Abstract
Here, we define an endothelial cell (EC) lumen signaling complex involving Cdc42, Par6b, Par3, junction adhesion molecule (Jam)–B and Jam-C, membrane type 1–matrix metalloproteinase (MT1-MMP), and integrin α2β1, which coassociate to control human EC tubulogenesis in 3D collagen matrices. Blockade of both Jam-B and Jam-C using antibodies, siRNA, or dominant-negative mutants completely interferes with lumen and tube formation resulting from a lack of Cdc42 activation, inhibition of Cdc42-GTP–dependent signal transduction, and blockade of MT1-MMP–dependent proteolysis. This process requires interdependent Cdc42 and MT1-MMP signaling, which involves Par3 binding to the Jam-B and Jam-C cytoplasmic tails, an interaction that is necessary to physically couple the components of the lumen signaling complex. MT1-MMP proteolytic activity is necessary for Cdc42 activation during EC tube formation in 3D collagen matrices but not on 2D collagen surfaces, whereas Cdc42 activation is necessary for MT1-MMP to create vascular guidance tunnels and tube networks in 3D matrices through proteolytic events. This work reveals a novel interdependent role for Cdc42-dependent signaling and MT1-MMP–dependent proteolysis, a process that occurs selectively in 3D collagen matrices and that requires EC lumen signaling complexes, to control human EC tubulogenesis during vascular morphogenesis.
Introduction
Recent work has lead to an increased understanding of how endothelial and epithelial cells make lumens and tubes in 3D extracellular matrices.1-9 Key regulators of lumen formation include Cdc42, which was first shown to regulate this process in endothelial cells (ECs),10-12 and later in epithelial cells.13,14 Components of the cell polarity machinery including Par3, Par6, and PKCζ control lumen formation of both cell types.6,11,13-15 Furthermore, we recently reported that Cdc42 activates a signaling cascade involving PKCϵ, Pak2, Pak4, Src, Yes, B-Raf, C-Raf, and Erk1/2 to control this process.12,16 In addition, EC-directed cell-surface proteolytic events through membrane type 1–matrix metalloproteinase (MT1-MMP)17,18 controls EC lumen and vascular guidance tunnel formation in 3D collagen matrices.19,20 A key question that has remained unresolved is how Cdc42-dependent signaling and MT1-MMP–dependent proteolysis are functionally coupled to regulate EC tube formation.2
Recent studies have revealed that MT1-MMP directs 3D matrix–specific events in relationship to tumor motility, cellular differentiation, and morphogenesis.2,21 We have shown that both tumor cell and EC invasion of 3D collagen matrices requires MT1-MMP, but not motility on 2D collagen substrates.19,22 Interestingly, adipocyte differentiation occurs in an MT1-MMP–dependent manner in 3D matrices but not on 2D matrix surfaces,23 and MT1-MMP controls the 3D-specific process of EC lumen and tube formation.18,19 Thus, MT1-MMP is functionally linked to critical cellular events that specifically occur in 3D matrix environments.
Here, we define the functional components of an EC lumen signaling complex that coordinates Cdc42- and MT1-MMP–dependent signal transduction events necessary for human ECs to form lumens and tubes in 3D collagen matrices. Junction adhesion molecule (Jam)–B and Jam-C are required components of these complexes as well as the polarity molecules, Par3 and Par6b, and the α2β1 integrin. Disruption of Jam-B and Jam-C function interferes with Cdc42 activation as well as its ability to form complexes with MT1-MMP to coordinate lumen and tunnel formation as well as kinase signaling necessary for EC tubulogenesis. In addition, MT1-MMP activity is necessary for Cdc42 activation in 3D collagen matrices but not on 2D collagen surfaces, and Cdc42 activation regulates MT1-MMP activity, demonstrating a critical interdependent role for these 2 molecules during EC tubulogenesis in 3D matrices.
Methods
EC lumen and tube formation in 3D collagen matrices
Human umbilical vein ECs (HUVECs) were purchased from Lonza and cultured as described.16 For vasculogenic assays, ECs were suspended as single cells or 10 to 15 cell-cell aggregates (preaggregated for 3 hours) within 3.75 mg/mL collagen type I matrices and allowed to undergo EC morphogenesis.16 A Nikon TE2000-E microscope was used for light and fluorescent microscopy with Pan-Fluor 10×, 20×, and 40× lenses with numeric apertures of 0.30, 0.45, and 0.60, respectively. A CoolSNAP HQ digital camera (Photometrics) was used with MetaMorph software (Molecular Devices) to acquire and process images.
Transfection of ECs with siRNAs
EC transfection with either siGENOME SMARTpool (Dharmacon) or Stealth Select RNAi (Invitrogen) siRNAs was carried out in growth media with 1% serum as described.16,24 Sequences of single siRNAs are shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
3D EC culture pull-down assays for components of the lumen signaling complex
EC lumen formation assays were established as described.16 Cultures were lysed at the indicated time points using cold detergent lysis buffer and incubated with S-protein agarose beads as described.11,16 In separate experiments, supernatants were incubated with GST-PAK-PBD or GST-RhoA-PBD protein beads (in the absence or presence of 100nM GDP) for 90 minutes at 4°C to assess the degree of Cdc42 or RhoA activation.11,16 The beads were washed 4 times with washing buffer. Bound active Cdc42 or RhoA proteins were detected by Western blots.
Results
Requirement for both Jam-B and Jam-C during EC lumen and tube formation in 3D collagen matrices
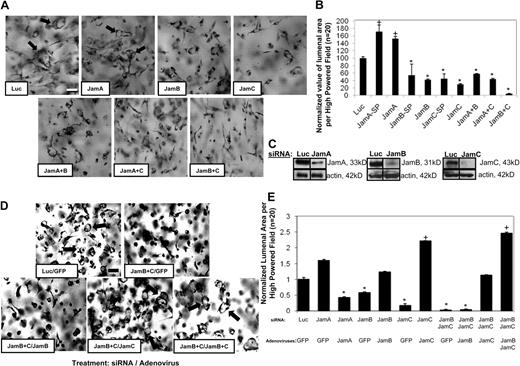
Members of the Jam family of adhesion receptors are known to interact through their cytoplasmic domains with Par3,25-28 a molecule that is required for EC lumen formation.11 We examined if siRNA suppression of Jams would affect this process. siRNA suppression of either Jam-B or Jam-C but not Jam-A (using both Smartpool mixtures and separate single siRNAs) resulted in inhibition of EC lumen formation (Figure 1) and suppression of protein expression (Figure 1C). Of interest is that Jam-A siRNA increases EC lumen formation, suggesting that Jam-A may be an inhibitor of this process. To address whether combinations of Jams might affect lumen formation, we used different combinations of Jam siRNAs. The combination of Jam-B and Jam-C siRNAs markedly blocked EC lumen and tube formation in a manner greater than either of the individual siRNAs (Figure 1B). In contrast, this did not occur when Jam-A siRNA was mixed with either Jam-B or Jam-C siRNA.
Jam-B and Jam-C are required for EC lumen and tube formation in 3D collagen matrices. (A) ECs were treated with the indicated siRNAs and were cultured in 3D collagen matrices for 24 hours before fixation, staining, and photography. Arrows indicate EC lumens. Bar equals 50 μm. SP indicates Smartpool siRNAs from Dharmacon; the other siRNAs are individual single siRNAs. Thus, we used 2 independent siRNAs for each target analyzed. In some cases, we mixed single siRNAs to assess the influence of more than 1 Jam family member. (B) EC lumenal areas were quantitated from the cultures in panel A were traced using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per high-powered field (HPF) ± SD. n = 20, P < .01. (C) siRNA-transfected EC lysates were analyzed using Western blots to assess Jam-A, Jam-B, Jam-C, or β-actin (control) expression. (D) ECs were treated with the indicated siRNAs followed by treatment with the indicated adenoviruses. Cultures were established to assess the ability of the treated ECs to form lumens and tubes for 24 hours. Arrows indicate EC lumens. Bar equals 50 μm. (E) EC lumenal areas were quantitated from the cultures in panel D were traced using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 20, P < .01. Panels B and E: * indicates a significant decrease; +, a significant increase.
Jam-B and Jam-C are required for EC lumen and tube formation in 3D collagen matrices. (A) ECs were treated with the indicated siRNAs and were cultured in 3D collagen matrices for 24 hours before fixation, staining, and photography. Arrows indicate EC lumens. Bar equals 50 μm. SP indicates Smartpool siRNAs from Dharmacon; the other siRNAs are individual single siRNAs. Thus, we used 2 independent siRNAs for each target analyzed. In some cases, we mixed single siRNAs to assess the influence of more than 1 Jam family member. (B) EC lumenal areas were quantitated from the cultures in panel A were traced using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per high-powered field (HPF) ± SD. n = 20, P < .01. (C) siRNA-transfected EC lysates were analyzed using Western blots to assess Jam-A, Jam-B, Jam-C, or β-actin (control) expression. (D) ECs were treated with the indicated siRNAs followed by treatment with the indicated adenoviruses. Cultures were established to assess the ability of the treated ECs to form lumens and tubes for 24 hours. Arrows indicate EC lumens. Bar equals 50 μm. (E) EC lumenal areas were quantitated from the cultures in panel D were traced using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 20, P < .01. Panels B and E: * indicates a significant decrease; +, a significant increase.
Rescue experiments were performed on siRNA-treated ECs using adenoviruses expressing wild-type Jam-B, Jam-C, and Jam-A (Figure 1D-E). As shown in Figure 1D-E, inhibition of lumen formation by siRNA suppression of Jam-B and Jam-C is rescued by increased expression of Jam-C or a combination of Jam-B and Jam-C expression. Jam-B alone, Jam-A alone, or green fluorescent protein (GFP) virus control fails to rescue this phenotype. Interestingly, while Jam-A siRNA treatment increases EC lumen formation, increasing expression of Jam-A leads to decreased lumen formation in these ECs. Thus, our data show that Jam-B and Jam-C control EC lumen formation, whereas Jam-A appears to be inhibitory. In this system, Jam-B and Jam-C act before multicellular EC tube assembly, and thus their influence in these assays, which mimic vasculogenesis, is not related to a lack of intercellular cell-cell junctional contacts (supplemental Videos 1-3). Time-lapse movies are shown of siRNA-treated ECs for luciferase control (supplemental Video 1), Jam-B and Jam-C (supplemental Video 2), and Jam-A (supplemental Video 3). siRNA suppression of Jam-B and Jam-C leads to complete blockade of EC lumen and tube formation in 3D collagen matrices before EC-EC contacts and multicellular tube assembly.
For comparison, we also demonstrate that siRNA suppression of Cdc42, MT1-MMP, Par3, and Par6b markedly block this process (supplemental Figure 1A) as we have previously reported.11,16,18,19 Western blot analysis of the expression of these genes over a 24-hour time course (supplemental Figure 1B) shows that Jam-B and Jam-C expression are up-regulated, while Jam-A expression is down-regulated (supplemental Figure 1C). MT1-MMP is strongly up-regulated, while the other genes show modest to no changes in expression compared with actin control.
Blocking antibodies to Jam-B and Jam-C markedly interfere with EC lumen and tube formation from single or preaggregated ECs in 3D collagen matrices
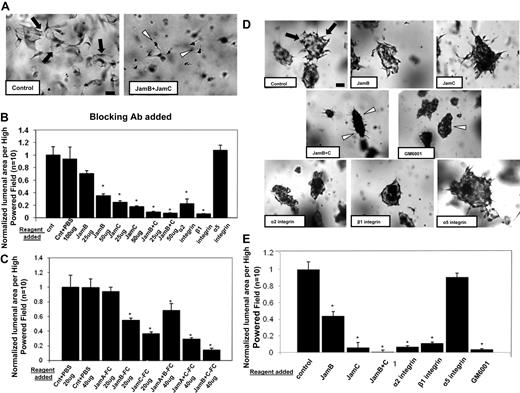
To further address the functional role of Jam-B and Jam-C during this process, blocking antibodies to these molecules were used. Neutralizing antibodies directed to Jam-B and Jam-C have a blocking influence individually, but when combined, they show dramatic blocking of EC lumen formation (Figure 2A-B). For comparison, blocking antibodies directed to the α2 and β1 integrin subunits also interfere with EC lumen formation,29 while those directed to the integrin α5 subunit have no influence (Figure 2A-B).
Blockade of Jam-B and Jam-C using neutralizing antibodies or soluble Jam-Fc proteins markedly inhibit EC lumen and tube formation from either single or preaggregated ECs suspended in 3D collagen matrices. (A) ECs were seeded within collagen matrices in the presence or absence of blocking antibodies to Jam-B and Jam-C each added at 50 μg/mL, and cultures were fixed, stained, and photographed after 24 hours. Bar equals 50 μm. Arrows indicate EC lumenal structures; arrowheads, individual ECs that do not have lumens. (B) Cultures were established as in panel A under the indicated conditions by adding the antibodies to the collagen matrices. EC lumenal areas were quantitated from the cultures using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 10, P < .01. (C) Cultures were established as in panel A except that recombinant Jam-Fc proteins were added at the indicated concentrations and conditions for 24 hours. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 10, P < .01. (D) EC-EC aggregates were suspended within 3D collagen matrices in the presence of the indicated neutralizing antibodies (50 μg/mL), the MMP inhibitor GM6001 at 5μM, or PBS control. ECs were preaggregated in a 35-mm polystyrene culture dish for 3 hours before their suspension in the collagen matrix. Arrows indicate lumen structures; arrowheads, EC aggregates with no apparent lumens. Bar equals 100 μm. (E) EC lumens were quantitated by tracing lumen areas using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 10, P < .01. * indicates a significant decrease.
Blockade of Jam-B and Jam-C using neutralizing antibodies or soluble Jam-Fc proteins markedly inhibit EC lumen and tube formation from either single or preaggregated ECs suspended in 3D collagen matrices. (A) ECs were seeded within collagen matrices in the presence or absence of blocking antibodies to Jam-B and Jam-C each added at 50 μg/mL, and cultures were fixed, stained, and photographed after 24 hours. Bar equals 50 μm. Arrows indicate EC lumenal structures; arrowheads, individual ECs that do not have lumens. (B) Cultures were established as in panel A under the indicated conditions by adding the antibodies to the collagen matrices. EC lumenal areas were quantitated from the cultures using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 10, P < .01. (C) Cultures were established as in panel A except that recombinant Jam-Fc proteins were added at the indicated concentrations and conditions for 24 hours. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 10, P < .01. (D) EC-EC aggregates were suspended within 3D collagen matrices in the presence of the indicated neutralizing antibodies (50 μg/mL), the MMP inhibitor GM6001 at 5μM, or PBS control. ECs were preaggregated in a 35-mm polystyrene culture dish for 3 hours before their suspension in the collagen matrix. Arrows indicate lumen structures; arrowheads, EC aggregates with no apparent lumens. Bar equals 100 μm. (E) EC lumens were quantitated by tracing lumen areas using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 10, P < .01. * indicates a significant decrease.
This experiment was also performed with preaggregated ECs that were suspended in 3D collagen matrices. These aggregates rapidly produce multicellular lumen structures (Figure 2D arrowheads, E), a process that is markedly blocked by the combination of Jam-B– and Jam-C–blocking antibodies. The α2β1 integrin is also required for lumen formation from aggregated cells (Figure 2D-E). Thus, Jam-B and Jam-C are required for EC lumen and tube formation from ECs suspended as single cells or as multicellular aggregates.
A further approach to address the functional role of Jam proteins is to use recombinant soluble Jam proteins, which have been reported to block various Jam-dependent cellular events.30 Soluble Jam-B or Jam-C had blocking effects, whereas soluble Jam-A had no influence on EC lumen formation (Figure 2C). Combining soluble Jam-B and Jam-C increased their blocking effects just like the influence of Jam-B and Jam-C siRNAs or blocking antibodies as shown.
siRNA suppression of Jam-B and Jam-C selectively blocks EC motility in 3D collagen matrices but not on 2D collagen substrates
Since the EC lumen and tube formation process involves EC motility,2,12 we examined if Jam-B and Jam-C affect motility during these events. Nuclear GFP-labeled ECs were treated with the indicated siRNAs and placed either in 3D collagen matrices or on 2D collagen-coated surfaces to assess motility over 24 hours. siRNA suppression of Jam-B and Jam-C selectively interferes with motility in 3D collagen matrices, whereas Jam-A siRNA treatment does not (supplemental Figure 2). In contrast, neither Jam-B and Jam-C nor Jam-A siRNA suppression affects EC motility on 2D collagen surfaces (supplemental Figure 2). Similarly, siRNA suppression of Cdc42, MT1-MMP, Par3, Par6b, and α2β1 integrin leads to selective blockade of EC motility in 3D collagen matrices and not on 2D collagen-coated surfaces. Since EC lumen and tube formation is a 3D matrix–specific process,31,32 it is of great interest that these molecules selectively affect EC behavior in 3D collagen matrices but not on 2D collagen surfaces.
The Jam-B and Jam-C cytoplasmic tails are required for EC lumen and tube formation and Cdc42 activation in 3D collagen matrices
To address the functional importance of the Jam-B and Jam-C cytoplasmic domains during these events, we generated adenoviral vectors carrying full-length and cytoplasmic tail–deleted versions of Jam-B and Jam-C. In addition, we generated a Jam-C construct with the distal-most 5 amino acids from the cytoplasmic domain deleted, which eliminates a Par3 binding site.25,26 ECs were induced to express these constructs and compared with control GFP vector (supplemental Figure 1D). Increased expression of full-length Jam-C leads to increased EC lumen formation compared with control, whereas increased full-length Jam-B expression has no effect (Figure 3A-B), and increased expression of Jam-A blocks lumen formation. Increased expression of cytoplasmic domain-deleted Jam-B or Jam-C leads to complete blockade of EC lumen formation (Figure 3A-B). A similar phenotype was observed using the 5–amino acid deleted Jam-C construct, which also strongly abrogates EC lumen formation events (Figure 3A-B), showing that the Par3-binding site of Jam-C is required for this process.
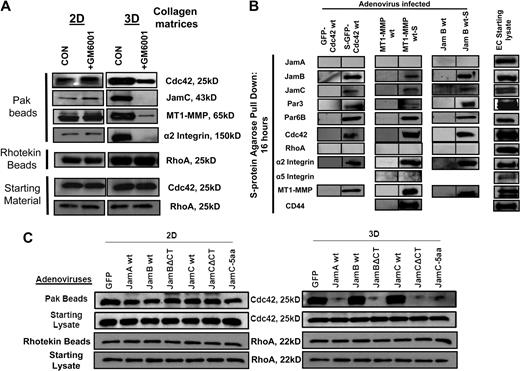
Jam-B and Jam-C cytoplasmic tails are required for EC lumen and tube formation as well as Cdc42 activation in 3D collagen matrices. (A) ECs were infected with the indicated recombinant adenoviruses overnight and then were resuspended in 3D collagen matrices. After 24 hours, cultures were fixed, stained, and photographed. Arrows indicate EC lumens. Bar equals 50 μm. (B) EC lumens were quantitated by tracing lumen areas using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 10, P < .01. (C) Cultures were established as in panel A, and after 16 hours, detergent lysates were prepared from the 3D cultures and incubated with GST-PAK-PBD protein beads as described16 to assess the degree of Cdc42 activation. Starting material lysates as well as the eluates from the Pak beads were assessed by Western blot analysis using anti-Cdc42 antibodies.
Jam-B and Jam-C cytoplasmic tails are required for EC lumen and tube formation as well as Cdc42 activation in 3D collagen matrices. (A) ECs were infected with the indicated recombinant adenoviruses overnight and then were resuspended in 3D collagen matrices. After 24 hours, cultures were fixed, stained, and photographed. Arrows indicate EC lumens. Bar equals 50 μm. (B) EC lumens were quantitated by tracing lumen areas using Metamorph software. Data shown are normalized to the control sample and presents values of lumenal area per HPF ± SD. n = 10, P < .01. (C) Cultures were established as in panel A, and after 16 hours, detergent lysates were prepared from the 3D cultures and incubated with GST-PAK-PBD protein beads as described16 to assess the degree of Cdc42 activation. Starting material lysates as well as the eluates from the Pak beads were assessed by Western blot analysis using anti-Cdc42 antibodies.
A key regulatory step in the EC lumen and tube formation process is activation of the Rho GTPase, Cdc42.11 To address whether Jam-B and Jam-C affect Cdc42 activation in 3D collagen matrices, we used the same Jam constructs to address their influence on Cdc42 activation during EC lumen formation in 3D collagen matrices. Expression of the Jam-B and Jam-C cytoplasmic tail–deleted constructs markedly block Cdc42 activation as assessed using EC lysates from 3D collagen matrix cultures at 16 hours (Figure 3C). Lysates were incubated with Pak beads to bind activated Cdc42-GTP from 3D cultures undergoing tube morphogenesis.11,16 Increased expression of Jam-A also blocks Cdc42 activation, whereas the other constructs have no effect compared with control. Starting material lysates show equal levels of Cdc42 in all cases. Thus, the cytoplasmic tails of Jam-B and Jam-C are necessary for Cdc42 activation in 3D collagen matrices, and this lack of activation directly correlates with the inability of these treated ECs to form lumens and tubes.
Jam-B and Jam-C are required for downstream kinase signaling events necessary to form EC lumens and tubes in 3D collagen matrices
We recently reported that a kinase cascade downstream of Cdc42 activation is required for EC lumen formation in 3D collagen matrices.12 siRNA suppression of Jam-B and Jam-C but not Jam-A or control luciferase results in marked blockade of signal transduction events necessary for EC lumen formation. Downstream Cdc42 effectors such as Src, Pak, Raf, and Erk kinases showed marked decreases in their phosphorylation in Jam-B and Jam-C siRNA-treated ECs during lumen formation events compared with controls (Figure 4A-B). In addition, strong increases in Erk phosphorylation occur during a time course of EC lumen and tube formation; this is markedly blocked after Jam-B and Jam-C siRNA knockdown compared with Jam-A knockdown and control.
Jam-B and Jam-C are required for Cdc42-dependent signal transduction events and vascular guidance tunnel formation during EC tubulogenesis in 3D collagen matrices. (A) ECs were treated with the indicated siRNAs and were then seeded within 3D collagen matrices to undergo tube morphogenesis. After 16 hours of culture, EC lysates were prepared and analyzed for the indicated molecules. In each case, we have analyzed for phosphorylated kinase as an indicator of kinase activation as well as the total levels for each kinase. (B) EC lysates from the different siRNA conditions were prepared at the indicated time points and were blotted for phospho-Erk1/2 as well as total Erk at the 24-hour point. (C) ECs were treated with the indicated siRNAs and were cultured in 3D collagen matrices for 24 hours. Cultures were fixed with paraformaldehyde and immunostained with an antibody to rat collagen-type I that selectively recognizes native collagen. Arrows indicate vascular guidance tunnels that represent physical spaces created by proteolysis during the EC lumen and tube formation process. Bar equals 50 μm. (D) Quantitation of vascular guidance tunnel formation from cultures described in panel C. Cultures were fixed at 16 and 24 hours, and tunnels were quantitated by tracing areas using Metamorph software. Data are shown as the average tunnel area per HPF ± SD. n = 6, P < .01. * indicates a significant decrease. (E) ECs were treated with the indicated siRNAs and cultures were established in 3D collagen matrices. After 16 hours, detergent lysates were prepared and incubated with GST-PAK-PBD beads as described.16 Eluates were analyzed for Cdc42 activation since these beads only bind Cdc42-GTP. Western blots were probed with antibodies to the indicated components of the lumen signaling complex. Starting material was probed for total Cdc42, which was equivalent in all cases.
Jam-B and Jam-C are required for Cdc42-dependent signal transduction events and vascular guidance tunnel formation during EC tubulogenesis in 3D collagen matrices. (A) ECs were treated with the indicated siRNAs and were then seeded within 3D collagen matrices to undergo tube morphogenesis. After 16 hours of culture, EC lysates were prepared and analyzed for the indicated molecules. In each case, we have analyzed for phosphorylated kinase as an indicator of kinase activation as well as the total levels for each kinase. (B) EC lysates from the different siRNA conditions were prepared at the indicated time points and were blotted for phospho-Erk1/2 as well as total Erk at the 24-hour point. (C) ECs were treated with the indicated siRNAs and were cultured in 3D collagen matrices for 24 hours. Cultures were fixed with paraformaldehyde and immunostained with an antibody to rat collagen-type I that selectively recognizes native collagen. Arrows indicate vascular guidance tunnels that represent physical spaces created by proteolysis during the EC lumen and tube formation process. Bar equals 50 μm. (D) Quantitation of vascular guidance tunnel formation from cultures described in panel C. Cultures were fixed at 16 and 24 hours, and tunnels were quantitated by tracing areas using Metamorph software. Data are shown as the average tunnel area per HPF ± SD. n = 6, P < .01. * indicates a significant decrease. (E) ECs were treated with the indicated siRNAs and cultures were established in 3D collagen matrices. After 16 hours, detergent lysates were prepared and incubated with GST-PAK-PBD beads as described.16 Eluates were analyzed for Cdc42 activation since these beads only bind Cdc42-GTP. Western blots were probed with antibodies to the indicated components of the lumen signaling complex. Starting material was probed for total Cdc42, which was equivalent in all cases.
Jam-B and Jam-C as well as Cdc42 are required for MT1-MMP–dependent vascular guidance tunnel formation during EC lumenogenesis
A fundamental step in EC lumen and tube formation is the creation of vascular guidance tunnels, which are MT1-MMP–generated matrix conduits controlling tube assembly and remodeling.19,20 siRNA suppression of Jam-B and Jam-C, Cdc42, and MT1-MMP markedly block vascular guidance tunnel formation, whereas siRNAs to Jam-A and control luciferase did not (Figure 4C-D). These data show that MT1-MMP–dependent lumen and vascular guidance tunnel formation directly involves Cdc42 as well as Jam-B and Jam-C. We show here that Jam-B and Jam-C (using cytoplasmic domain deletion constructs) are required for Cdc42 activation during lumen formation in 3D collagen matrices (Figure 3C).
To assess whether we could detect coassociated Jam or other proteins in these Pak bead eluates along with Cdc42-GTP, we performed Western blot analysis. Luciferase control culture eluates contained Jam-B and Jam-C, Par3, MT1-MMP, and the α2β1 integrin along with Cdc42-GTP, suggesting that these proteins, which control EC lumen and tube formation, are coassociated in a multicomponent complex (Figures 4E, 5B). siRNA suppression of Jam-B and Jam-C, Par3, or MT1-MMP all markedly decrease Cdc42 activation. The starting levels of Cdc42 were equivalent in all cases. Furthermore, no binding of the other components of the complex was observed, showing that their ability to be captured by Pak beads depends on Cdc42-GTP. siRNA suppression of MT1-MMP, which blocks EC lumen and tube formation, also resulted in markedly decreased Cdc42 activation, suggesting that MT1-MMP proteinase activity might be required for EC Cdc42 activation in 3D collagen matrices. The reverse situation appears to be true in that knockdown of Cdc42 strongly blocks vascular guidance tunnel formation, a process that requires MT1-MMP proteinase activity (Figure 4C-D).19
MT1-MMP activity and the Jam-B and Jam-C cytoplasmic tails are necessary for Cdc42 activation in 3D collagen matrices but not on 2D collagen surfaces. (A) EC cultures were established on 2D collagen substrates or within 3D collagen matrices in the presence or absence of 5μM GM6001 and otherwise were cultured under the same conditions. The ECs on 2D collagen substrates were seeded in a subconfluent state to mimic the lack of cell-cell contacts observed in our assays that mimic vasculogenesis in 3D collagen matrices. Cell lysates were prepared at 16 hours in such a manner that the volume of lysis buffer used in relation to total cell number was identical in both cases. Lysates were incubated from the different culture conditions with either GST-PAK-PBD (to bind Cdc42-GTP) or Rhotekin beads (to bind RhoA-GTP) to assess the level of either activated Cdc42 or RhoA. Starting material lysates were also probed on Western blots for either Cdc42 or RhoA. (B) ECs were infected with the indicated recombinant adenoviruses carrying GFP-Cdc42, S-GFP-Cdc42, MT1-MMP, MT1-MMP-S, Jam-B wt, and Jam-B wt-S. ECs were then cultured in 3D collagen matrices for 16 hours. At this time point, detergent lysates from the 3D cultures were prepared and incubated with S-protein agarose beads to selectively bind S-tagged recombinant proteins and their associated proteins as described.16 Eluates were probed on Western blots for the indicated proteins. EC starting lysates were from control 3D cultures at 16 hours to assess whether the proteins examined were present at this stage of tube formation. (C) ECs were infected with the indicated wild-type and mutant Jam viruses as well as control GFP virus, and the cells were seeded on 2D collagen surfaces or within 3D collagen matrices for 16 hours. At this time, detergent lysates were prepared and incubated with GST-PAK-PBD beads to assess the degree of Cdc42 activation. Eluates were probed on Western blots for Cdc42 and starting lysates were similarly examined for Cdc42 levels.
MT1-MMP activity and the Jam-B and Jam-C cytoplasmic tails are necessary for Cdc42 activation in 3D collagen matrices but not on 2D collagen surfaces. (A) EC cultures were established on 2D collagen substrates or within 3D collagen matrices in the presence or absence of 5μM GM6001 and otherwise were cultured under the same conditions. The ECs on 2D collagen substrates were seeded in a subconfluent state to mimic the lack of cell-cell contacts observed in our assays that mimic vasculogenesis in 3D collagen matrices. Cell lysates were prepared at 16 hours in such a manner that the volume of lysis buffer used in relation to total cell number was identical in both cases. Lysates were incubated from the different culture conditions with either GST-PAK-PBD (to bind Cdc42-GTP) or Rhotekin beads (to bind RhoA-GTP) to assess the level of either activated Cdc42 or RhoA. Starting material lysates were also probed on Western blots for either Cdc42 or RhoA. (B) ECs were infected with the indicated recombinant adenoviruses carrying GFP-Cdc42, S-GFP-Cdc42, MT1-MMP, MT1-MMP-S, Jam-B wt, and Jam-B wt-S. ECs were then cultured in 3D collagen matrices for 16 hours. At this time point, detergent lysates from the 3D cultures were prepared and incubated with S-protein agarose beads to selectively bind S-tagged recombinant proteins and their associated proteins as described.16 Eluates were probed on Western blots for the indicated proteins. EC starting lysates were from control 3D cultures at 16 hours to assess whether the proteins examined were present at this stage of tube formation. (C) ECs were infected with the indicated wild-type and mutant Jam viruses as well as control GFP virus, and the cells were seeded on 2D collagen surfaces or within 3D collagen matrices for 16 hours. At this time, detergent lysates were prepared and incubated with GST-PAK-PBD beads to assess the degree of Cdc42 activation. Eluates were probed on Western blots for Cdc42 and starting lysates were similarly examined for Cdc42 levels.
MT1-MMP activity as well as the Jam-B and Jam-C cytoplasmic tails are required for Cdc42 activation in 3D collagen matrices but not on 2D collagen surfaces
To address the apparent interdependent role of MT1-MMP and Cdc42 signaling during these events, we performed an experiment whereby ECs were seeded within 3D collagen matrices or on 2D collagen surfaces to determine whether MT1-MMP activity influences the degree of Cdc42 activation. We show that MT1-MMP activity is only required for EC motility and lumen formation in 3D collagen matrices,18,19 and does not affect EC migratory behavior on 2D collagen surfaces (supplemental Figure 2). siRNA suppression of Cdc42 as well as Jam-B and Jam-C show the same phenotype with a 3D matrix–specific effect (supplemental Figure 2). EC cultures were established in 3D collagen matrices or on 2D collagen surfaces in the presence or absence of GM6001, a broad-spectrum MMP inhibitor that blocks MT1-MMP (Figure 5A). Lysates were prepared at 16 hours of culture to assess Cdc42 activation, which controls EC lumen formation, and RhoA activation, which does not control this process.2,10,11 As shown in Figure 5A, Cdc 42 activation is observed at higher levels in the 3D versus 2D cultures, and a strong association of Jam-C, MT1-MMP, and the α2β1 integrin are detected in the Pak bead eluates. In the presence of GM6001, there is a strong decrease in Cdc42 activation (and markedly decreased capture of Jam-C, MT1-MMP, and α2β1 integrin), which selectively occurs in the 3D cultures but not the 2D cultures, an effect mimicked by siRNA suppression of MT1-MMP (Figure 4E). This does not occur when RhoA activation is assessed. There is increased RhoA activation in 3D versus 2D culture, but there is no influence of GM6001 addition. These data strongly suggest that MT1-MMP activity controls Cdc42 activation and vice versa in 3D collagen matrices, revealing a critical interdependent signaling role for these molecules during these events.
As we show in Figure 3, the Jam-B and Jam-C cytoplasmic tails are necessary for both EC lumen formation and Cdc42 activation. To address if this effect occurred selectively in 3D collagen matrices, we directly compared the influence of wild-type and Jam mutant expression in ECs that were either seeded on 2D collagen surfaces or within 3D collagen matrices (Figure 5C). After 16 hours of culture, lysates were prepared and Cdc42 activation was assessed. Expression of cytoplasmic tail–deleted Jam-B and Jam-C blocks Cdc42 activation selectively in 3D collagen matrices but not on 2D collagen surfaces compared with control conditions (Figure 5C), whereas these constructs have no influence on RhoA activation in either case.
To further address the role of MT1-MMP activity in Cdc42 activation within 3D collagen matrices, we expressed wild-type or catalytically dead (EA mutant) MT1-MMP full-length constructs19 as well as cytoplasmic domain deletion mutants and performed EC lumen formation and Cdc42 activation assays. Each of the constructs also contains a C-terminal S-tag, which allows the recombinant protein to bind S-protein agarose beads from lysates (Figure 5B). As shown in Figure 6A, a time-lapse movie analysis of EC lumen formation over 24 hours reveals that expression of full-length wild-type MT1-MMP increases the rate of lumen formation, whereas expression of the cytoplasmic domain-deleted wild-type construct markedly increases this rate. We analyzed surface expression of MT1-MMP under these conditions by immunostaining (2D vs 3D) and immunoprecipition of biotinylated surface proteins (supplemental Figure 3). The wild-type cytoplasmic domain-deleted construct shows increased cell-surface expression compared with the wild-type full-length construct, which accounts in part for the increased lumen formation observed. The EA mutant full-length construct does not enhance formation compared with control GFP, whereas the EA mutant cytoplasmic tail–deleted construct strongly inhibits EC lumen and tube formation (Figure 6B). This latter mutant shows dominant-negative functions in blocking lumen formation. Each of these constructs is expressed as they bind S-protein agarose beads from lysates obtained from 3D cultures at 16 hours (Figure 5C). These results were confirmed by assessing vascular guidance tunnel formation where increasing wild-type MT1-MMP expression increases tunnel formation, which was further increased using the wild-type cytoplasmic tail–deleted construct (Figure 6B). In contrast, the EA mutant cytoplasmic tail–deleted construct blocked vascular guidance tunnel formation consistent with its ability to block EC lumen formation and to serve as a dominant-negative MT1-MMP mutant.
MT1-MMP activity controls Cdc42 activation as well as EC lumen and vascular guidance tunnel formation. (A) ECs were infected with the indicated recombinant adenoviruses, which were GFP control, MT1-MMP full-length (FL), MT1-MMP FL with the EA mutant which makes the enzyme catalytically inactive, MT1-MMP without its cytoplasmic tail (tailless or ΔCT), and MT1-MMP EA mutant without its cytoplasmic tail. ECs were seeded in 3D collagen matrices, and real-time video analysis was performed to quantitatively measure lumen formation over the indicated time course. EC lumen areas were determined at each time point (n = 10, ± SD). (B) Cultures at 24 hours were fixed with paraformaldehyde and stained with anti–collagen type I antibodies to assess vascular guidance tunnel formation. Arrows indicate vascular guidance tunnels. Bar equals 100 μm. (C) ECs were infected with the indicated recombinant adenoviruses and cultures were established in 3D collagen matrices. After 16 hours, detergent lysates were prepared and incubated with S-protein agarose beads as described16 to assess whether the indicated components of the lumen signaling complex could be found to be coassociated with MT1-MMP. In addition, the lysates were incubated with GST-PAK-PBD beads to assess the degree of Cdc42 activation. In each case, the starting material lysates were examined for total Cdc42 levels.
MT1-MMP activity controls Cdc42 activation as well as EC lumen and vascular guidance tunnel formation. (A) ECs were infected with the indicated recombinant adenoviruses, which were GFP control, MT1-MMP full-length (FL), MT1-MMP FL with the EA mutant which makes the enzyme catalytically inactive, MT1-MMP without its cytoplasmic tail (tailless or ΔCT), and MT1-MMP EA mutant without its cytoplasmic tail. ECs were seeded in 3D collagen matrices, and real-time video analysis was performed to quantitatively measure lumen formation over the indicated time course. EC lumen areas were determined at each time point (n = 10, ± SD). (B) Cultures at 24 hours were fixed with paraformaldehyde and stained with anti–collagen type I antibodies to assess vascular guidance tunnel formation. Arrows indicate vascular guidance tunnels. Bar equals 100 μm. (C) ECs were infected with the indicated recombinant adenoviruses and cultures were established in 3D collagen matrices. After 16 hours, detergent lysates were prepared and incubated with S-protein agarose beads as described16 to assess whether the indicated components of the lumen signaling complex could be found to be coassociated with MT1-MMP. In addition, the lysates were incubated with GST-PAK-PBD beads to assess the degree of Cdc42 activation. In each case, the starting material lysates were examined for total Cdc42 levels.
Lysates prepared from the 3D cultures were incubated with either Pak beads to assess the degree of Cdc42 activation or separately with S-protein beads to assess the association of Cdc42, Jam-C, or α2β1 with the different MT1-MMP constructs (Figure 6C). Expression of the dominant-negative MT1-MMP construct shows no lumen or vascular guidance tunnel formation and, importantly, blocks Cdc42 activation compared with the other MT1-MMP constructs. This result provides additional key evidence that MT1-MMP activity is required for Cdc42 activation in 3D collagen matrices to coordinate EC lumen formation. In addition, the dominant-negative MT1-MMP mutant does not associate with Cdc42, Jam-C, or α2β1 in contrast to the other constructs, where clear binding of these molecules is demonstrated. The binding of these associated molecules to S-protein beads depends on the S-tag, because MT1-MMP constructs without this tag do not show these associations (Figure 5B). Increasing MT1-MMP activity using the wild-type cytoplasmic tail–deleted mutant reveals an even greater degree of association of MT1-MMP with α2β1, Jam-C, and Cdc42 (Figure 6C), which correlates with increased lumen and vascular guidance tunnel formation (Figure 6A-B). In addition, this result demonstrates that the cytoplasmic tail of MT1-MMP is not required for its association with this lumen signaling complex of proteins.
Jam-B and Jam-C coordinate a lumen signaling complex involving Cdc42, Par6b, Par3, MT1-MMP, and the α2β1 integrin to control EC lumen and tube formation in 3D collagen matrices
To address how Jam-B and Jam-C control EC lumen and tube formation, we have performed experiments to assess whether these adhesion receptors are critical regulatory components of a multimolecular lumen signaling complex. Cdc42, Jam-B and Jam-C, MT1-MMP, and α2β1 coassociate in complexes that directly correlate with the ability of ECs to form lumens and tubes (Figures 4E,5A). S-tagged versions of Cdc42, MT1-MMP, and Jam B are able to bind each of the key components of the lumen signaling complex, including Cdc42, Par3, Par6b, Jam-B, Jam-C, MT1-MMP, and α2β1, using lysates prepared from 3D collagen matrix cultures at 16 hours (Figure 5B). We do not observe binding of Jam-A, RhoA, or the α5β1 integrin within this complex (Figure 5B). Previous work showed that MT1-MMP was found to be associated with the hyaluronate receptor, CD44,33 and we confirm this interaction (Figure 5B). These associations are only observed using the S-tagged recombinant proteins and not with expressed untagged protein using S-protein agarose beads (Figure 5B).
To further address the specificity of these binding experiments, they were performed under control conditions or with GDP, which does not allow Cdc42 to interact with its effectors. The indicated recombinant proteins were expressed in ECs, and lysates were made from 3D collagen cultures at 16 hours and were incubated with Pak beads, Rhotekin beads, or S-protein agarose beads. The binding of each of the members of the lumen signaling complex to Pak beads (which captures active Cdc42-GTP) is blocked by the presence of GDP, showing the specificity of the interactions shown (supplemental Figure 4). Similar results are shown with RhoA, where its binding to Rhotekin beads also does not occur in the presence of GDP. Under control conditions where RhoA binds, there is no coassociation of MT1-MMP or Cdc42 (supplemental Figure 4). The specificity of S-protein agarose beads is shown in supplemental Figure 4, where cocapture of the lumen signaling complex only occurs when constructs are expressed with S-tags; these interactions do not occur with GDP either. One exception to this result is that there is equivalent binding of α2β1 integrin to MT1-MMP-S in the presence of GDP, showing that this interaction does not depend on GTP. These data demonstrate that the lumen signaling complex consisting of Cdc42-GTP, Par6b, Par3, Jam-B, Jam-C, MT1-MMP, and α2β1 integrin shows appropriate binding specificity to the input beads and demonstrate that coassociation of these molecules directly correlates with the ability of ECs to form lumens and tubes in 3D collagen matrices. To further confirm this conclusion, we performed experiments showing that MT1-MMP and Jam-C are colocalized in ECs with lumens as assessed by staining nonpermeabilized ECs in 3D collagen matrices (supplemental Figure 5).
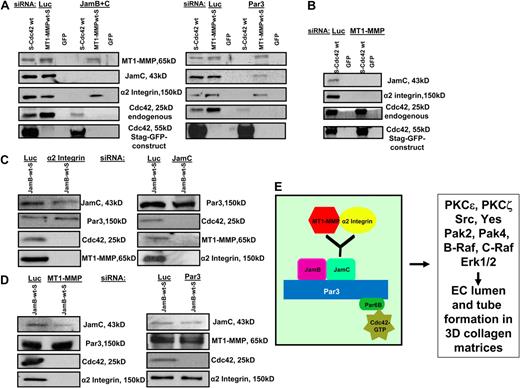
An additional series of experiments was performed to elucidate how this complex is organized to control EC lumen and tube formation. We combined siRNA suppression with expression of recombinant S-tagged Cdc42, Jam-B, and MT1-MMP or control GFP virus to identify how the lumen signaling complex is organized. Jam-B and Jam-C siRNA suppression compared with control luciferase allows for the binding of the expressed S-tagged constructs to S-protein agarose beads as expected, but no binding of Jam-C, MT1-MMP, or α2β1 integrin to S-GFP-Cdc42 (Figure 7A). Similarly, Jam-B and Jam-C knockdown eliminates the coassociation of MT1-MMP-S to endogenous Cdc42, but does not affect its binding to α2β1. There is no binding of these molecules to S-protein agarose beads when control GFP virus is used. siRNA suppression of Par3 eliminates the binding of Jam-C, MT1-MMP, and α2β1 integrin to S-GFP-Cdc42, but has no influence on the interaction of these molecules to MT1-MMP-S, showing that Par3 is proximal to Cdc42 in this multiprotein complex. siRNA suppression of MT1-MMP eliminated the ability of S-GFP-Cdc42 to associate with either Jam-C or α2β1 integrin, which provides further support for the concept that MT1-MMP activity is necessary for Cdc42 activation and its ability to couple to this complex (Figure 7B). Additional experiments used Jam-B S-tag to investigate its associations during the lumen formation process. As shown in Figure 7C, siRNA suppression of the α2 integrin subunit allows for the cocapture of Jam-C and Par3 with Jam-B, but there is no cocapture of Cdc42 or MT1-MMP. These interesting data indicate that despite association of Jam-B and Jam-C with Par3, there is still uncoupling of the MT1-MMP with Cdc42, resulting in blockade of EC lumen formation. siRNA suppression of MT1-MMP shows the same effect as α2 integrin siRNA suppression, where the Jam-B S-tag is able to pull down Jam-C and Par3 but not Cdc42 or α2β1 integrin. These data suggest that MT1-MMP and the α2β1 integrin appear functionally interdependent, and siRNA suppression of either molecule does not allow for coupling of either of these 2 molecules with Jam-C. In support of this conclusion, siRNA suppression of Jam-C completely eliminates the association of Jam-B with MT1-MMP and α2β1 integrin (Figure 7C), suggesting that Jam-C is coupled to this proteinase/integrin complex and is in between Jam-B and this complex. In addition, siRNA suppression of Par3 disconnects Cdc42 from the Jam-B S-tag, but does not affect Jam-B's association with Jam-C, MT1-MMP, or α2β1 integrin (Figure 7D). This further supports the idea that Jam-C interacts with MT1-MMP and α2β1 complexes. A schematic diagram illustrating the organization of the EC lumen signaling complex is shown in Figure 7E. This EC lumen signaling complex, which contains Jam-B and Jam-C, coordinates integrin signaling, MT1-MMP proteolytic events, and Cdc42-GTP–dependent signaling events to control vascular tube morphogenesis in 3D collagen matrices.
Lumen signaling complexes containing Cdc42, Par3, Par6b, Jam-B, Jam-C, MT1-MMP, and α2β1 integrin control EC lumen and tube formation in 3D collagen matrices. (A-D) ECs were siRNA-treated for Luciferase (control), Jam-B plus Jam-C, Par3, MT1-MMP, α2 integrin, or Jam-C alone and transfected with MT1-MMP-S, S-GFP-Cdc42, or JamB wt-S adenoviral vectors. They were suspended in 3D collagen matrices and allowed to undergo morphogenesis for 16 hours, followed by the preparation of detergent lysates and incubation with S-protein agarose beads as described.16 Eluates were analyzed for MT1-MMP, Cdc42, Jam-B, Jam-C, α2 integrin, Par3, and Par6b. (E) Schematic diagram showing the interactive relationships of the lumen signaling complex which controls EC lumen and tube formation in 3D collagen matrices.
Lumen signaling complexes containing Cdc42, Par3, Par6b, Jam-B, Jam-C, MT1-MMP, and α2β1 integrin control EC lumen and tube formation in 3D collagen matrices. (A-D) ECs were siRNA-treated for Luciferase (control), Jam-B plus Jam-C, Par3, MT1-MMP, α2 integrin, or Jam-C alone and transfected with MT1-MMP-S, S-GFP-Cdc42, or JamB wt-S adenoviral vectors. They were suspended in 3D collagen matrices and allowed to undergo morphogenesis for 16 hours, followed by the preparation of detergent lysates and incubation with S-protein agarose beads as described.16 Eluates were analyzed for MT1-MMP, Cdc42, Jam-B, Jam-C, α2 integrin, Par3, and Par6b. (E) Schematic diagram showing the interactive relationships of the lumen signaling complex which controls EC lumen and tube formation in 3D collagen matrices.
Discussion
In this work, we present the novel concept that a lumen signaling complex exists controlling the ability of ECs to form tubular networks in 3D collagen matrices. This complex of proteins consists of Cdc42, Par6b, Par3, Jam-B, Jam-C, MT1-MMP, and the α2β1 integrin, and together they control the lumen and tube formation process by coordinating integrin recognition and localized MT1-MMP–dependent degradation of collagen matrix with Cdc42 activation-dependent signaling necessary for EC tube and network formation (Figure 7E). This complex stimulates a 3D matrix–specific lumen formation process in collagen matrices that reveals a fundamental interdependent role for Cdc42- and MT1-MMP–dependent signaling during these events. MT1-MMP activity is required for Cdc42 activation in 3D collagen matrices but not on 2D collagen surfaces, while Cdc42 is required for MT1-MMP–dependent proteolytic events in 3D matrices, a necessary step for lumen and coincident vascular guidance tunnel formation.
Jam-B and Jam-C control EC lumen and tube formation in 3D collagen matrices
We report the novel observation that Jam-B and Jam-C are critical regulators of EC lumen and tube formation in 3D collagen matrices. Previous work with Jam proteins suggest that they are primary regulators of tight junctional contacts and coordinate polarity signaling through interactions with Par3 and their cytoplasmic tails.25,34 Each of the Jams, Jam-A, Jam-B, and Jam-C, can bind Par3 through a binding site on the C-terminus of the tail. The ability of Par3 to bind Par6 allows for coupling of these receptors to Cdc42, a key GTPase known to affect cell polarization events, including apical-basal polarity and directed cell motility.35,36 Importantly, we first reported that Cdc42 controlled lumen formation of ECs in 3D matrices.10 Our work and that of others has shown that Par3 is involved in lumen formation of both endothelial cells and epithelial cells in 3D matrices.10,13 A key distinction in our systems versus the epithelial cell models is that the blocking influence of Par3 can be observed before multicellular tube assembly. During vasculogenesis, individual ECs interconnect through sprouting and early lumen formation events,1,9,37 such as intracellular vacuolation, to assemble into multicellular tube networks. Knockdown of Par3 or Par6b markedly blocks the ability of ECs to undergo these critical early events that are necessary to reach their neighbors and assemble into multicellular tube networks.11 Our results reveal the same phenomenon with blockade of Jam-B and Jam-C. However, it is likely that the Jams as well as Par3 and Par6 proteins also play a role in the process of EC multicellular tube assembly in order for ECs to be properly spatially organized within a tube structure. There is precedent for our results with single ECs in that Jam-A knockout neutrophils are unable to directionally migrate as single cells in response to chemotactic cues.38,39 To address the issue of multicellular tube assembly, we showed that blocking antibodies to Jam-B and Jam-C markedly block lumen formation from either single ECs or preaggregated ECs in 3D collagen matrices (Figure 2), suggesting that the signaling mechanisms controlling lumen formation under these different conditions appear similar.
There is prior work revealing a role for Jam-C in angiogenic responses in that Jam-C blocking antibodies, when administered to mice, were able to interfere with angiogenesis.40 The mechanisms underlying the influence of these antibodies were unclear. Our studies here show a direct role for both EC Jam-B and Jam-C in controlling human EC tubular morphogenesis in 3D collagen matrices. The systems used here have previously identified critical regulators of EC lumen and tube formation1,10-12,18-20,29,31,32,41 that have been demonstrated to be relevant during vessel morphogenesis in vivo. This includes the identification of MT1-MMP as a critical regulator of angiogenesis in adult mice;17,42 the identification of Pak4, a Cdc42-specific effector, and Rac1 as both required for developmental vascularization;43,44 the finding that Src, Pak, and Raf regulate angiogenesis;45 the identification of CCM2 as a regulator of early aortic arch lumen development and EC lumen formation in vitro;46 and an important role for α2β1 and β1 integrins and Par3 during developmental and adult vascularization events.15,47,48
Lumen signaling complexes involving Cdc42, Par3, Jam-B and Jam-C, and MT1-MMP control EC tubulogenesis in a 3D matrix–specific manner
In this study, we present data showing that a lumen signaling complex exists in ECs to control the 3D matrix–specific process of EC lumen and tube formation. We make the novel observation that Cdc42 activation is controlled by MT1-MMP proteolytic activity in 3D collagen matrices but not on 2D collagen matrix surfaces. The coupling of MT1-MMP to Cdc42 activation and signaling through the lumen signaling complex provides an explanation for why MT1-MMP is so critical for EC tubulogenesis in 3D collagen matrices. In addition, Cdc42 activation is necessary for MT1-MMP to proteolyze collagen matrices, a required step during EC tube and vascular guidance tunnel formation. Thus, Cdc42 and MT1-MMP are interdependent molecules controlling EC lumen and tube formation, and Jam-B and Jam-C as well as integrins represent bridging molecules that facilitate their organization so that they are physically and functionally coupled during vascular tube morphogenesis in 3D extracellular matrices.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants HL79460, HL59373, and HL87308 to G.E.D.
National Institutes of Health
Authorship
Contribution: A.S. designed and performed research, analyzed data, and wrote the paper; W.K. performed research and analyzed data; A.N.S. performed research, analyzed data, and wrote the paper; A.M.M. analyzed data; K.E.F. performed research; and G.E.D. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: George E. Davis, Mulligan Professor of Medical Research, University of Missouri School of Medicine, MA415 Medical Sciences Bldg, Columbia, MO 85212; e-mail: davisgeo@health.missouri.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal