Abstract
Quebec platelet disorder (QPD) is an autosomal dominant bleeding disorder linked to a region on chromosome 10 that includes PLAU, the urokinase plasminogen activator gene. QPD increases urokinase plasminogen activator mRNA levels, particularly during megakaryocyte differentiation, without altering expression of flanking genes. Because PLAU sequence changes were excluded as the cause of this bleeding disorder, we investigated whether the QPD mutation involved PLAU copy number variation. All 38 subjects with QPD had a direct tandem duplication of a 78-kb genomic segment that includes PLAU. This mutation was specific to QPD as it was not present in any unaffected family members (n = 114), unrelated French Canadians (n = 221), or other persons tested (n = 90). This new information on the genetic mutation will facilitate diagnostic testing for QPD and studies of its pathogenesis and prevalence. QPD is the first bleeding disorder to be associated with a gene duplication event and a PLAU mutation.
Introduction
Quebec platelet disorder (QPD) is an autosomal dominant bleeding disorder with a unique, gain-of-function defect in fibrinolysis.1,2 The disorder is characterized by: delayed onset bleeding3 ; markedly increased production and storage of urokinase plasminogen activator (uPA) in platelets4,5 without systemic fibrinolysis or markedly increased plasma or urinary uPA1,2,4,6,7 ; and uPA-triggered, plasmin-mediated proteolysis of diverse platelet α-granule proteins.5,7-11 QPD is strongly linked to PLAU.12 Furthermore, QPD increases uPA mRNA levels, from the disease chromosome, by approximately 2- to 4-fold in saliva and CD34+ hematopoietic progenitor cells, and more than 150-fold in megakaryocytes and platelets.12 Mice that overexpress uPA in megakaryocytes have QPD-like abnormalities.13 However, extensive sequencing and Southern blot analysis of PLAU, and its regulatory elements, failed to identify QPD-specific mutations.12 This led us to investigate whether the QPD mutation is a PLAU copy number variation (CNV).
Methods
Studies were carried out in accordance with the revised Declaratoin of Helsinki protocol for research on human subjects, with ethics review board approval of McMaster University and Hamilton Health Sciences, and Center Hospitalier Universitaire Sainte-Justine. Samples were obtained with written informed consent from adult subjects and parents of subjects less than 18 years of age.
All available persons with QPD were studied (38 affected, 114 unaffected; all descendants of a common ancestor), including 14 new subjects (10 affected, based on increased platelet uPA and degraded platelet fibrinogen3,4,8 ). Unrelated participants included 221 French-Canadians and 90 persons from HapMap Centre d'Etude du Polymorphisme Humain (CEPH) pedigrees of 30 white trio families from Utah.14
Molecular methods
Supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article) contain the details of the sample preparation and molecular methods. Samples from distantly related, QPD persons were selected for CNV screening, fluorescence in situ hybridization, and G-band karyotyping of blood lymphocytes. Chromosome 10 locations were expressed as Build 36.3 positions from the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/RefSec/). Results were analyzed using the Kruskall-Wallis nonparametric test and the Fisher exact test for disease associations. MatInspector 8.0 (Genomatrix Software) was used to analyze sequences in silico for transcription factor binding motifs.15
Results and discussion
QPD-1 (T/G single nucleotide polymorphism [SNP] ∼ 13.7 kb 5′ of PLAU)12 genotyping of previously unstudied family members confirmed that the G allele segregates with the disease, consistent with linkage to PLAU.
To screen for CNV mutations,16,17 2 QPD subjects were genotyped using Illumina 1M BeadArray and analyzed using BeadStudio. The assays failed to classify genotypes for SNPs immediately 3′ of PLAU for QPD subjects (rs2461863, rs2675666, rs2688621, and rs2633321), but not for 1304 whites tested with the same assays for another study. Visual inspection of the SNP allele signal intensity plots near PLAU in QPD (example shown in supplemental Figure 1) suggested that there were copy number abnormalities (summarized in supplemental Table 1). For example, at rs2461863, QPD samples had A-allele signal intensities similar to AA homozygous persons, but G-allele signal intensities similar to heterozygous persons (supplemental Figure 1). Real-time analysis of more QPD family members for SNPs rs2461863, rs4065, and rs1815076 (supplemental Table 2) estimated that there was one additional copy of PLAU in QPD. Real-time, dosage analyses of nonpolymorphic sequences estimated that the duplication boundaries on chromosome 10 were probably between nt positions 75 328 946 and 75 329 067 at the centromeric end and between 75 406 671 and 75 407 151 at the telomeric end (supplemental Table 3). Fluorescence in situ hybridization analysis suggested that this was a local duplication as chromosomes from both QPD subjects (who had normal karyotypes) showed normal, single-position, hybridization patterns with PLAU (10q22.2) and control chromosome 10 probes (supplemental Figure 2).
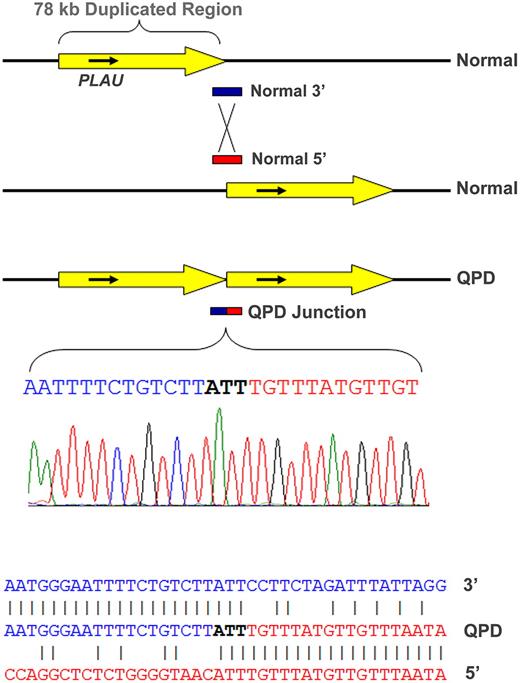
Polymerase chain reaction assays, designed to assess the duplicated segment orientation (see strategy diagram in Supplemental data), amplified a novel, 930-bp-sized fragment from QPD, but not control samples using a primer set specific for direct, tandem, PLAU duplication. The breakpoint junction endpoints (Figure 1; determined by bidirectional sequencing of this fragment from 2 representative QPD subjects and confirmed for 2 additional subjects; GenBank accessions GQ246945-8) were at nt positions 75 329 022-4 (∼ 11.87 kb 5′ of PLAU transcription start site) and 75 406 960-2 (∼ 59.69 kb 3′ of PLAU), in agreement with real-time dosage estimates (supplemental Tables 1 and 3). Alignments of normal and breakpoint sequences on chromosome 10 indicated that nonhomologous recombination had produced a direct (head-to-tail) tandem repeat of a 77 938-nt region, containing PLAU (Figure 1) and C10orf55 (unknown function; antisense to PLAU), but not other genes. There were no additional sequence changes at the junction (Figure 1) or duplication ends (not shown). Southern blotting (using probes for a region larger than previously evaluated12 ) confirmed the local duplication (supplemental Figure 3).
Tandem duplication of PLAU in subjects with QPD and alignment of the junction sequence. The duplicated, approximately 78-kb region (yellow arrow) on chromosome 10 in QPD includes PLAU and all of its characterized regulatory elements (top diagram, not to scale) and C10orf55 (not shown) on the antisense strand to PLAU, a gene of unknown function. Alignment of the breakpoint junction between the copies (below) with the normal gene sequence indicates that the QPD duplication resulted from nonhomologous recombination.
Tandem duplication of PLAU in subjects with QPD and alignment of the junction sequence. The duplicated, approximately 78-kb region (yellow arrow) on chromosome 10 in QPD includes PLAU and all of its characterized regulatory elements (top diagram, not to scale) and C10orf55 (not shown) on the antisense strand to PLAU, a gene of unknown function. Alignment of the breakpoint junction between the copies (below) with the normal gene sequence indicates that the QPD duplication resulted from nonhomologous recombination.
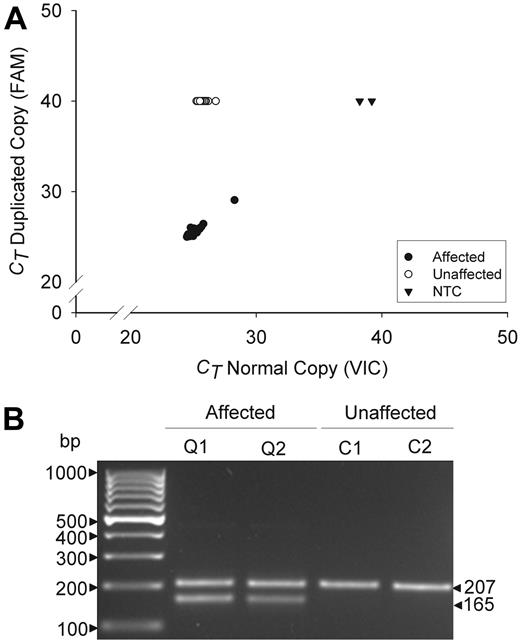
The specificity of the mutation was evaluated by multiplex, real-time, and gel fragment size assays (Figure 2). These assays detected the duplication in all 38 QPD subjects, but not in any others tested (n = 321, real time assay; n = 425, fragment size assay; P < .0001), including all unaffected family members, suggesting that the tandem duplication is probably the causative mutation.
Genotyping assays for the tandem duplication of PLAU in QPD. Panels compare data for affected (Q) and unaffected (C) QPD family members. (A) Real-time assay findings compare the cycle thresholds (CT) of the normal (VIC) and duplicated (FAM) allele copies for 38 affected and 54 unaffected family members, tested on the same run. CT values of 40 indicate an undetectable genotype. NTC indicates no template controls. (B) Multiplex fragment size assays show the 207-bp and 165-bp fragments that correspond to the normal and duplicated copies of PLAU (representative subjects shown).
Genotyping assays for the tandem duplication of PLAU in QPD. Panels compare data for affected (Q) and unaffected (C) QPD family members. (A) Real-time assay findings compare the cycle thresholds (CT) of the normal (VIC) and duplicated (FAM) allele copies for 38 affected and 54 unaffected family members, tested on the same run. CT values of 40 indicate an undetectable genotype. NTC indicates no template controls. (B) Multiplex fragment size assays show the 207-bp and 165-bp fragments that correspond to the normal and duplicated copies of PLAU (representative subjects shown).
At present, QPD is the only bleeding disorder to be associated with a gene duplication event and a PLAU mutation. Our observation that QPD is associated with one additional copy of PLAU is consistent with the modest (2- to 4-fold) but significant increases in PLAU transcripts from the disease chromosome in QPD CD34+ hematopoietic progenitor cells and saliva.12 It may prove challenging to determine why an extra copy of PLAU leads to greater increases (> 150-fold) in the disease chromosome PLAU transcripts during megakaryocyte differentiation12 and whether these increases come from the centromeric and/or telomeric copies of PLAU. The breakpoint (located upstream of all characterized PLAU regulatory elements1 and ∼11.87 kb upstream of the transcription start site of the telomeric copy) introduces motifs for Forkhead domain factors, which have roles in megakaryopoiesis,18,19 and removes binding motifs for heat shock transcription factors20 and for some transcription factors not known to influence megakaryopoiesis (supplemental Figure 4). The duplication also alters the sequences further upstream of the telomeric copy of PLAU on the QPD chromosome, and this region on the QPD and normal chromosomes contains many motifs for binding megakaryocyte-expressed transcription factors, albeit in different locations (supplemental Figure 5). It is possible that the QPD duplication mutation further increases PLAU expression during megakaryopoiesis by other mechanisms, perhaps involving alternative transcription, splicing, and/or histone binding. Whereas determining the mechanism(s) is beyond the scope of this study, the information on the genetic mutation will facilitate diagnostic testing for QPD and studies of its pathogenesis and prevalence as a cause of bleeding.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ms Francine Derome for sample collections and Mr Barry Eng, Mr Andrew McFarlane, and Dr Shilun Zheng for technical help.
This work was supported by the Canadian Institutes of Health Research (grant MOP 97942; C.P.M.H.), Heart and Stroke Foundation of Ontario (grant T5888 and Career Investigator Award; C.P.M.H.), Bayer Canada (G.E.R.), Ontario Graduate Student Scholarship (M.D.), and Canada Research Chairs in Molecular Hemostasis (C.P.M.H.) and Genetics of Complex Diseases (A.D.P.).
Authorship
Contribution: A.D.P. and C.P.M.H. supervised the project, designed experiments, interpreted results, and wrote the manuscript; C.P.M.H. and G.E.R. recruited subjects; J.M.R., B.B., J.B., I.W., M.D., J.S.W., and G.E.R. contributed to study design, result interpretation, and manuscript writing; and B.B., J.B., I.W., and M.D. also performed experimental work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine P. M. Hayward, McMaster University, MDCL 2235, 1200 Main St West, Hamilton, ON L8N 3Z5 Canada; e-mail: haywrdc@mcmaster.ca.
References
Author notes
A.D.P. and C.P.M.H. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal