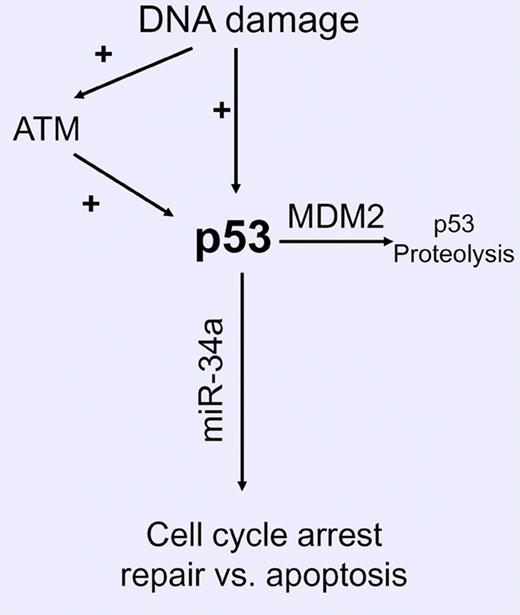
There has been considerable progress in analyzing p53 pathway integrity in CLL. Routine fluorescent in situ hybridization (FISH) analysis detects deletion of the region of chromosome 17 (17p13−), which includes the gene (TP53) coding for p53. 17p13− is a powerful predictor of resistance to therapy with purine analogues and alkylating agents and poor prognosis in CLL. Approximately 80% of CLL patients with 17p13- have loss of p53 function because of mutation in the remaining TP53 allele.1 In contrast, a minority of patients with 17p13- have a more indolent clinical course, which suggests that they retain some p53 function. CLL patients with mutant TP53 without 17p13- also have a poor prognosis.1 However, the biological consequences of monoallelic mutations of TP53 and the different types of mutations in TP53 on p53 function have still to be fully defined.2 In addition, defects in other components of the p53 pathway can affect function. Deletion of the ATM gene on chromosome 11 (11q22-) and increased MDM2 activity are known to decrease p53 pathway activity in CLL (see figure) and other as-yet-unknown defects could occur. Thus, there is a clear need to develop a simple test to measure p53 pathway function in CLL cells.
miR-34a expression and p53 pathway activity in CLL. Deletion (17p13) or mutation of TP53, deletion (11q22−) or mutation of ATM, and increased MDM2 activity (SNP309 GG) impair p53 pathway function in CLL, resulting in decreased expression of miR-34a.
miR-34a expression and p53 pathway activity in CLL. Deletion (17p13) or mutation of TP53, deletion (11q22−) or mutation of ATM, and increased MDM2 activity (SNP309 GG) impair p53 pathway function in CLL, resulting in decreased expression of miR-34a.
TP53 mutations can be detected by screening assays and defined by direct sequencing of the coding exons 2-11.1,2 However, these analyses will not detect mutations in the noncoding or regulatory regions that affect gene expression and are not informative about other functional defects in the p53 pathway (see figure). p53 pathway function can be assayed following exposure to ionizing radiation or with yeast culture–based methods that overcome these limitations, but are difficult to perform in routine clinical laboratories.2 In this issue of Blood, Asslaber et al3 report the results of a well-designed series of experiments that provide both confirmatory and novel data about the relationship of p53 pathway function, MDM2 single nucleotide polymorphism (SNP)309, and expression of microRNA-34a (miR-34a) that could contribute to the development of a clinically useful assay of p53 pathway function in CLL.
miR-34a is up-regulated by p53 and mediates some of the p53 pathway effects on cell cycling and apoptosis.4 CLL patients with defective p53 function have decreased miR-34a expression.5,,–8 In addition, decreased levels of miR-34a have been shown to predict poor prognosis and failure to respond to fludarabine-based therapy.5,,–8 Thus, measurement of expression of miR-34a could be used to determine p53 function. However, there are few data on the effect of defects of other components of the p53 pathway on miR-34a expression in CLL.
Asslaber et al provide additional data on the effects of MDM2 and ATM on expression of miR-34a.3 Because MDM2 accelerates proteolysis of p53, increased expression by the promoter region SNP309 GG genotype, compared with the TT variant, decreases p53 pathway function. In CLL patients, this results in decreased overall and treatment-free survival.9 Asslaber et al now show that patients with MDM2 SNP309 GG have lower levels of miR-34a.3 This is both additional evidence that miR-34a expression is a useful measurement of p53 pathway function and supportive of testing inhibitors of MDM2 such as nutlin-3a for treatment of CLL.10 Asslaber et al also provide evidence that deletion of ATM (11q22−) could result in lower levels of miR-34a. These data demonstrate that a miR-34a assay could measure activity of components of the p53 pathway other than p53.
These findings by Asslaber et al will need to be confirmed in larger and prospective studies with longer follow-up using response to treatment and overall survival rather than the surrogate end points of treatment-free survival and lymphocyte doubling time. In addition, further studies need to be done to better define the effects on miR-34a expression of mono- versus biallelic TP53 mutations and TP53 deletion with and without mutation in the remaining allele. The sensitivity of the assay, especially for small CLL subclones with p53 pathway dysfunction, will also need to be determined. miR-34a expression has previously been reported to be lower in CLL cells from patients with fludarabine refractory CLL in absence of TP53 mutation/17p13−.8 Asslaber et al now report that 10% of patients with low levels of miR-34a did not have known defects in the p53 pathway.3 This suggests that additional defects in the p53 pathway or miR-34a regulation and metabolism could be important in CLL and this will also require further investigation.
Asslaber et al introduce the intriguing possibility that therapeutic interventions could be directed at increasing the levels of miR-34a in CLL. They show that overexpression of miR-34a in CLL cells induced apoptosis. However, this occurred only in cells derived from patients with wild-type p53, and they suggest that this paradoxical result is compatible with previous reports that an intact p53 pathway is required for miR-34a effects. Unfortunately, this result does not offer any new opportunities to develop novel treatment for those patients with p53 pathway dysfunction who are most in need of more effective therapy.
Conflict-of-interest disclosure: C.S.Z. has research funding from Genentech, Genzyme, and Novartis. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal