of Slp65-deficient tumors expresses p53 protein
The p53 response also can be activated by oncogenic signaling. In particular, it has been shown that Myc-induced lymphomagenesis strongly selects for spontaneous inactivation of the p19Arf-Mdm2-p53 tumor suppressor pathway.23 Therefore, we next investigated whether malignant transformation of Slp65−/− pre-B cells involves disruption of this pathway.
Loss of p53 function can occur through deletions or point mutations. We did not detect gross deletions in the p53 gene by Southern blot analyses in a panel of 23 Slp65−/− and Btk−/−Slp65−/− tumors (supplemental Figure 5A). Next, we sequenced p53 transcripts and genomic DNA encompassing exons 2 to 8 (encoding the DNA-binding domain where p53 most frequently undergoes mutation in tumors) from a panel of 29 Slp65−/− and Btk−/−Slp65−/− tumors (supplemental Table 3A-B). This analysis revealed a mutation only in a single Slp65−/− tumor (S13): a R270C missense mutation. This mutation has previously been identified in Eμ-myc Tg lymphoma and corresponds with a mutation hot-spot of p53 found in human tumors.38 Missense mutations often result in accumulation of mutant p53 protein to supraphysiologic levels because mutant p53 is often more stable than wild type.39 Western blot analyses confirmed very high p53 protein expression in the tumor with the R270C p53 mutation (sample S13; Figure 4A).
Aberrations of the p19Arf-Mdm2-p53 pathway in Slp65-deficient leukemias. (A) Western blot analysis of total cell lysates from pre-B cell tumors from Slp65−/− mice for the expression of p53, p19Arf, Mdm2, and Syk (loading control). (B) Quantitative RT-PCR analyses of p21Cip1 expression in Slp65−/− pre-B cell tumors without or with detectable p53 protein, normalized to GAPDH. Values in sorted WT BM B220+ cells were set to 1. Horizontal lines represent mean values of the relative expression levels. (C) Western blot analysis of total cell lysates from pre-B cell tumors from Btk−/−Slp65−/− mice for the expression of p53, p19Arf, Mdm2, and Syk (loading control). (D) Western blot analysis of total cell lysates from pre-B cell tumors from p53−/−Slp65−/− mice for the expression of p19Arf, Mdm2, and Syk (loading control). Protein of p53 was not detected in p53−/−Slp65−/− tumors.
Aberrations of the p19Arf-Mdm2-p53 pathway in Slp65-deficient leukemias. (A) Western blot analysis of total cell lysates from pre-B cell tumors from Slp65−/− mice for the expression of p53, p19Arf, Mdm2, and Syk (loading control). (B) Quantitative RT-PCR analyses of p21Cip1 expression in Slp65−/− pre-B cell tumors without or with detectable p53 protein, normalized to GAPDH. Values in sorted WT BM B220+ cells were set to 1. Horizontal lines represent mean values of the relative expression levels. (C) Western blot analysis of total cell lysates from pre-B cell tumors from Btk−/−Slp65−/− mice for the expression of p53, p19Arf, Mdm2, and Syk (loading control). (D) Western blot analysis of total cell lysates from pre-B cell tumors from p53−/−Slp65−/− mice for the expression of p19Arf, Mdm2, and Syk (loading control). Protein of p53 was not detected in p53−/−Slp65−/− tumors.
In 7 of 17 Slp-65−/− tumors analyzed we found low-to-moderate p53 protein levels (Figure 4A; Table 1). Sequencing of 3 of these tumors (S96, S100, and S102) did not reveal point mutations in exons 2 to 8 (supplemental Table 3A). Furthermore, the p53 protein was not detected in any of the 32 tumors from Btk−/−Slp65−/− mice (Figure 4C; Table 1). To examine p53 functional activity in these tumors, we compared transcription of the p53 target p21Cip1 in Slp65−/− tumors without detectable p53 protein expression and tumors with p53 protein (Figure 4B). We did not observe increased p21Cip1 transcription in tumors expressing p53, indicating that p53 protein in these tumors has lost part of its normal function. Taken together, in 8 of 49 Slp65−/− or Btk−/−Slp65−/− tumors we found p53 protein expression, whereby only in a single case we were able to identify a p53 missense mutation.
Arf-Mdm2-p53 protein expression in Slp65-deficient leukemias
| Tumor . | p53 . | p19ARF . | Mdm2 . |
|---|---|---|---|
| Aberrant p53 | |||
| S13 | High* | High | Expressed |
| S92 | Expressed | Low | Expressed |
| S96 | Expressed | Expressed | Expressed |
| S100 | Low | High | Expressed |
| S102 | Low | High | Expressed |
| S116 | Low | High | Expressed |
| S203 | Low | High | Expressed |
| S208 | Low | High | Expressed |
| p53 wild-type, Arf expressed/ high | |||
| S14 | Undetected | High | Expressed |
| S25 | Undetected | High | Expressed |
| S75 | Undetected | High | Undetected |
| S99 | Undetected | High | Expressed |
| S101 | Undetected | High | Expressed |
| S107 | Undetected | High | Expressed |
| S109 | Undetected | High | Expressed |
| S114 | Undetected | High | Expressed |
| BS27 | Undetected | High | Expressed |
| BS29 | Undetected | High | Expressed |
| BS30 | Undetected | Expressed | Expressed |
| BS36 | Undetected | High | Low |
| BS45 | Undetected | High | Low |
| BS50 | Undetected | High | Low |
| BS53 | Undetected | High | Expressed |
| BS70 | Undetected | High | Expressed |
| BS91 | Undetected | High | Expressed |
| BS103 | Undetected | High | Expressed |
| BS105 | Undetected | High | Expressed |
| BS108 | Undetected | High | Expressed |
| BS115 | Undetected | High | Expressed |
| BS118 | Undetected | High | Low |
| BS119 | Undetected | High | Expressed |
| BS123 | Undetected | High | Expressed |
| BS124 | Undetected | High | Expressed |
| BS301 | Undetected | High | Expressed |
| BS303 | Undetected | High | Expressed |
| BS304 | Undetected | High | Expressed |
| BS305 | Undetected | High | Expressed |
| BS306 | Undetected | High | Expressed |
| BS309 | Undetected | High | Expressed |
| BS310 | Undetected | High | Expressed |
| BS311 | Undetected | Expressed | Expressed |
| BS312 | Undetected | Expressed | Expressed |
| BS316 | Undetected | High | Expressed |
| BS318 | Undetected | Expressed | Expressed |
| BS326 | Undetected | High | Expressed |
| BS327 | Undetected | High | Expressed |
| Arf low/ undetected | |||
| S98 | Undetected | Low | Expressed |
| BS72 | Undetected | Undetected | Low |
| BS93 | Undetected | Undetected | Expressed |
| p53-deficient tumors, Arf expressed | |||
| PS15 | Undetected | High | Expressed |
| PS16 | Undetected | High | Expressed |
| PS18 | Undetected | High | Expressed |
| PS19 | Undetected | Expressed | Expressed |
| PS21 | Undetected | High | Expressed |
| PS23 | Undetected | High | Expressed |
| PS24 | Undetected | High | Expressed |
| p53-deficient tumors, Arf undetected | |||
| PS17 | Undetected | Undetected | Expressed |
| PS22 | Undetected | Undetected | Low |
| PS27 | Undetected | Undetected | Expressed |
| Tumor . | p53 . | p19ARF . | Mdm2 . |
|---|---|---|---|
| Aberrant p53 | |||
| S13 | High* | High | Expressed |
| S92 | Expressed | Low | Expressed |
| S96 | Expressed | Expressed | Expressed |
| S100 | Low | High | Expressed |
| S102 | Low | High | Expressed |
| S116 | Low | High | Expressed |
| S203 | Low | High | Expressed |
| S208 | Low | High | Expressed |
| p53 wild-type, Arf expressed/ high | |||
| S14 | Undetected | High | Expressed |
| S25 | Undetected | High | Expressed |
| S75 | Undetected | High | Undetected |
| S99 | Undetected | High | Expressed |
| S101 | Undetected | High | Expressed |
| S107 | Undetected | High | Expressed |
| S109 | Undetected | High | Expressed |
| S114 | Undetected | High | Expressed |
| BS27 | Undetected | High | Expressed |
| BS29 | Undetected | High | Expressed |
| BS30 | Undetected | Expressed | Expressed |
| BS36 | Undetected | High | Low |
| BS45 | Undetected | High | Low |
| BS50 | Undetected | High | Low |
| BS53 | Undetected | High | Expressed |
| BS70 | Undetected | High | Expressed |
| BS91 | Undetected | High | Expressed |
| BS103 | Undetected | High | Expressed |
| BS105 | Undetected | High | Expressed |
| BS108 | Undetected | High | Expressed |
| BS115 | Undetected | High | Expressed |
| BS118 | Undetected | High | Low |
| BS119 | Undetected | High | Expressed |
| BS123 | Undetected | High | Expressed |
| BS124 | Undetected | High | Expressed |
| BS301 | Undetected | High | Expressed |
| BS303 | Undetected | High | Expressed |
| BS304 | Undetected | High | Expressed |
| BS305 | Undetected | High | Expressed |
| BS306 | Undetected | High | Expressed |
| BS309 | Undetected | High | Expressed |
| BS310 | Undetected | High | Expressed |
| BS311 | Undetected | Expressed | Expressed |
| BS312 | Undetected | Expressed | Expressed |
| BS316 | Undetected | High | Expressed |
| BS318 | Undetected | Expressed | Expressed |
| BS326 | Undetected | High | Expressed |
| BS327 | Undetected | High | Expressed |
| Arf low/ undetected | |||
| S98 | Undetected | Low | Expressed |
| BS72 | Undetected | Undetected | Low |
| BS93 | Undetected | Undetected | Expressed |
| p53-deficient tumors, Arf expressed | |||
| PS15 | Undetected | High | Expressed |
| PS16 | Undetected | High | Expressed |
| PS18 | Undetected | High | Expressed |
| PS19 | Undetected | Expressed | Expressed |
| PS21 | Undetected | High | Expressed |
| PS23 | Undetected | High | Expressed |
| PS24 | Undetected | High | Expressed |
| p53-deficient tumors, Arf undetected | |||
| PS17 | Undetected | Undetected | Expressed |
| PS22 | Undetected | Undetected | Low |
| PS27 | Undetected | Undetected | Expressed |
Immunoblotting experiments were performed twice, and a summary of those experiments is shown.
All Slp65-deficient tumors manifest aberrations of the p19Arf-Mdm2-p53 pathway
Consistent with disruption of the p53-p19Arf feedback loop, p19Arf expression was significantly increased in the pre-B cell tumor with the R270C p53 mutation (S13; Figure 4A) and in 6 of 7 additional Slp65−/− samples with detectable p53. Remarkably, substantial levels of p19ARF were also present in 38 of 41 Slp65−/− or Btk−/−Slp65−/− tumors in which p53 protein was undetectable. The presence of p19Arf protein indicates inactivation of the p53 response because p53 is a negative regulator of p19ARF.
In only 2 samples p19ARF was undetectable (BS72, BS93; Figure 4C; Table 1). Inactivation of the p53 pathway also can occur through loss of p19ARF, which requires deletion of both Arf alleles.40 The Ink4a-Arf locus encodes 2 linked tumor-suppressor genes: p16Ink4a and p19ARF. PCR amplification of exon 1α (Ink4a-specific) and exon 1β (Arf-specific) in combination with Southern blot analyses for exon 1α, 2, and 3 did not reveal any deletions in the Ink4a-Arf locus in these 2 samples.
Elevated expression of the Mdm2 oncogene augments proliferation, reduces susceptibility to p53-dependent apoptosis, and induces chromosomal instability.41 High levels of Mdm2 protein were found in all 8 tumors with detectable p53 and in 32 of 38 tumors that expressed p19ARF. In 6 p19ARF-expressing tumors, Mdm2 levels were low (BS30, BS36, BS45, BS50, BS118) or undetectable (S75; Figure 4AC, Table 1). From those 3 tumors that had low or undetectable p19ARF, 2 exhibited significant Mdm2 expression.
In summary, we found that all Slp65−/− (n = 17) and Btk−/−Slp65−/− (n = 32) pre-B cell leukemias analyzed expressed either p19ARF or Mdm2, perhaps with the exception one sample, BS72, with no detectable expression of p53 or p19ARFand only low Mdm2 expression. These results provide evidence for the involvement of disruption of the Arf-Mdm2-p53 tumor suppressor pathway in malignant transformation of Slp65-deficient pre-B cells.
Additional aberrations in p19Arf and Mdm2 in p53-deficient pre-B cell tumors
The finding that most Slp65-deficient pre-B cell tumors expressed Mdm2 raised the possibility that Mdm2 might be induced even in the absence of functional p53. To investigate this issue, we analyzed p19ARF and Mdm2 expression in a panel of 10 p53−/−Slp65−/− pre-B cell tumors and found that 7 expressed both p19ARF and Mdm2 (Figure 4D; Table 1). In 3 samples, p19ARF was not detected. Because in cells lacking p53 the levels if p19ARF are normally increased, this finding indicates that the p53-p19ARF feedback loop was interrupted. This finding did not appear to result from genomic Arf loss, because no aberrations were found in the Ink4a-Arf locus by genomic PCR and Southern blotting analyses (supplemental Figure 5BD). In these 3 tumors without detectable p19ARF, we found that Mdm2 expression was high in 2 cases and low in 1 case (Table 1). In summary, p53-deficient pre-B cell tumors have additional aberrations in p19ARF and Mdm2, suggesting that Mdm2 promotes transformation of Slp65-deficient pre-B cells by targeting effectors other than p53.
Discussion
In this study we provide evidence for disruption of the p19Arf-Mdm2-p53 tumor suppressor pathway in malignant transformation of Slp65−/− pre-B cells. We demonstrated that combined loss of Slp65 and p53 in mice transformed pre-B ells very efficiently. Furthermore, we found aberrations in p19Arf, Mdm2, or p53 in all 49 Slp65−/− and Btk−/−Slp65−/− pre-B cell leukemias analyzed.
Our findings reveal striking parallels in pre-B cell tumor formation between Slp65−/− and Eμ-myc Tg mice. First, in both models the expression of a prerearranged IgH chain transgene in early B-cell differentiation reduces oncogenic transformation. Second, malignant transformation of both Slp65−/− and Eμ-myc Tg pre-B cells involves disruption of the p19Arf-Mdm2-p53 tumor suppressor pathway. Third, Aid-induced DNA damage is not required for tumor development in either of the 2 models. Also the reported cooperation of Btk as a tumor suppressor in both Slp65−/− and Eμ-myc Tg pre-B cells point to parallel transformation mechanisms. In addition to the failure of Slp65-deficient pre-B cells to efficiently down-regulate pre-BCR and IL-7R signaling, it was recently shown that Slp65 has the capacity to inhibit IL-7R signaling by direct inhibition of Jak3 and that Slp65−/− tumors acquire autonomous IL-7R signaling by autocrine IL-7 production.15 Because c-Myc is induced by IL-7R signaling, it is very well possible that sustained IL-7R signaling results in constitutive high levels of c-Myc expression in Slp65-deficient pre-B cells.
The finding that Slp65-deficient pre-B cell tumors display disruptions of the Arf-Mdm2-p53 pathway implies that oncogenic transformation of Slp65−/− pre-B cells does not exclusively result from sustained IL-7R signaling and endocrine IL-7 production.15 Instead, it is conceivable that mutations have accumulated before this stage, which would be supported by the finding that expression of the prerearranged VH81X IgH chain transgene in early B-cell differentiation stages prevents malignant transformation. Because early expression of a functional IgH chain is known to considerably shorten or even bypass pro-B cell development,42 acceleration of the passage of B-cell precursors through the pro-B cell stage may prevent oncogenic transformation by reducing the size of the cell population most susceptible to transformation by eg c-Myc, N-myc, or Abl.31 However, there are alternative explanations for the tumor suppressive effect of VH81X IgH chain. We cannot exclude that VH81X IgH chain transgene prevents tumor formation by limiting the time window for V(D)J recombination activity. Several reported findings indicate that V(D)J recombination is ongoing in Slp65−/− large pre-B cells. First, Slp65−/− pre-B cell tumors mostly coexpress IgH chain, SLC, IgL chain,8 and the recombination activation genes Rag1 and Rag2 (V.B.T.T., unpublished data, October 2007). Second, pre-B cell fractions from CD19−/−Slp65−/− mice show increased expression of Rag2 protein.9 Third, In human pre-B cell leukemia, SLP65 deficiency correlates with RAG expression and ongoing VH gene rearrangement activity.43 However, recently it has been found that Rag1 does not contribute to lymphomagenesis in Eμ-myc Tg mice,44 suggesting that malignant transformation of Slp65−/− pre-B cells mediated by Myc does not require deregulated Rag activity. Furthermore, it is possible that transformation of Slp65−/− pre-B cells is dependent on the expression of IgH chains with specific structural properties that induce strong autonomous pre-BCR signaling. Recently, Kohler et al45 have described functional similarities between the pre-BCR and autoreactive BCRs, indicating that recognition of self-antigens by the pre-BCR might play a role in the initiation of pre-B cell proliferation. This explanation requires further investigation of the properties of IgH chains in Slp65-deficient pre-B cell tumors. Finally, it is unlikely that the VH81X IgH chain is signaling incompetent because it can pair efficiently with IgL chains, it induces strong proliferation and Syk phosphorylation, and its presence does not significantly affect pre-B cell differentiation.
We found high levels of p19Arf protein, in the absence of detectable p53 gene mutations or deletions, indicating that these tumors have inactivated p53 through some other mechanism. In this context, we found by RT-PCR that Slp65−/− and Btk−/−Slp65−/− pre-B cell tumors contain high levels of p53 transcripts and that p53 protein was induced upon 10 gray irradiation in 4 pre-B cell tumors tested (V.B.T.T., unpublished data, August 2008). Thus, it is possible that p53 protein stability is low in these tumors, for instance, because of the high levels of Mdm2 that was present in the majority of tumors. Nevertheless, 6 Slp65−/− or Btk−/−Slp65−/− pre-B cell leukemias with high p19Arf levels (Table 1) and without detectable p53 did not exhibit Mdm2 overexpression. Importantly, p19Arf has p53-independent functions, for example, it promotes the progression of lymphomas by mediating autophagy, a process of lysosome-mediated self-digestion that occurs during periods of nutrient deprivation.40,46 Silencing of p19Arf inhibits the progression of Myc-driven lymphoma cells containing mutant or no p53.46 Thus, Slp65−/− tumors may retain p19Arf to promote survival under metabolic stress.
Slp65 regulates the activity of the forkhead-box transcription factors Foxo3a and Foxo1, which do not only promote IgL chain recombination but also suppress Myc-driven lymphomagenesis via direct p19Arf activation.2,47 This leads us to propose the following mechanism for malignant transformation of Slp65−/− pre-B cells. The absence of Slp65 results in sustained expression of the pre-BCR and the IL-7R in large cycling pre-B cells. Because IL-7R signaling induces c-Myc, this also results in constitutively high levels of c-Myc. At this stage, FoxO transcription factors that normally suppress c-Myc–driven lymphomagenesis via direct activation of p19Arf are not properly activated because of the absence of Slp65. Activation of c-Myc in combination with sporadic alterations in the p19Arf-Mdm2-p53 pathway may finally lead to oncogenic transformation of Slp65−/− pre-B cells.
Our mouse model links defective pre-BCR signaling to oncogenic c-Myc activation and disruption of the p19Arf-Mdm2-p53 tumor suppressor pathway, which also play an essential role in the pre-B cell transformation process mediated by Abelson murine leukemia virus.48 Although defective expression of Btk or Slp65 may only be involved in a limited fraction of childhood ALL, and IL-7 signaling does not play a role in human pre-B cell development, our findings have relevance for human ALL. Deletion of CDKN2A (INK4A-ARF) is a significant secondary abnormality in childhood ALL, which strongly correlates with phenotype and genotype.49,50 Furthermore, overexpression of the MDM2 gene was found in childhood acute lymphoblastic leukemia cells expressing wild-type p53.51
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank H. Jumaa, J. F. Kearney, and T. Honjo for providing us with, respectively, the Slp65-deficient, VH81X Tg, and AID-deficient mice. We thank G. Dingjan, S. Middendorp, B. Beverloo, and E. van Drunen for their assistance at various stages of the project.
This work was supported by the Netherlands Organization for Scientific Research, the Dutch Cancer Society, and the Association for International Cancer Research.
Authorship
Contribution: V.B.T.T. and R.W.H. designed the research, analyzed the data, and wrote the paper; and V.B.T.T., P.J.t.B., J.P.v.H., M.J.W.d.B., R.K., P.F.v.L., and H.J.A.D. performed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rudi W. Hendriks, Department of Pulmonary Medicine, Erasmus MC Rotterdam, PO Box 2040, NL-3000 CA Rotterdam, The Netherlands; e-mail: r.hendriks@erasmusmc.nl.