Abstract
The impact of WT1 mutations in acute myeloid leukemia (AML) is not completely settled. We aimed to determine the clinical implication of WT1 mutation in 470 de novo non-M3 AML patients and its stability during the clinical course. WT1 mutations were identified in 6.8% of total patients and 8.3% of younger patients with normal karyotype (CN-AML). The WT1 mutation was closely associated with younger age (P < .001), French-American-British M6 subtype (P = .006), and t(7;11)(p15;p15) (P = .003). Multivariate analysis demonstrated that the WT1 mutation was an independent poor prognostic factor for overall survival and relapse-free survival among total patients and the CN-AML group. A scoring system incorporating WT1 mutation, NPM1/FLT3-ITD, CEBPA mutations, and age into survival analysis proved to be very useful to stratify CN-AML patients into different prognostic groups (P < .001). Sequential analyses were performed on 133 patients. WT1 mutations disappeared at complete remission in all WT1-mutated patients studied. At relapse, 3 of the 16 WT1-mutated patients who had paired samples lost the mutation and 2 acquired additional mutations, whereas 3 of 110 WT1-wild patients acquired novel mutations. In conclusion, WT1 mutations are correlated with poor prognosis in AML patients. The mutation status may be changed in some patients during AML progression.
Introduction
The Wilms' Tumor 1 (WT1) gene, encoding a zinc-finger transcription factor, is physiologically expressed in embryonic kidney cells1 and hematopoietic stem cells.2,3 WT1 was first identified as a tumor suppressor gene in patients with the WAGR (Wilms tumor, aniridia, genitourinary abnormalities, and mental retardation) tumor predisposition syndrome.4 The WT1 gene is demonstrated to be overexpressed in various leukemias, particularly acute myeloid leukemia (AML), as well as other cancers,5,6 and thus is suggested to be an oncogene.7,8 On the other hand, mutations in WT1 gene are found in approximately 10% of AML patients with hotspots in the 4 Cys-His zinc finger domains on exons 7 and 9.6,9 As a result, WT1 as a tumor suppressor gene is suggested.6,9 The precise role of WT1 in leukemogenesis remains to be defined.10
Several reports addressed that WT1 mutation was an independent poor risk factor for overall survival (OS) in cytogenetically normal AML (CN-AML) patients.11-13 Accompanied with other mutations, such as FLT3-ITD, WT1 mutations were associated with failure of standard induction chemotherapy.14 However, different results were also reported by other groups.15,16 Most studies on WT1 mutations were restricted to CN-AML, and sequential analyses to evaluate the stability of this gene mutation during the clinical course were limited to a small number of patients. In this study, we delineated WT1 mutation in 470 de novo non-M3 AML patients, both cytogenetically normal and abnormal, and investigated its association with other gene alterations. Sequential analysis of WT1 mutation during the clinical course was also performed on 133 patients to investigate the stability and pathogenic role of this mutation in AML. Further, to better stratify CN-AML patients into different risk groups,15,17 a simple scoring system integrating WT1 mutations with other gene alterations in survival analysis was proposed.
Methods
From November 1995 to March 2007, a total of 470 adult patients 15 years of age or older who were newly diagnosed as having de novo non-M3 AML at National Taiwan University Hospital (NTUH) and had cryopreserved bone marrow samples for genetic studies were enrolled. Patients with antecedent hematologic diseases, therapy-related AML, or acute promyelocytic leukemia (M3 subtype) were excluded because the pathogenesis of their leukemias and survival of these patients differ from other AML patients significantly. Among 470 patients, 330 (70.2%) patients received intensive induction chemotherapy (idarubicin 12 mg/m2 per day on days 1-3 and cytarabine 100 mg/m2 per day on days 1-7) and then consolidation chemotherapy with 2 to 4 courses of high-dose cytarabine (2000 mg/m2 every 12 hours on days 1-4, total 8 doses), with or without an anthracycline if complete remission (CR) was achieved. The remaining 140 patients received low-dose chemotherapy and/or supportive care because of the poor performance status or the patients' will. Ninety-six patients received allogeneic hematopoietic stem cell transplantation (HSCT). This study was approved by the Institutional Review Board of the NTUH, and written informed consent was obtained from all participants in accordance with the Declaration of Helsinki.
Cytogenetics
Bone marrow cells were harvested directly or after 1- to 3-day unstimulated culture as previously described.18 Metaphase chromosomes were banded by the trypsin-Giemsa method and karyotyped according to International System for Human Cytogenetic Nomenclature.
Mutation analysis
Mutation analysis of WT1 exons 7 and 9 was performed by genomic DNA polymerase chain reaction (PCR) and direct sequencing as previously reported.14 The primers used were as follows: 7F 5′ GACCTACGTGAATG TTCACATG-3′ and 7R 5′-ACCAACACCTGGATCAGACCT-3′; 9F 5′-TGCA GACATTGCAGGCATGGCAGG-3′ and 9R 5′-GCACTATTCCTTCTCTAACT GAG-3′. Abnormal sequencing results were confirmed by at least 2 repeated analyses. Sequential analysis of WT1 mutation during the clinical course was performed in 309 samples from 133 patients. Mutation analyses of 11 other relevant molecular marker genes (NPM1,19 CEBPA,20 FLT3/ITD and FLT3/TKD,19 N-RAS,21 K-RAS,21 JAK2,21 KIT,22 MLL/PTD,23 AML1/RUNX1,24 and PTPN1125 ) were performed as previously described.
GeneScan analysis
Percentage of WT1 mutant was determined using PCR and fragment analysis as previously reported.26 Two pairs of primers covering exon 7 and exon 9 were designed using PrimerQuest software (Integrated DNA Technologies). Each one of the primer pairs was labeled with appropriate fluorescence. The primers used for exon 7 were FAM-5′-ttactctctgcctgcaggatgtgcgac-3′ and 5′-agcgggcacacttaccagt-3′, and those for exon 9 were VIC-5′-gccgaggctagaccttctct-3′ and 5′-tccaatccctctcatcacaa-3′, which would produce amplicons with sizes of 183 bp and 203 bp, respectively, for wild-type WT1. The amplified products were subsequently diluted in distilled water and mixed with deionized formamide and GeneScan 500 LIZ Size Standard (Applied Biosystems). These mixtures were then electrophoresed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). After electrophoresis, the fluorescence signals were analyzed using GeneScan, Volume 3.1 software (Applied Biosystems).
Taq polymerase–amplified cloning analysis
For patients with double WT1 mutations, Taq polymerase–amplified cloning was performed to determine whether the 2 mutations were in the same or different alleles as previously described.20 Briefly, the DNA was amplified to cover both mutations, the PCR products were then cloned into the Taq polymerase-amplified cloning vector pGEM-T Easy (Promega), and 10 clones were selected for sequencing.
Immunophenotype analysis
A panel of monoclonal antibodies to myeloid-associated antigens, including CD13, CD33, CD11b, CD15, CD14, and CD41a, lymphoid-associated antigens, including CD2, CD5, CD7, CD19, CD10, and CD20, and lineage-nonspecific antigens HLA-DR, CD34, and CD56 was used to characterize the phenotypes of the leukemia cells, as previously described.19
Statistical analysis
The discrete variables of patients with and without gene mutation were compared using the χ2 tests; but if the expected values of contingency tables were smaller than 5, Fisher exact test was used. Mann-Whitney tests were used to compare continuous variables and medians of distributions if the continuous data were not normally distributed. OS was measured from the date of first diagnosis to death from any cause, and relapse-free survival (RFS) was calculated from the time of CR until relapse, death, or end of study. Kaplan-Meier estimation was used to plot survival curves, and log-rank tests were used to test the difference between groups. Multivariate Cox proportional hazard regression analysis was used to investigate independent prognostic factors for OS and RFS. The proportional hazards assumption (constant hazards assumption) was examined using time-dependent covariate Cox regression before conducting multivariate Cox proportional hazard regression. The variables, including age, white blood cell (WBC), karyotype, and mutations of CEBPA, WT1, and NPM1/FLT3-ITD, were used as covariates. Those patients who received HSCT were censored at the time of HSCT in survival analysis to ameliorate the influence of this treatment. A P value less than .05 was considered statistically significant. All statistical analyses were performed with the SPSS, Version 15 software (SPSS Inc) and Statsdirect.
Results
WT1 mutations in 470 adult patients with de novo non-M3 AML
Mutation screening of the coding and intron-exon boundaries of WT1 exons 7 and 9 was performed using a direct sequencing method on DNA samples from 470 de novo AML patients. Overall, 29 different kinds of WT1 mutations were detected in 32 patients (6.8%, Table 1), including 26 on exon 7 and 3 on exon 9. Of them, 4 mutations (V382fsx385, N381fsx450, Q380fsx384, and S381X) occurred in more than one patient (nos. 1 and 7, 4 and 5, 6 and 16, and 9, 28, and 31, respectively).
Mutation patterns in 32 patients with WT1 mutations at diagnosis
| UPN . | Age, y/sex . | FAB . | Karyotype . | WT1 mutation . | Other gene mutations detected . | ||
|---|---|---|---|---|---|---|---|
| Location . | DNA change . | Protein change . | |||||
| 1 | 30/M | 2 | 46XY | Exon 7 | nt1340/1341 (+TCGG), | V382fsX385 | FLT3/ITD |
| 2 | 64/M | 1 | 46XY | Exon 7 | nt1338/1339 (+A), nt1337/1338 (+ACGGT) | G382fsX384, Y381fsX450 | CEBPA |
| 3 | 59/F | 2 | 47,XX, t(7;11)(p15;p15),+8 | Exon 7 | nt1333 (−A) | G380fsX448 | KRAS |
| 4 | 61/M | 1 | 46,XY | Exon 7 | nt1336/1337 (+AACGG) | N381fsX450 | NPM1 |
| 5 | 69/M | 2 | NM | Exon 7 | nt1336/1337 (+AACGG) | N381fsX450 | CEBPA |
| 6 | 35/F | 2 | 46,XY,t(8;21)(q22;q22) | Exon 7 | nt1334/1335 (+A) | Q380fsX384 | — |
| 7 | 25/M | 2 | 46,XY,t(7;11)(p15;p15) | Exon 7 | nt1340/1341 (+TCGG) | V382fsX385 | — |
| 8 | 31/M | 2 | 46,XY,t(8;21)(q21;q22) | Exon 7 | nt1420/1421 (+GAGCAGCCAA) | E409fsX421 | — |
| 9 | 15/M | 2 | 46,XY,t(2;22)(p21;q11) | Exon 7 | nt1338 (C>A) | S381X | FLT3/ITD |
| 10 | 38/F | 2 | 45,XX,t(4;12;11)(q21;q13;p15),-7 | Exon 9 | nt1590-1600 (−ACCTGAAGACC) | P465fsX472 | — |
| 11 | 20/F | 6 | 46,XX | Exon 9 | nt1574/1575 (+AAGT) | F460X | CEBPA |
| 12 | 26/M | 6 | 46,XY | Exon 7 | nt1326/1327 (+AAGAGAC) | R378fsX386 | — |
| 13 | 25/F | 2 | 46,XX,t(8;21)(q22;q22) | Exon 9 | nt1536 (G>T) | G447V | — |
| 14 | 43/M | 6 | 46,XY | Exon 7 | nt1331/1332 (+ACTCTTG) | D379fsX386 | — |
| 15 | 28/F | 1 | 46,XX | Exon 7 | nt1392/1393 (+A) | K399fsX400 | CEBPA |
| 16 | 85/M | 5 | 46,XY | Exon 7 | nt1334/1335(+A) | Q380fsX384 | FLT3/ITD, NPM1 |
| 17 | 80/M | 6 | 46,XY | Exon 7 | nt1271-1275 (−CGAGG, +GCTCAGTGGGGGGAC) | A359fsX387 | RUNX1 |
| 18 | 23/F | 2 | 46,XX,t(7;11)(p15;p15) | Exon 7 | nt1332-1339 (−TACGGTCG) | G379fsX381 | KRAS |
| 19 | 27/F | 2 | 46,XX,t(7;11)(p15;p15) | Exon 7 | nt1336/1337 (+G) | V381fsX384 | FLT3/ITD |
| 20 | 43/M | 2 | 46,XY,del(20)(q11q13)[15]/ 46,idem,del(11)(p13p15)[3] | Exon 7 | nt1302-1306 (−GACGT, +AAGA) | Q369fsX374 | — |
| Exon 7 | nt1398 (G>A) | R401K | |||||
| 21 | 31/F | 2 | 46,XX | Exon 7 | nt1332-1333(−TA) | A379fsX383 | FLT3/ITD,NPM1, NRAS |
| 22 | 53/F | 2 | 46,X,-X,+21 | Exon 7 | nt1323/1324 (+C) | D377fsX384 | CEBPA |
| 23 | 39/F | 4 | 46,XX | Exon 7 | nt1338/1339 (+TCTTGTACGGTCG) | S382fsX388 | FLT3/TKD, NPM1 |
| 24 | 31/F | 1 | 46,XX | Exon 7 | nt1334 (−C, +GG) | G380fsX384 | FLT3/ITD |
| 25 | 16/M | 4 | 46,XY | Exon 7 | nt1328-1331 (−CTTG) | Y378fsX447 | FLT3/TKD, RUNX1 |
| 26 | 48/M | 4 | NM | Exon 7 | nt1326/1327 (+AAGAC) | R378fsX450 | FLT3/ITD, NRAS |
| 27 | 43/F | 8 | Cplx* | Exon 7 | nt1303-1304 (−AC, +GCCTCCTATA) | P370fsX377 | — |
| 28 | 70/F | 4 | 46,XX, t(11;12)(q23;q13), del(9)(q12q33) | Exon 7 | nt1338 (C>A) | S381X | FLT3/ITD, NPM1 |
| 29 | 40/F | 2 | 46,XX | Exon 7 | nt1302-1303 (−GA, +CGG) | P369fsX384 | MLL/PTD |
| 30 | 45/F | 1 | 48,XX,+4,+10[6]/ 47,XX,del(9)(q13q22),+10[4] | Exon 7 | nt1400/1401 (+GAGAT) | Y402X | CEBPA |
| 31 | 26/F | 2 | 46,XX | Exon 7 | nt1338 (C>A) | S381X | FLT3/ITD, MLL/PTD |
| 32 | 40/M | 1 | 46,XY | Exon 7 | nt1393 (−T) | K399fsX448 | CEBPA |
| UPN . | Age, y/sex . | FAB . | Karyotype . | WT1 mutation . | Other gene mutations detected . | ||
|---|---|---|---|---|---|---|---|
| Location . | DNA change . | Protein change . | |||||
| 1 | 30/M | 2 | 46XY | Exon 7 | nt1340/1341 (+TCGG), | V382fsX385 | FLT3/ITD |
| 2 | 64/M | 1 | 46XY | Exon 7 | nt1338/1339 (+A), nt1337/1338 (+ACGGT) | G382fsX384, Y381fsX450 | CEBPA |
| 3 | 59/F | 2 | 47,XX, t(7;11)(p15;p15),+8 | Exon 7 | nt1333 (−A) | G380fsX448 | KRAS |
| 4 | 61/M | 1 | 46,XY | Exon 7 | nt1336/1337 (+AACGG) | N381fsX450 | NPM1 |
| 5 | 69/M | 2 | NM | Exon 7 | nt1336/1337 (+AACGG) | N381fsX450 | CEBPA |
| 6 | 35/F | 2 | 46,XY,t(8;21)(q22;q22) | Exon 7 | nt1334/1335 (+A) | Q380fsX384 | — |
| 7 | 25/M | 2 | 46,XY,t(7;11)(p15;p15) | Exon 7 | nt1340/1341 (+TCGG) | V382fsX385 | — |
| 8 | 31/M | 2 | 46,XY,t(8;21)(q21;q22) | Exon 7 | nt1420/1421 (+GAGCAGCCAA) | E409fsX421 | — |
| 9 | 15/M | 2 | 46,XY,t(2;22)(p21;q11) | Exon 7 | nt1338 (C>A) | S381X | FLT3/ITD |
| 10 | 38/F | 2 | 45,XX,t(4;12;11)(q21;q13;p15),-7 | Exon 9 | nt1590-1600 (−ACCTGAAGACC) | P465fsX472 | — |
| 11 | 20/F | 6 | 46,XX | Exon 9 | nt1574/1575 (+AAGT) | F460X | CEBPA |
| 12 | 26/M | 6 | 46,XY | Exon 7 | nt1326/1327 (+AAGAGAC) | R378fsX386 | — |
| 13 | 25/F | 2 | 46,XX,t(8;21)(q22;q22) | Exon 9 | nt1536 (G>T) | G447V | — |
| 14 | 43/M | 6 | 46,XY | Exon 7 | nt1331/1332 (+ACTCTTG) | D379fsX386 | — |
| 15 | 28/F | 1 | 46,XX | Exon 7 | nt1392/1393 (+A) | K399fsX400 | CEBPA |
| 16 | 85/M | 5 | 46,XY | Exon 7 | nt1334/1335(+A) | Q380fsX384 | FLT3/ITD, NPM1 |
| 17 | 80/M | 6 | 46,XY | Exon 7 | nt1271-1275 (−CGAGG, +GCTCAGTGGGGGGAC) | A359fsX387 | RUNX1 |
| 18 | 23/F | 2 | 46,XX,t(7;11)(p15;p15) | Exon 7 | nt1332-1339 (−TACGGTCG) | G379fsX381 | KRAS |
| 19 | 27/F | 2 | 46,XX,t(7;11)(p15;p15) | Exon 7 | nt1336/1337 (+G) | V381fsX384 | FLT3/ITD |
| 20 | 43/M | 2 | 46,XY,del(20)(q11q13)[15]/ 46,idem,del(11)(p13p15)[3] | Exon 7 | nt1302-1306 (−GACGT, +AAGA) | Q369fsX374 | — |
| Exon 7 | nt1398 (G>A) | R401K | |||||
| 21 | 31/F | 2 | 46,XX | Exon 7 | nt1332-1333(−TA) | A379fsX383 | FLT3/ITD,NPM1, NRAS |
| 22 | 53/F | 2 | 46,X,-X,+21 | Exon 7 | nt1323/1324 (+C) | D377fsX384 | CEBPA |
| 23 | 39/F | 4 | 46,XX | Exon 7 | nt1338/1339 (+TCTTGTACGGTCG) | S382fsX388 | FLT3/TKD, NPM1 |
| 24 | 31/F | 1 | 46,XX | Exon 7 | nt1334 (−C, +GG) | G380fsX384 | FLT3/ITD |
| 25 | 16/M | 4 | 46,XY | Exon 7 | nt1328-1331 (−CTTG) | Y378fsX447 | FLT3/TKD, RUNX1 |
| 26 | 48/M | 4 | NM | Exon 7 | nt1326/1327 (+AAGAC) | R378fsX450 | FLT3/ITD, NRAS |
| 27 | 43/F | 8 | Cplx* | Exon 7 | nt1303-1304 (−AC, +GCCTCCTATA) | P370fsX377 | — |
| 28 | 70/F | 4 | 46,XX, t(11;12)(q23;q13), del(9)(q12q33) | Exon 7 | nt1338 (C>A) | S381X | FLT3/ITD, NPM1 |
| 29 | 40/F | 2 | 46,XX | Exon 7 | nt1302-1303 (−GA, +CGG) | P369fsX384 | MLL/PTD |
| 30 | 45/F | 1 | 48,XX,+4,+10[6]/ 47,XX,del(9)(q13q22),+10[4] | Exon 7 | nt1400/1401 (+GAGAT) | Y402X | CEBPA |
| 31 | 26/F | 2 | 46,XX | Exon 7 | nt1338 (C>A) | S381X | FLT3/ITD, MLL/PTD |
| 32 | 40/M | 1 | 46,XY | Exon 7 | nt1393 (−T) | K399fsX448 | CEBPA |
The nucleotide numberings are according to NCBI Reference Sequence: NM_024426.
FAB indicates French-American-British subtype; NM, no mitosis; and —, negative (no other mutation found).
Cplx: 46,XX,add(5)(p15),del(5)(q11q35),add(6)(p23),add(8)(p21),add(10)(q25),del(12)(p11p13),der(14)t(7;14)(q11;p12),add(19)(q13).
The mutations in 2 patients (nos. 2 and 20) were double heterozygous with 2 exon 7 mutations in different alleles. The remaining 30 patients showed only one mutation; all are heterozygous. Of the 29 kinds of mutations, 3 were base substitutions, 19 with 1- to 13-bp insertions, and 7 with 1- to 11-bp deletions. Of them, 2 of the 3 base substitutions caused amino acid changes, the remaining one base substitution and 2 insertion mutations (patient nos. 11 and 30) created a stop codon, and the others resulted in reading frame shift, which was predicted to generate nonfunctional truncated proteins with loss of their DNA-binding sites (aa391-aa506), RNA recognition sites (zinc finger 1), and nuclear localization signal (aa359-aa418). Among 26 mutations on exon 7, 23 (88.5%) were frame-shift mutations, compared with one of the 3 exon 9 mutations (P = .068).
Correlation of WT1 mutations with clinical features and immunophenotypes of leukemic cells
Among the 470 AML patients recruited, 266 were males and 204 were females with a median age of 52 years (range, 15-90 years). The comparison of clinical characteristics and laboratory data between patients with and without WT1 mutation is summarized in Table 2. WT1-mutated patients were younger than WT1-wild patients (median, 38.5 years vs 53 years, P < .001). Patients with French-American-British (FAB) M6 subtype of AML had the highest incidence (33.3%) of WT1 mutation, whereas those with M0 subtype had the lowest incidence (P = .006). There was no difference in other clinical parameters, including sex, WBC count, hemoglobin, platelet count, and lactate dehydrogenase value between patients with and without the mutation. The WT1 mutation did not correlate with the expression of the antigens studied (data not shown).
Comparison of clinical manifestations and laboratory features between AML patients with and without WT1 mutation
| Variable . | Total (n = 470) . | WT1-mutated (n = 32, 6.8%) . | WT1-wild (n = 438, 93.2%) . | P . |
|---|---|---|---|---|
| Sex, no. (%) of patients | .250 | |||
| Male | 266 | 15 (46.9) | 251 (57.3) | — |
| Female | 204 | 17 (53.1) | 187 (42.7) | — |
| Median age, y (range) | 52.0 (15-90) | 38.5 (15-85) | 53 (15-90) | < .001 |
| Laboratory data, median (range) | ||||
| WBCs, per μL | 21 850 (120-627 800) | 20 290 (1200-227 700) | 21 850 (120-627 800) | .785 |
| Hemoglobin, g/dL | 8.0 (2.9-16.2) | 7.65 (3-13.6) | 8 (2.9-16.2) | .468 |
| Platelets, × 1000/μL | 45.0 (3-802) | 46.5 (7-802) | 44 (3-802) | .653 |
| Blasts, per μL | 9863 (0-456 725) | 9927 (120-154 836) | 9863 (0-456 725) | .928 |
| LDH, U/L | 890 (206-15 000) | 1228 (301-7734) | 856 (206-15 000) | .107 |
| FAB, no. (%) of patients | .017 | |||
| M0 | 10 | 0 (0.0) | 10 (100.0) | — |
| M1 | 114 | 6 (5.3) | 108 (94.7) | — |
| M2 | 174 | 16 (9.2) | 158 (90.8) | — |
| M4 | 126 | 4 (3.2) | 122 (96.8) | — |
| M5 | 25 | 1 (4.0) | 24 (96.0) | — |
| M6 | 12 | 4 (33.3) | 8 (66.7) | — |
| Undetermined | 9 | 1 (11.1) | 8 (88.9) | — |
| Induction response* | 330 | 28 | 302 | .516 |
| CR | 248 (75.2) | 21 (75.0) | 227 (75.2) | — |
| Refractory | 59 (17.8) | 6 (21.4) | 53 (17.5) | — |
| Induction death | 23 (7.0) | 1 (3.6) | 22 (7.3) | — |
| Relapse | 134 (54.0) | 18 (85.7) | 116 (51.1) | .002 |
| Variable . | Total (n = 470) . | WT1-mutated (n = 32, 6.8%) . | WT1-wild (n = 438, 93.2%) . | P . |
|---|---|---|---|---|
| Sex, no. (%) of patients | .250 | |||
| Male | 266 | 15 (46.9) | 251 (57.3) | — |
| Female | 204 | 17 (53.1) | 187 (42.7) | — |
| Median age, y (range) | 52.0 (15-90) | 38.5 (15-85) | 53 (15-90) | < .001 |
| Laboratory data, median (range) | ||||
| WBCs, per μL | 21 850 (120-627 800) | 20 290 (1200-227 700) | 21 850 (120-627 800) | .785 |
| Hemoglobin, g/dL | 8.0 (2.9-16.2) | 7.65 (3-13.6) | 8 (2.9-16.2) | .468 |
| Platelets, × 1000/μL | 45.0 (3-802) | 46.5 (7-802) | 44 (3-802) | .653 |
| Blasts, per μL | 9863 (0-456 725) | 9927 (120-154 836) | 9863 (0-456 725) | .928 |
| LDH, U/L | 890 (206-15 000) | 1228 (301-7734) | 856 (206-15 000) | .107 |
| FAB, no. (%) of patients | .017 | |||
| M0 | 10 | 0 (0.0) | 10 (100.0) | — |
| M1 | 114 | 6 (5.3) | 108 (94.7) | — |
| M2 | 174 | 16 (9.2) | 158 (90.8) | — |
| M4 | 126 | 4 (3.2) | 122 (96.8) | — |
| M5 | 25 | 1 (4.0) | 24 (96.0) | — |
| M6 | 12 | 4 (33.3) | 8 (66.7) | — |
| Undetermined | 9 | 1 (11.1) | 8 (88.9) | — |
| Induction response* | 330 | 28 | 302 | .516 |
| CR | 248 (75.2) | 21 (75.0) | 227 (75.2) | — |
| Refractory | 59 (17.8) | 6 (21.4) | 53 (17.5) | — |
| Induction death | 23 (7.0) | 1 (3.6) | 22 (7.3) | — |
| Relapse | 134 (54.0) | 18 (85.7) | 116 (51.1) | .002 |
— indicates not applicable.
Only the 330 patients (including 28 with the WT1 mutation and 302 without) who received conventional intensive induction chemotherapy and then consolidation chemotherapy if CR was achieved, were included in the analysis.
Association of WT1 mutations with cytogenetic abnormalities
Chromosome data were available in 452 patients at diagnosis, including 30 WT1-mutated and 422 WT1-wild patients (Table 3). WT1 mutations were highly associated with t(7;11)(p15;p15) (P = .003), a chromosomal translocation resulting in fusion between NUP98 on 11p15 and HOXA9 on 7P15.27,28 Four (40%) of the 10 patients with this cytogenetic abnormality showed concurrent WT1 mutation. There was no association of WT1 mutation with other chromosomal abnormalities, including +8, +11, +13, +21, −5/del(5q), and −7/del(7q). The incidences of WT1 mutation were not significantly different between patients with normal karyotype (16 of 230, 7.0%) and those with abnormal cytogenetics (14 of 222, 6.3%; P = .851), and among patients with favorable (5.1%), intermediate (7.0%), and unfavorable karyotypes (6.1%). In younger patients (< 60 years of age) with normal karyotype (n = 121), the incidence of WT1 mutation was 8.3%.
Association of WT1 mutation with chromosomal abnormalities*
| Karyotype . | Total . | WT1-mutated . | WT1-wild . | P . |
|---|---|---|---|---|
| Favorable | 59 | 3 (5.1) | 56 (94.9) | .953 |
| Intermediate | 327 | 23 (7.0) | 304 (93.0) | — |
| Unfavorable | 66 | 4 (6.1) | 62 (93.9) | — |
| Unknown | 18 | 2 (11.1) | 16 (88.9) | — |
| Normal | 230 | 16 (7.0) | 214 (93.0) | > .999 |
| Simple | 169 | 11 (6.5) | 158 (93.5) | — |
| Complex | 53 | 3 (5.7) | 50 (94.3) | — |
| t(8;21) | 40 | 3 (7.5) | 37 (92.5) | .740 |
| inv(16) | 19 | 0 (0.0) | 19 (100.0) | .628 |
| t(11q23) | 16 | 1 (6.3) | 15 (93.7) | > .999 |
| t(7;11) | 10 | 4 (40.0) | 6 (60.0) | .003 |
| −5/5q−† | 1 | 0 (0.0) | 1 (100.0) | > .999 |
| −7/7q−† | 10 | 1 (10.0) | 9 (90.0) | .500 |
| +8† | 22 | 1 (4.5) | 21 (95.5) | > .999 |
| +11† | 3 | 0 (0.0) | 3 (100.0) | > .999 |
| +13† | 1 | 0 (0.0) | 1 (100.0) | > .999 |
| +21† | 11 | 1 (9.1) | 10 (90.9) | > .999 |
| Karyotype . | Total . | WT1-mutated . | WT1-wild . | P . |
|---|---|---|---|---|
| Favorable | 59 | 3 (5.1) | 56 (94.9) | .953 |
| Intermediate | 327 | 23 (7.0) | 304 (93.0) | — |
| Unfavorable | 66 | 4 (6.1) | 62 (93.9) | — |
| Unknown | 18 | 2 (11.1) | 16 (88.9) | — |
| Normal | 230 | 16 (7.0) | 214 (93.0) | > .999 |
| Simple | 169 | 11 (6.5) | 158 (93.5) | — |
| Complex | 53 | 3 (5.7) | 50 (94.3) | — |
| t(8;21) | 40 | 3 (7.5) | 37 (92.5) | .740 |
| inv(16) | 19 | 0 (0.0) | 19 (100.0) | .628 |
| t(11q23) | 16 | 1 (6.3) | 15 (93.7) | > .999 |
| t(7;11) | 10 | 4 (40.0) | 6 (60.0) | .003 |
| −5/5q−† | 1 | 0 (0.0) | 1 (100.0) | > .999 |
| −7/7q−† | 10 | 1 (10.0) | 9 (90.0) | .500 |
| +8† | 22 | 1 (4.5) | 21 (95.5) | > .999 |
| +11† | 3 | 0 (0.0) | 3 (100.0) | > .999 |
| +13† | 1 | 0 (0.0) | 1 (100.0) | > .999 |
| +21† | 11 | 1 (9.1) | 10 (90.9) | > .999 |
— indicates not applicable.
A total of 452 patients, including 30 WT1-mutated and 422 WT1-wild patients, had chromosome data at diagnosis.
Only including simple chromosomal abnormalities with 2 or fewer changes, but not those with complex abnormalities with 3 or more aberrations.
Association of WT1 mutation with other molecular abnormalities
To investigate the interaction of gene mutations in the pathogenesis of adult AML, a complete mutational screening of 11 other genes was performed in all 470 patients. Among the 32 patients with WT1 mutations, 23 (72%) showed additional molecular abnormalities at diagnosis (Table 4); 16 had one additional change, 6 had 2, and 1 had 3. Sixteen (69.6%) of them had at least one concurrent class 2 mutation that would impair differentiation of hematopoietic cells,29 and 13 (56.5%) had a class 1 mutation that would confer proliferation advantage (Table 4). The most frequently associated molecular event was FLT3/ITD (9 cases), followed by CEBPA (7 cases), and NPM1 (5 cases). However, there was no difference in the incidence of any mutation between patients with and without WT1 mutation (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Cooperative gene mutations in the 32 AML patients with WT1 mutations*
| UPNs . | Class 1 mutations . | Class 2 mutations . | NPM1 mutation . | |||||
|---|---|---|---|---|---|---|---|---|
| FLT3/ITD . | FLT3/TKD . | NRAS . | KRAS . | RUNX1 . | CEBPA . | MLL/PTD . | ||
| 1, 9, 19, 24 | + | — | — | — | — | — | — | — |
| 16, 28 | + | — | — | — | — | — | — | + |
| 21 | + | — | + | — | — | — | — | + |
| 26 | + | — | + | — | — | — | — | — |
| 31 | + | — | — | — | — | — | + | — |
| 25 | — | + | — | — | + | — | — | — |
| 23 | — | + | — | — | — | — | — | + |
| 18 | — | — | — | + | — | — | — | — |
| 3 | — | — | — | + | — | — | — | — |
| 2, 5, 11, 15, 22, 30, 32 | — | — | — | — | — | + | — | — |
| 17 | — | — | — | — | + | — | — | — |
| 29 | — | — | — | — | — | — | + | — |
| 4 | — | — | — | — | — | — | — | + |
| UPNs . | Class 1 mutations . | Class 2 mutations . | NPM1 mutation . | |||||
|---|---|---|---|---|---|---|---|---|
| FLT3/ITD . | FLT3/TKD . | NRAS . | KRAS . | RUNX1 . | CEBPA . | MLL/PTD . | ||
| 1, 9, 19, 24 | + | — | — | — | — | — | — | — |
| 16, 28 | + | — | — | — | — | — | — | + |
| 21 | + | — | + | — | — | — | — | + |
| 26 | + | — | + | — | — | — | — | — |
| 31 | + | — | — | — | — | — | + | — |
| 25 | — | + | — | — | + | — | — | — |
| 23 | — | + | — | — | — | — | — | + |
| 18 | — | — | — | + | — | — | — | — |
| 3 | — | — | — | + | — | — | — | — |
| 2, 5, 11, 15, 22, 30, 32 | — | — | — | — | — | + | — | — |
| 17 | — | — | — | — | + | — | — | — |
| 29 | — | — | — | — | — | — | + | — |
| 4 | — | — | — | — | — | — | — | + |
A total of 23 of the 32 WT1-mutated patients concurrently had other gene mutations, including 13 (56.5%) with class 1 mutations and 16 (69.6%) with class 2 mutations or NPM1 mutations. None of these patients had KIT, PTPN11, or JAK2 mutation.
UPN indicates unique patient number; and —, negative (not mutated).
Sequential studies of WT1 mutations in AML patients
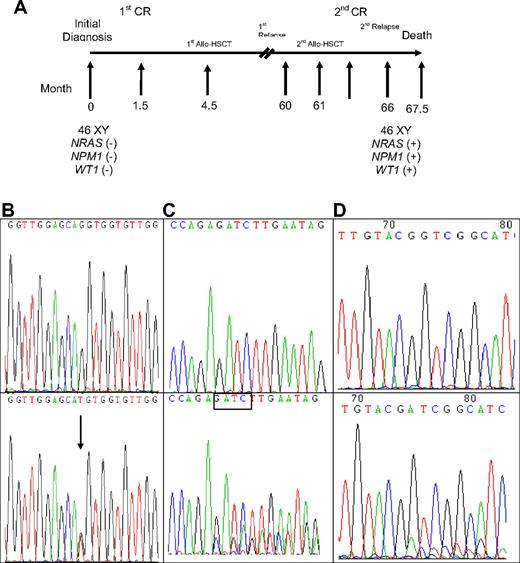
A total of 309 samples from 133 patients were serially studied for WT1 mutations using both direct sequencing and GeneScan analysis, including 23 patients with WT1 mutations and 110 without mutation at diagnosis. Among the 23 patients with WT1 mutations at diagnosis, all lost their mutations at remission status (Table 5). Among the 16 patients who had paired samples for serial study both at diagnosis and at relapse, 13 patients retained the original mutation at first relapse, but one of them (no. 26) lost the mutation at second relapse (Table 5). Two patients (nos. 21 and 23) each acquired an additional WT1 mutation on a different allele. In these 13 patients, the percentage of mutant at relapse, compared with that at diagnosis, was increased in 4 patients (nos. 12, 16, 22, and 29), decreased in 6 patients (nos. 7, 18, 23, 25, 26, and 32), had no much change in 2 patients (nos. 3 and 21), and cannot be evaluated in one patient (no. 13) because the mutation is single base substitution (Table 5). The remaining 3 patients (nos. 4, 11, and 19) lost WT1 mutations at relapse. Among the 110 patients who had no WT1 mutation at diagnosis, 3 acquired WT1 mutation at relapse (Table 5). Patient 33 acquired WT1 mutation at second relapse, 66 months after the initial study at diagnosis and 5 months after second human leukocyte antigen-matched sibling allogeneic HSCT. He also gained concurrently novel mutations of NPM1 and NRAS at that time (Table 5; Figure 1). No bone marrow sample was available for mutation analysis at first relapse. Patient 34 acquired WT1 mutation at first relapse, which disappeared at second CR but reappeared at second relapse; the same NPM1 mutation detected at diagnosis was retained at first and second relapse. Patient 35 acquired novel WT1 mutation and karyotypic evolution at first relapse, whereas the original CEBPA mutant remained the same (Table 5).
Sequential studies of WT1 mutations in AML patients
| UPN* . | Date . | Status . | Chromosomal abnormality . | WT1 mutation . | Other genetic mutations . | |
|---|---|---|---|---|---|---|
| Amino acid change . | Mutant by gene scan, percentage . | |||||
| 1 | 7/29/1994 | Initial | — | V382fsx385 | 54.02 | FLT3/ITD |
| 2/13/1995 | CR1 | — | — | — | ||
| 3 | 4/9/1999 | Initial | t(7;11)(p15;p15),+8 | G380fsX448 | 39.53 | KRAS |
| 7/9/1999 | CR1 | — | — | — | ||
| 5/3/2000 | Relapse1 | t(7;11)(p15;p15),+8 | G380fsX448 | 35.10 | KRAS | |
| 4 | 10/30/1995 | Initial | — | N381fsX450 | 41.55 | NPM1 |
| 1/15/1996 | CR1 | — | — | — | ||
| 10/22/1996 | Relapse1 | ND | — | 0 | NPM1 | |
| 12/21/1999 | CR2 | — | ||||
| 8/1/2000 | Relapse2 | —7,t(12;18)(p11;q11) | — | 0 | — | |
| 6 | 12/12/1995 | Initial | t(8;21)(q22;q22) | Q380fsX384 | 8.71 | — |
| 11/26/2002 | CR1 | — | — | — | ||
| 7 | 10/8/1996 | Initial | t(7;11)(p15;p15) | V382fsX385 | 59.24 | — |
| 11/8/1996 | CR1 | — | — | — | ||
| 8/22/1997 | Relapse1 | t(7;11)(p15;p15) | V382fsX385 | 40.45 | — | |
| 8 | 4/6/1999 | Initial | t(8;21)(q21;q22) | E409fsX421 | 44.27 | — |
| 7/23/2002 | CR1 | — | — | — | ||
| 10 | 6/30/2000 | Initial | t(4;12;11)(q21;q13;p15),—7 | P465fsX472 | 46.43 | — |
| 9/22/2000 | CR1 | — | — | — | ||
| 11 | 9/1/2000 | Initial | — | F460X | 21.07 | CEBPA |
| 9/22/2000 | CR1 | — | — | — | ||
| 9/24/2001 | Relapse1 | NM | — | 0 | CEBPA | |
| 1/25/2005 | CR2 | — | — | — | ||
| 12 | 3/15/2001 | Initial | 46,XY | R378fsX386 | 47.27 | — |
| 11/16/2001 | CR1 | — | — | — | ||
| 10/8/2002 | Relapse1 | add(19)(q13) | R378fsX386 | 88.36 | FLT3/ITD | |
| 13 | 6/26/2001 | Initial | t(8;21)(q22;q22) | G447V | NA | — |
| 7/31/2001 | CR1 | — | — | — | ||
| 5/10/2002 | Relapse1 | t(8;21)(q22;q22),t(3;12)(p21;q24) | G447V | NA | — | |
| 14 | 4/3/2002 | Initial | — | D379fsX386 | 21.93 | — |
| 5/14/2002 | CR1 | — | — | — | ||
| 16 | 5/29/2003 | Initial | — | Q380fsX384 | 11.9 | FLT3/ITD, NPM1 |
| 7/17/2003 | CR1 | — | — | — | ||
| 12/26/2003 | Relapse1 | ND | Q380fsX384 | 37.76 | FLT3/ITD, NPM1 | |
| 18 | 8/7/2003 | Initial | t(7;11)(p15;p15) | G379fsX381 | 34.16 | KRAS |
| 11/20/2003 | CR1 | — | — | — | ||
| 1/4/2005 | Relapse1 | t(7;11)(p15;p15) | G379fsX381 | 8.65 | — | |
| 19 | 8/17/2007 | Initial | t(7;11)(p15;p15) | V381fsX384 | 47.43 | FLT3/ITD |
| 9/28/2007 | CR1 | — | — | — | ||
| 2/14/2008 | Relapse1 | t(7;11)(p15;p15) | — | 0 | — | |
| 21 | 3/8/2005 | Initial | — | A379fsX383 | 35.63 | FLT3/ITD, NRAS, NPM1 |
| 5/10/2005 | CR1 | — | — | — | ||
| 12/2/2005 | Relapse1 | — | A379fsX383 | 40.46 | FLT3/ITD, NPM1 | |
| S378fsX387 | 39.56 | |||||
| 22 | 2005/5/20 | Initial | −X,+21 | D377fsX384 | 11.71 | CEBPA |
| 6/22/2005 | CR1 | — | — | — | ||
| 5/16/2006 | Relapse1 | −X,+21 | D377fsX384 | 61.55 | CEBPA | |
| 23 | 6/15/2005 | Initial | — | S382fsX388 | 42.20 | NPM1, FLT3/TKD |
| 9/13/2005 | CR1 | — | — | — | ||
| 12/9/2005 | Relapse1 | — | S382fsX388 | 33.32 | NPM1, FLT3/TKD | |
| V382fsX385 | 27.44 | |||||
| 25 | 7/21/2006 | Initial | — | Y378fsX447 | 49.63 | FLT3/TKD, RUNX1 |
| 9/21/2006 | CR1 | — | — | — | ||
| 7/15/2008 | Relapse1 | — | Y378fsX447 | 6.32 | FLT3/TKD | |
| 26 | 10/19/2006 | Initial | NM | R378fsX450 | 53.90 | FLT3/ITD, NRAS |
| 1/30/2007 | CR1 | — | — | — | ||
| 6/5/2007 | Relapse1 | ND | R378fsX450 | 36.44 | FLT3/ITD, NRAS | |
| 11/2/2007 | CR2 | ND | — | — | ||
| 12/3/2007 | Relapse 2 | t(8;11)(q24;p15),del(12)(p13),add(19)(p13), t(2;17)(q21;q25) | — | 0 | FLT3/ITD | |
| 29 | 1/2/2007 | Initial | — | P369fsX384 | 11.38 | MLL/PTD |
| 7/13/2007 | CR1 | — | — | — | ||
| 1/7/2008 | Relapse1 | — | P369fsX384 | 29.56 | MLL/PTD | |
| 30 | 7/9/2007 | Initial | +4, del(9)(q13q22),+10 | Y402X | 47.04 | CEBPA |
| 8/21/2007 | CR1 | — | — | — | ||
| 31 | 9/11/2007 | Initial | — | S381X | NA | FLT3/ITD, MLL/PTD |
| 11/19/2007 | CR1 | — | — | — | ||
| 32 | 12/4/2007 | Initial | — | K399fsX448 | 28.70 | CEBPA |
| 12/27/2007 | CR1 | — | — | — | ||
| 4/1/2008 | Relapse1 | — | K399fsX448 | 7.20 | CEBPA | |
| 33 | 12/8/1996 | Initial | — | — | 0 | |
| 1/21/1997 | CR1 | — | — | — | ||
| 9/11/2001 | CR2 | — | — | — | ||
| 6/10/2002 | Relapse 2 | ND | L381fsX452 | 11.09 | NRAS, NPM1 | |
| 34 | 7/27/2000 | Initial | — | — | 0 | NPM1 |
| 11/10/2000 | CR1 | — | — | — | ||
| 7/17/2001 | Relapse1 | — | D379fsX386 | 20.70 | NPM1 | |
| 10/23/2001 | CR2 | — | — | |||
| 5/7/2002 | Relapse 2 | del(6)(p21) | D379fsX386 | 28.06 | NPM1 | |
| 35 | 4/18/2002 | Initial | — | — | NA | CEBPA |
| 5/13/2002 | CR1 | — | — | — | ||
| 11/19/2002 | Relapse1 | t(1;3)(p36;q21),del(3)(p13p21) | R471T | NA | CEBPA | |
| 12/16/2002 | CR2 | — | — | — | ||
| UPN* . | Date . | Status . | Chromosomal abnormality . | WT1 mutation . | Other genetic mutations . | |
|---|---|---|---|---|---|---|
| Amino acid change . | Mutant by gene scan, percentage . | |||||
| 1 | 7/29/1994 | Initial | — | V382fsx385 | 54.02 | FLT3/ITD |
| 2/13/1995 | CR1 | — | — | — | ||
| 3 | 4/9/1999 | Initial | t(7;11)(p15;p15),+8 | G380fsX448 | 39.53 | KRAS |
| 7/9/1999 | CR1 | — | — | — | ||
| 5/3/2000 | Relapse1 | t(7;11)(p15;p15),+8 | G380fsX448 | 35.10 | KRAS | |
| 4 | 10/30/1995 | Initial | — | N381fsX450 | 41.55 | NPM1 |
| 1/15/1996 | CR1 | — | — | — | ||
| 10/22/1996 | Relapse1 | ND | — | 0 | NPM1 | |
| 12/21/1999 | CR2 | — | ||||
| 8/1/2000 | Relapse2 | —7,t(12;18)(p11;q11) | — | 0 | — | |
| 6 | 12/12/1995 | Initial | t(8;21)(q22;q22) | Q380fsX384 | 8.71 | — |
| 11/26/2002 | CR1 | — | — | — | ||
| 7 | 10/8/1996 | Initial | t(7;11)(p15;p15) | V382fsX385 | 59.24 | — |
| 11/8/1996 | CR1 | — | — | — | ||
| 8/22/1997 | Relapse1 | t(7;11)(p15;p15) | V382fsX385 | 40.45 | — | |
| 8 | 4/6/1999 | Initial | t(8;21)(q21;q22) | E409fsX421 | 44.27 | — |
| 7/23/2002 | CR1 | — | — | — | ||
| 10 | 6/30/2000 | Initial | t(4;12;11)(q21;q13;p15),—7 | P465fsX472 | 46.43 | — |
| 9/22/2000 | CR1 | — | — | — | ||
| 11 | 9/1/2000 | Initial | — | F460X | 21.07 | CEBPA |
| 9/22/2000 | CR1 | — | — | — | ||
| 9/24/2001 | Relapse1 | NM | — | 0 | CEBPA | |
| 1/25/2005 | CR2 | — | — | — | ||
| 12 | 3/15/2001 | Initial | 46,XY | R378fsX386 | 47.27 | — |
| 11/16/2001 | CR1 | — | — | — | ||
| 10/8/2002 | Relapse1 | add(19)(q13) | R378fsX386 | 88.36 | FLT3/ITD | |
| 13 | 6/26/2001 | Initial | t(8;21)(q22;q22) | G447V | NA | — |
| 7/31/2001 | CR1 | — | — | — | ||
| 5/10/2002 | Relapse1 | t(8;21)(q22;q22),t(3;12)(p21;q24) | G447V | NA | — | |
| 14 | 4/3/2002 | Initial | — | D379fsX386 | 21.93 | — |
| 5/14/2002 | CR1 | — | — | — | ||
| 16 | 5/29/2003 | Initial | — | Q380fsX384 | 11.9 | FLT3/ITD, NPM1 |
| 7/17/2003 | CR1 | — | — | — | ||
| 12/26/2003 | Relapse1 | ND | Q380fsX384 | 37.76 | FLT3/ITD, NPM1 | |
| 18 | 8/7/2003 | Initial | t(7;11)(p15;p15) | G379fsX381 | 34.16 | KRAS |
| 11/20/2003 | CR1 | — | — | — | ||
| 1/4/2005 | Relapse1 | t(7;11)(p15;p15) | G379fsX381 | 8.65 | — | |
| 19 | 8/17/2007 | Initial | t(7;11)(p15;p15) | V381fsX384 | 47.43 | FLT3/ITD |
| 9/28/2007 | CR1 | — | — | — | ||
| 2/14/2008 | Relapse1 | t(7;11)(p15;p15) | — | 0 | — | |
| 21 | 3/8/2005 | Initial | — | A379fsX383 | 35.63 | FLT3/ITD, NRAS, NPM1 |
| 5/10/2005 | CR1 | — | — | — | ||
| 12/2/2005 | Relapse1 | — | A379fsX383 | 40.46 | FLT3/ITD, NPM1 | |
| S378fsX387 | 39.56 | |||||
| 22 | 2005/5/20 | Initial | −X,+21 | D377fsX384 | 11.71 | CEBPA |
| 6/22/2005 | CR1 | — | — | — | ||
| 5/16/2006 | Relapse1 | −X,+21 | D377fsX384 | 61.55 | CEBPA | |
| 23 | 6/15/2005 | Initial | — | S382fsX388 | 42.20 | NPM1, FLT3/TKD |
| 9/13/2005 | CR1 | — | — | — | ||
| 12/9/2005 | Relapse1 | — | S382fsX388 | 33.32 | NPM1, FLT3/TKD | |
| V382fsX385 | 27.44 | |||||
| 25 | 7/21/2006 | Initial | — | Y378fsX447 | 49.63 | FLT3/TKD, RUNX1 |
| 9/21/2006 | CR1 | — | — | — | ||
| 7/15/2008 | Relapse1 | — | Y378fsX447 | 6.32 | FLT3/TKD | |
| 26 | 10/19/2006 | Initial | NM | R378fsX450 | 53.90 | FLT3/ITD, NRAS |
| 1/30/2007 | CR1 | — | — | — | ||
| 6/5/2007 | Relapse1 | ND | R378fsX450 | 36.44 | FLT3/ITD, NRAS | |
| 11/2/2007 | CR2 | ND | — | — | ||
| 12/3/2007 | Relapse 2 | t(8;11)(q24;p15),del(12)(p13),add(19)(p13), t(2;17)(q21;q25) | — | 0 | FLT3/ITD | |
| 29 | 1/2/2007 | Initial | — | P369fsX384 | 11.38 | MLL/PTD |
| 7/13/2007 | CR1 | — | — | — | ||
| 1/7/2008 | Relapse1 | — | P369fsX384 | 29.56 | MLL/PTD | |
| 30 | 7/9/2007 | Initial | +4, del(9)(q13q22),+10 | Y402X | 47.04 | CEBPA |
| 8/21/2007 | CR1 | — | — | — | ||
| 31 | 9/11/2007 | Initial | — | S381X | NA | FLT3/ITD, MLL/PTD |
| 11/19/2007 | CR1 | — | — | — | ||
| 32 | 12/4/2007 | Initial | — | K399fsX448 | 28.70 | CEBPA |
| 12/27/2007 | CR1 | — | — | — | ||
| 4/1/2008 | Relapse1 | — | K399fsX448 | 7.20 | CEBPA | |
| 33 | 12/8/1996 | Initial | — | — | 0 | |
| 1/21/1997 | CR1 | — | — | — | ||
| 9/11/2001 | CR2 | — | — | — | ||
| 6/10/2002 | Relapse 2 | ND | L381fsX452 | 11.09 | NRAS, NPM1 | |
| 34 | 7/27/2000 | Initial | — | — | 0 | NPM1 |
| 11/10/2000 | CR1 | — | — | — | ||
| 7/17/2001 | Relapse1 | — | D379fsX386 | 20.70 | NPM1 | |
| 10/23/2001 | CR2 | — | — | |||
| 5/7/2002 | Relapse 2 | del(6)(p21) | D379fsX386 | 28.06 | NPM1 | |
| 35 | 4/18/2002 | Initial | — | — | NA | CEBPA |
| 5/13/2002 | CR1 | — | — | — | ||
| 11/19/2002 | Relapse1 | t(1;3)(p36;q21),del(3)(p13p21) | R471T | NA | CEBPA | |
| 12/16/2002 | CR2 | — | — | — | ||
UPN indicates unique patient number; CR, complete remission; ND, not done; NM, no mitosis; —, negative; and NA, not applicable because the mutations are point mutations that cannot be determined by GeneScan.
Patients 1 through 31 had WT1 mutations at diagnosis; patients 32 through 35 had no WT1 mutations at diagnosis but acquired the mutation at relapse. The data of serial studies in another 107 patients, who did not have WT1 mutation either at diagnosis or at relapse, are not shown in this table.
Sequential analyses of NRAS, NPM1, and WT1 mutations in patient 33 showing acquisition of novel mutations in these 3 genes at relapse. (A) Sequential follow-up of mutation pattern at Patient 33. (B) Wild-type NRAS at diagnosis (top) and GGT>TGT at NRAS codon 12 at relapse (bottom). Arrows indicate the location of the mutation. (C) Wild-type NPM1 at diagnosis (top); NPM1 mutation with a TCTG insertion at relapse (bottom). The boxed tetranucleotides indicate the location of the mutation. (D) Wild-type WT1 at diagnosis (top) and nt1335/1336 (+11 bp) on exon 7 at relapse (bottom).
Sequential analyses of NRAS, NPM1, and WT1 mutations in patient 33 showing acquisition of novel mutations in these 3 genes at relapse. (A) Sequential follow-up of mutation pattern at Patient 33. (B) Wild-type NRAS at diagnosis (top) and GGT>TGT at NRAS codon 12 at relapse (bottom). Arrows indicate the location of the mutation. (C) Wild-type NPM1 at diagnosis (top); NPM1 mutation with a TCTG insertion at relapse (bottom). The boxed tetranucleotides indicate the location of the mutation. (D) Wild-type WT1 at diagnosis (top) and nt1335/1336 (+11 bp) on exon 7 at relapse (bottom).
Impact of WT1 mutation on response to therapy and clinical outcome
Of the 330 AML patients undergoing conventional intensive induction chemotherapy, 248 (75.2%) patients achieved CR. The probability of achieving a first CR was similar between patients with and without WT1 mutations (75% vs 75.2%, P = .985). However, the patients with WT1 mutations had a higher incidence of relapse than those without (85.7% vs 51.1%, P = .002; Table 2).
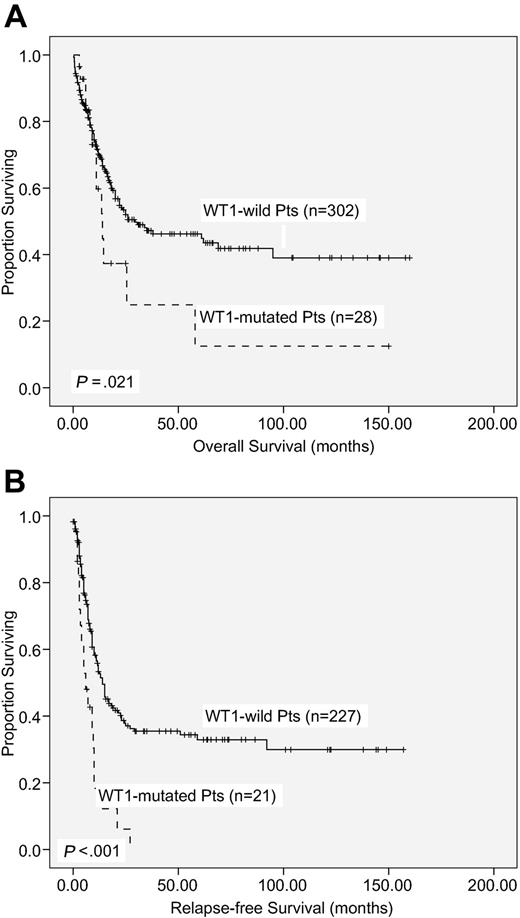
With a median follow-up time of 53 months (range, 1.0-160 months), patients with WT1 mutation had a significantly poorer OS and RFS than those without WT1 mutation (median, 14.0 months vs 29.5 months, P = .021; and 6 months vs 14 months, P < .001, respectively; supplemental Table 2; Figure 2). There was no significant difference in OS and RFS between patients with frame-shift mutations and other mutations (P < .987 and P < .99, respectively).
Kaplan-Meier survival curves. OS (A) and RFS (B) according to WT1 mutation status in 330 AML patients receiving standard intensive chemotherapy.
Kaplan-Meier survival curves. OS (A) and RFS (B) according to WT1 mutation status in 330 AML patients receiving standard intensive chemotherapy.
In multivariate analysis (Table 6), the independent poor risk factors for OS were age more than 50 years, WBC count more than 50 000/μL, unfavorable karyotype, and WT1 mutation. CEBPAdouble-mutation and NPM1mut/FLT3-ITD− were independent favorable prognostic factors. The independent poor risk factors for RFS included unfavorable karyotype and WT1 mutation. CEBPAdouble-mutation and NPM1mut/FLT3-ITD− were independent favorable factors for RFS. Double CEBPA mutations (CEBPAdouble-mutation) instead of all CEBPA mutations were used as a variable for survival analysis in this study because recent studies showed that only CEBPAdouble-mutation, but not single mutation, were associated with favorable prognosis.30,31
Multivariate analysis (Cox regression) on the RFS and OS
| Variable . | RFS . | OS . | ||||
|---|---|---|---|---|---|---|
| RR . | 95% CI . | P . | RR . | 95% CI . | P . | |
| Whole cohort (n = 330) | ||||||
| Age† | 1.334 | 0.929-1.915 | .119 | 2.724 | 1.902-3.902 | < .001* |
| WBC‡ | 1.377 | 0.941-2.014 | .100 | 1.863 | 1.297-2.677 | .001* |
| Karyotype§ | 1.851 | 1.017-3.368 | .044* | 2.684 | 1.626-4.432 | < .001* |
| NPM1/FLT3-ITD‖ | 0.400 | 0.208-0.770 | .006* | 0.344 | 0.165-0.714 | .004* |
| CEBPA¶ | 0.516 | 0.300-0.889 | .017* | 0.371 | 0.187-0.737 | .005* |
| WT1 | 2.601 | 1.562-4.333 | < .001* | 2.141 | 1.188-3.859 | .011* |
| Patients with normal karyotype and age < 60 y (n = 121) | ||||||
| Age† | 2.340 | 1.114-4.787 | .020* | 2.515 | 1.065-5.939 | .035* |
| WBC‡ | 2.533 | 1.231-5.210 | .012* | 5.508 | 2.536-11.964 | < .001* |
| NPM1/FLT3-ITD‖ | 0.151 | 0.059-0.387 | < .001* | 0.246 | 0.079-0.768 | .016* |
| CEBPA¶ | 0.170 | 0.070-0.416 | < .001* | 0.146 | 0.045-0.475 | .001* |
| WT1 | 3.806 | 1.588-9.122 | .003* | 3.752 | 1.195-11.783 | .024* |
| Variable . | RFS . | OS . | ||||
|---|---|---|---|---|---|---|
| RR . | 95% CI . | P . | RR . | 95% CI . | P . | |
| Whole cohort (n = 330) | ||||||
| Age† | 1.334 | 0.929-1.915 | .119 | 2.724 | 1.902-3.902 | < .001* |
| WBC‡ | 1.377 | 0.941-2.014 | .100 | 1.863 | 1.297-2.677 | .001* |
| Karyotype§ | 1.851 | 1.017-3.368 | .044* | 2.684 | 1.626-4.432 | < .001* |
| NPM1/FLT3-ITD‖ | 0.400 | 0.208-0.770 | .006* | 0.344 | 0.165-0.714 | .004* |
| CEBPA¶ | 0.516 | 0.300-0.889 | .017* | 0.371 | 0.187-0.737 | .005* |
| WT1 | 2.601 | 1.562-4.333 | < .001* | 2.141 | 1.188-3.859 | .011* |
| Patients with normal karyotype and age < 60 y (n = 121) | ||||||
| Age† | 2.340 | 1.114-4.787 | .020* | 2.515 | 1.065-5.939 | .035* |
| WBC‡ | 2.533 | 1.231-5.210 | .012* | 5.508 | 2.536-11.964 | < .001* |
| NPM1/FLT3-ITD‖ | 0.151 | 0.059-0.387 | < .001* | 0.246 | 0.079-0.768 | .016* |
| CEBPA¶ | 0.170 | 0.070-0.416 | < .001* | 0.146 | 0.045-0.475 | .001* |
| WT1 | 3.806 | 1.588-9.122 | .003* | 3.752 | 1.195-11.783 | .024* |
RR indicates relative risk; and CI, confidence interval.
Statistically significant (P < .05).
Age more than 50 years relative to age less than or equal to 50 years (the reference).
WBC greater than 50 000/μL versus less than 50 000/μL.
Unfavorable cytogenetics versus others.
NPM1mut/FLT3-ITD− versus other subtypes.
CEBPAdouble-mutation versus others.
Further, we made survival analysis in a relatively homogeneous population of younger patients (< 60 years) with normal karyotype (CN-AML). WT1 mutation was still an independent poor prognosis for OS and RFS (relative risk = 3.752, 95% confidence interval 1.195-11.783, P = .024; and relative risk = 3.806, 95% confidence interval 1.588-9.112, P = .003, respectively, Table 6).
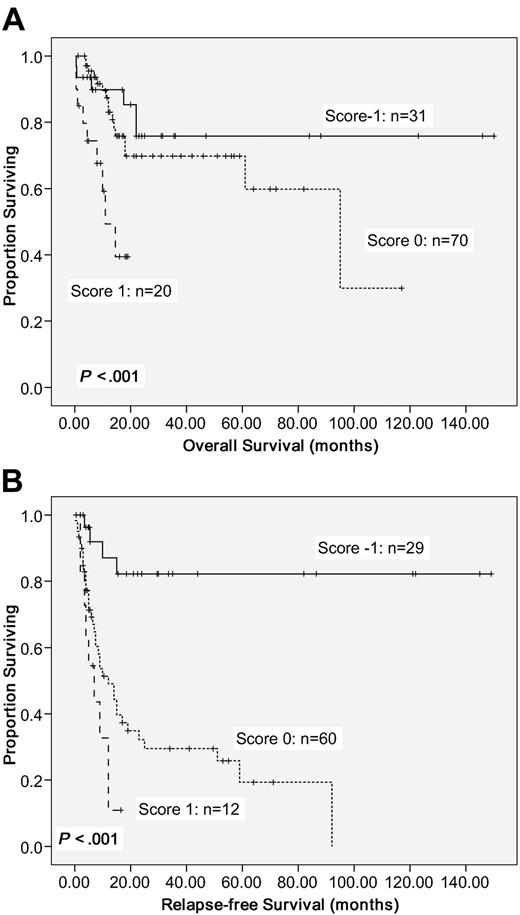
To better stratify the CN-AML patients into different risk groups according to gene mutations, a scoring system was formulated based on the results of our Cox proportional hazards model. Positivity of WT1 mutations or older age was scored +1 individually, whereas NPM1mut/FLT3-ITD− or CEBPAdouble-mutation was scored −1. The algebraic summation of these scores of each patient was the final score. This score system divided these CN-AML patients into 3 groups with different survival (P < .001 for both OS and RFS; Figure 3).
Kaplan-Meier survival curves. OS (A) and RFS (B) in 121 CN-AML patients based on scoring system (P < .001 for both OS and RFS). CN-AML patients were grouped according to scoring system based on 4 prognostic markers (CEBPAdouble-mutation, NPM1/FLT3-ITD, WT1 mutation, and age). A score of −1 was assigned for each parameter associated with a favorable outcome (CEBPAdouble-mutation and NPM1mut/FLT3-ITD−) and a score of 1 for each factor associated with an adverse outcome (WT1 mutation and older age). The algebraic summation of these scores of each patient was the final score.
Kaplan-Meier survival curves. OS (A) and RFS (B) in 121 CN-AML patients based on scoring system (P < .001 for both OS and RFS). CN-AML patients were grouped according to scoring system based on 4 prognostic markers (CEBPAdouble-mutation, NPM1/FLT3-ITD, WT1 mutation, and age). A score of −1 was assigned for each parameter associated with a favorable outcome (CEBPAdouble-mutation and NPM1mut/FLT3-ITD−) and a score of 1 for each factor associated with an adverse outcome (WT1 mutation and older age). The algebraic summation of these scores of each patient was the final score.
Discussion
In this study, we showed the incidence of WT1 mutations was 6.8% for total de novo non-M3 AML patients, and 7.0% and 8.3%, respectively, for patients with CN-AML of all age groups and those younger than 60 years. Further, WT1 mutations were distinctly associated with poor prognosis in AML patients.
Most studies concerning WT1 mutations in AML were focused on patients with normal karyotype. In this study, we recruited both patients with CN-AML and those with chromosomal aberrations. We found that WT1 mutations occurred with similar frequencies in patients with normal karyotype and those with abnormal cytogenetics. The mutation could be detected in patients with t(8;21), t(15;17) (not recruited in this study), and t(7;11). But none of the patients with inv(16) showed this gene mutation. In the limited reports in literature regarding WT1 mutation in AML patients afflicted with abnormal cytogenetics, the mutation was also found in patients with t(15;17), t(8;21), t(6;9), and 11q23 abnormalities.6,32-34
To the best of our knowledge, this study recruited the largest number of WT1-mutated patients with abnormal cytogenetics to date. We demonstrated that t(7;11)(p15;15), a translocation resulting in NUP98/HOXA9 fusion, was closely associated with the WT1 mutation. This chromosomal translocation occurs more frequently in the East than in the West35 and is associated with poor prognosis.36 It is interesting that NUP98 (on 11p15) and WT1 (on p13) involved in these 2 gene alterations are located in proximity on chromosome 11p. Whether the occurrence of t(7;11) would cause any specific conformational change closed to WT1 conferring its vulnerability to mutation warrants further study.
An association of WT1 mutation with FAB M6 subtype and younger age was also found in this study (Table 2). The WT1 mutation has been demonstrated in AML-M6 patients,11,37 but no correlation of this gene mutation with FAB subtype was demonstrated in previous reports.11,34 Four of 12 AML-M6 patients in this study had WT1 mutations. Clinical profiles of these patients were reviewed, and the diagnosis was confirmed by the overwhelming percentage of erythroblasts (> 50% of total nucleated cells) in bone marrow aspirate smears and/or the expression of glycophorin-A on blast cells. The cause of the close association between M6 and WT1 mutations is not clear. Kirschner et al38 revealed the permissive role of WT1 gene on the expression of erythropoietin receptor in human erythroid progenitors. This indirectly offered a connection between WT1 gene and erythroblasts. But additional elaborate studies are needed to clarify this point. In this study, patients with WT1 mutations were much younger than those without the mutation (38.5 years vs 53 years), a result similar to the reports of Renneville et al11 and Gaidzik et al,16 but not others.12,13 In another study focused on pediatric AML patients, the patients older than or equal to 3 years and less than 10 years of age had higher frequency of WT1 mutations than those younger than 3 years or those 10 years or older.34
Most patients (23 of 32, 72%) with WT1 mutations showed concurrent mutations of other genes, more frequently class 2 (16 of 23), but also class 1 mutations (13 of 23, Table 4). Although FLT3/ITD and CEBPA mutations were the most frequent gene alterations accompanied with WT1 mutation, there was no difference in the incidence of these 2 gene mutations between patients with and without WT1 mutation (supplemental Table 1). Significantly higher incidences of FLT3/ITD11,16 and CEBPA mutation in patients with WT1 mutations16 were reported by some researchers, but not others.12,13
The percentage of WT1 mutations in exons 7 and 9 in whole cohort in this study was 6.8%. Focused on 121 younger CN-AML, the percentage of WT1 mutations was 8.3%, similar to larger cohort studies that analyzed exons 7 and 9 mutations in younger adults with CN-AML (8.5%–10.7%).11-13 A slightly higher incidence (12.6%) of WT1 mutations was reported by Gaidzik el al who analyzed all exons 1 to 10 but found the mutations were clustered in exons 7 and 9.16 Hollink et al reported a high incidence (22%) of WT1 mutations in pediatric CN-AML patients,34 a finding compatible with the close association of WT1 mutation with younger age11,16 (and current study). Furthermore, they found that approximately half of WT1-mutated patients had double heterozygous mutations (15 of 35) or a homozygous mutation (2 of 35), in contrast to adults in whom most patients showed single heterozygous mutation11-13,16 (and current study). Whether there is a true difference in WT1 mutation pattern between childhood and adult AML and its significance needs to be clarified by further studies.
To the best of our knowledge, this study recruited the largest number of de novo adult AML patients for sequential analysis of WT1 mutations during clinical follow-ups. All 23 WT1-mutated patients studied lost their WT1 mutations at remission status, suggesting that these mutations were somatically acquired in the leukemogenesis. In contrast to NPM1 mutations, which are quite stable during disease progression,19 WT1 mutation status may change at relapse. Three of the 16 patients with WT1 mutations at diagnosis lost the mutations at relapse, whereas 2 others acquired additional mutations. On the other side, among the 110 patients who had no WT1 mutation at diagnosis, 3 acquired a novel WT1 mutation at relapse. These results were confirmed by reexamining the paired samples with GeneScan, a technique capable of detecting mutant DNA down to 5% of total DNA.26 This phenomenon cast doubts on the appropriateness of mutated WT1 as a surrogate marker in monitoring minimal residual disease. Only one report addressed the stability of WT1 mutations in the literature34 ; they found that 4 of the 28 WT1-wild patients gained WT1 mutations at relapse, but none of the 11 WT1-mutated patients lost the mutations. WT1 mutations may play a role in the disease progression in some patients.
By multivariate analysis using the Cox regression model, we demonstrated that WT1 mutation and older age were independent poor prognostic factors, whereas CEBPAdouble-mutation and NPM1mut/FLT3-ITD− were good prognostic factors in both total cohort and CN-AML patients. Based on these results, a survival scoring system incorporated these 3 molecular markers and age into survival analysis was designed to better stratify CN-AML patients into different risk groups. Indeed, this scoring system was more powerful than single marker to separate patients into different prognostic groups; and unlike the sophisticated gene-expression profiling, it could be easily implemented in routine clinical laboratories.
In conclusion, this study demonstrated that WT1 mutations occurred with similar frequencies in patients with normal karyotype and those with abnormal cytogenetics. The mutation was closely associated with younger age, FAB M6 subtype, and t(7;11)(p15;15), but inversely related to M0 subtype. Furthermore, the WT1 mutation was an independent poor risk factor for OS and RFS among total cohort and CN-AML patients. Sequential study during the clinical course showed the instability of WT1 mutations during disease progression. One should be cautious to use this mutation as a marker for minimal residual disease monitoring. Incorporation of the 3 gene mutations, including NPM1/FLT3-ITD, CEBPAdouble-mutation, and WT1, and age at diagnosis, which are closely associated with prognosis, into survival analyses can better stratify patients into different risk groups.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the National Science Council (NSC 97-2314-B002-015-MY3), Taiwan, Republic of China, and Department of Medical Research (NTUH.99P14), National Taiwan University Hospital.
Authorship
Contribution: H.-A.H. was responsible for literature collection, data management and interpretation, and manuscript writing; T.-C.H. was responsible for data management and manuscript writing; L.-I.L. was responsible for mutation analysis and interpretation; C.-Y.L. was responsible for statistical analysis and interpretation of the statistical findings; C.-Y.C., W.-C.C., M.Y., S.-Y.H., J.-L.T., B.-S.K., S.-C.H., S.-J.W., W.T., and Y.-C. Chen contributed patient samples and clinical data; M.-H.T., C.-F.H., Y.-C. Chiang, F.-Y.L., and M.-C.L. performed the gene mutation and chromosomal studies; and H.-F.T. planned, designed, and wrote the manuscript and coordinated the study over the entire period.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Hwei-Fang Tien, Department of Internal Medicine, National Taiwan University Hospital, No. 7 Chung-Shan South St, 100 Taipei, Taiwan, Republic of China; e-mail: hftien@ntu.edu.tw.
References
Author notes
H.-A.H. and T.-C.H. contributed equally to this study and should be considered as joint first authors.