Abstract
Factor H autoantibodies have been reported in approximately 10% of patients with atypical hemolytic uremic syndrome (aHUS) and are associated with deficiency of factor H–related proteins 1 and 3. In this study we examined the prevalence of factor H autoantibodies in the Newcastle cohort of aHUS patients, determined whether the presence of such autoantibodies is always associated with deficiency of factor H–related proteins 1 and 3, and examined whether such patients have additional susceptibility factors and/or mutations in the genes encoding complement regulator/activators. We screened 142 patients with aHUS and found factor H autoantibodies in 13 individuals (age 1-11 years). The presence of the autoantibodies was confirmed by Western blotting. By using multiplex ligation-dependent probe amplification we measured complement factor H–related (CFHR)1 and CFHR3 copy number. In 10 of the 13 patients there were 0 copies of CFHR1, and in 3 patients there were 2. In 3 of the patients with 0 copies of CFHR1 there was 1 copy of CFHR3, and these individuals exhibited a novel deletion incorporating CFHR1 and CFHR4. In 5 patients mutations were identified: 1 in CFH, 1 in CFI, 1 in CD46, and 2 in C3. The latter observation emphasizes that multiple concurrent factors may be necessary in individual patients for disease manifestation.
Introduction
It is now well established that atypical hemolytic uremic syndrome (aHUS) is a disease of complement dysregulation.1 Mutations have been found in the genes encoding both complement regulators (factor H, factor I, and membrane cofactor protein) and complement activators (factor B and C3) in approximately 50% of patients.2-6 In addition, factor H autoantibodies have been described in a further approximately 10% of patients.7-9 These antibodies have been shown to block the C-terminal recognition domain of factor H,8 an area in which it is known that complement factor H (CFH) mutations are associated with aHUS cluster.10 In addition it has been shown that the majority of patients with factor H autoantibodies have associated complete deficiency of factor H–related proteins 1 and 39 secondary to a homozygous deletion of the genes (complement factor H–related [CFHR]1 and CFHR3) that encode these proteins. This deletion occurs secondary to nonallelic homologous recombination (NAHR) in segmental duplications in the regulators of complement activation gene cluster at chromosome 1q32 and is known to be associated with an increased risk of aHUS,11 especially in particular subgroups.12 NAHR is also known to predispose to the formation of a hybrid complement gene associated with aHUS.13
Recent studies have shown that patients with aHUS may have mutations in more than one complement regulator/activator, but such mutations have not been described in association with factor H autoantibodies. It is also well established that there are additional susceptibility factors (single nucleotide polymorphisms [SNPs] and haplotype blocks) in CFH and CD46 that increase the risk of developing aHUS.14 In this study we have therefore examined the prevalence of factor H autoantibodies in the Newcastle cohort of aHUS patients, determined whether the presence of such autoantibodies is always associated with deficiency of factor H–related proteins 1 and 3, and examined whether such patients have additional susceptibility factors and/or mutations in the genes encoding complement regulators/activators.
Methods
Subjects
Patients from the Newcastle cohort of aHUS (n = 308) were included in this study. Serum samples at the time of or shortly after presentation were available from 142 aHUS patients, and of these paired DNA samples were available for 128. Control paired serum and DNA samples were available from 100 local blood donors. The study was approved by the Northern and Yorkshire Multi-Center Research Ethics Committee and informed consent obtained in accordance with the Declaration of Helsinki.
Factor H autoantibody assay
This assay was undertaken in 142 aHUS patients and 100 control subjects. Flexible 96-well plates were coated with 5 μg/mL of purified factor H (Merck Chemicals Ltd) or bovine serum albumin (BSA; Sigma) or molar equivalents of factor H fragments (short consensus repeats [SCRs] 1-4, 8-15, 19-20)15,16 or molar equivalents of a factor H–related protein 1 fragment (SCRs 4-5)16 in 0.1M carbonate buffer, pH 9.6, and incubated overnight at 4°C. Plates were then washed thrice with phosphate-buffered saline (PBS)/Tween 0.05%, followed by blocking in PBS/Tween 0.05%/BSA 1% for 1 hour at room temperature. After blocking, a 1/50 dilution of sera in PBS/Tween 0.05% was loaded in triplicate and incubated for 1 to 2 hours. Plates were washed thrice and then blocked as described previously. Goat anti-human IgG horse radish peroxidase (HRP, Stratech Scientific) at 1/4000 was then added and incubated for 1 hour at room temperature. Plates were then washed twice with PBS/Tween 0.05% and twice with PBS. OPD solution was prepared according to the manufacturer's instructions (Merck Chemicals Ltd) and added to each well for exactly 15 minutes before the reaction was stopped by the use of 10% sulfuric acid. Plates were then analyzed with the use of a MULTISKAN Ascent plate reader (Thermo-Scientific). Triplicate data were analyzed and mean BSA readings subtracted from mean factor H readings to control for anti-BSA/false-positive results.
SDS-PAGE and Western blotting
Detection of factor H autoantibody.
Purified complement factor H (Comptech) was diluted in solubilizing buffer and loaded onto a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) preparative gel. After transfer to nitrocellulose and blocking as described previously, the nitrocellulose was cut into 0.5- to 1-cm wide strips. These strips were then incubated with individual sera samples (1/100 in 5% dried milk/PBS) for 1 to 2 hours at room temperature. After washing as described previously, bound autoantibody was detected by the use of goat anti-human IgG-HRP incubated for 1 to 2 hours at room temperature. Blots were then washed twice with PBS/0.1% Tween 20 and with PBS only. All blots were developed by the use of an enhanced chemiluminescence (ECL) substrate (Pierce) according to the manufacturer's specifications.
Detection of factor H and factor H–related protein 1.
Sera was diluted 1/100 in solubilizing buffer and electrophoresed on 10% SDS-PAGE gels, blotted onto nitrocellulose, and blocked with 5% dried milk/PBS. Sections of the blots were incubated overnight at 4°C with either polyclonal goat anti-human factor H (1/10 000 or 1/50 000 as appropriate; Comptech) or C18/3 (1/2000; Santa Cruz Biotechnology Inc) in 5% dried milk/PBS. Blots were washed 3 times in PBS/0.1% Tween 20 and subsequently incubated with donkey anti-goat-HRP or goat anti-mouse-HRP (1/3000 in 5% dried milk/PBS), as appropriate. After 1 to 2 hours at room temperature, blots were washed twice with PBS/0.1% Tween 20 and with PBS only.
Detection of factor H–related protein 3.
This step was similar to factor H and factor H–related protein 1, but sera also was incubated with anti-HSA beads (Sartorious) for 30 minutes before analysis to remove albumin. After this step, a 1/500 dilution of a rabbit polyclonal anti–factor H–related protein 3 (a generous gift from Professor P. Zipfel, Leibniz Institute for Natural Product Research and Infection Biology, Jena, Germany) with a goat anti–rabbit–HRP as secondary was used to probe the nitrocellulose.
Measurement of CFHR1, CFHR3, and CFHR4 copy number
CFHR1 and CFHR3 copy number was measured in 196 aHUS patients (including the 128 patients with paired serum and DNA samples) and 505 control subjects. The normal control subjects comprised 405 DNA samples obtained from the Health Protection Agency Culture Collections17 and 100 DNA samples obtained from the aforementioned local blood donors. The samples from the Health Protection Agency also were originally obtained from a control population of randomly selected, nonrelated United Kingdom white blood donors. CFHR1 and CFHR3 copy number was measured by the use of multiplex ligation-dependent probe amplification18 with a kit from MRC Holland (SALSA MLPA kit P236-A1 ARMD). In this kit there are 5 probes for CFHR1 and 6 for CFHR3. Details of these probes are given in Table 1.
MLPA probes used to determine CFHR1 and CFHR3 copy number
| Gene, exon . | Ligation site . | Partial sequence (20 nucleotides adjacent to ligation site) . |
|---|---|---|
| CFHR3, exon 1 | Intron 1 | AGGTAAGTTA-AAAGAGATCT |
| CFHR3, exon 2 | Intron 1 | CATTTTCTTG-TGGAATTACA |
| CFHR3, exon 3 | Intron 3 | CGGACGACAG-TCTCAGACTT |
| CFHR3, exon 4 | Intron 4 | GGGTTATATG-AATTCCTACA |
| CFHR3, exon 6 | Intron 5 | TTCCCCAACA-TCACAGCAGA |
| CFHR3, exon 6 | 1003-1002 reverse | TCCCTTCCCG-ACACACTGCT |
| CFHR1, exon 1 | Intron 1 | GGATAATTCA-ATTGAAATGG |
| CFHR1, exon 3 | Intron 3 | AGAGTTTCAG-GTCCATGTGT |
| CFHR1, exon5 | Intron 5 | AATCTGTGAT-TATTTTGTTA |
| CFHR1, exon 6 | 945-946 | CCTGTTCTCA-AATAAAAGCTT |
| CFHR1, exon 6 | 1246-1247 | TTTTCCAAGT-TTTAATATGG |
| Gene, exon . | Ligation site . | Partial sequence (20 nucleotides adjacent to ligation site) . |
|---|---|---|
| CFHR3, exon 1 | Intron 1 | AGGTAAGTTA-AAAGAGATCT |
| CFHR3, exon 2 | Intron 1 | CATTTTCTTG-TGGAATTACA |
| CFHR3, exon 3 | Intron 3 | CGGACGACAG-TCTCAGACTT |
| CFHR3, exon 4 | Intron 4 | GGGTTATATG-AATTCCTACA |
| CFHR3, exon 6 | Intron 5 | TTCCCCAACA-TCACAGCAGA |
| CFHR3, exon 6 | 1003-1002 reverse | TCCCTTCCCG-ACACACTGCT |
| CFHR1, exon 1 | Intron 1 | GGATAATTCA-ATTGAAATGG |
| CFHR1, exon 3 | Intron 3 | AGAGTTTCAG-GTCCATGTGT |
| CFHR1, exon5 | Intron 5 | AATCTGTGAT-TATTTTGTTA |
| CFHR1, exon 6 | 945-946 | CCTGTTCTCA-AATAAAAGCTT |
| CFHR1, exon 6 | 1246-1247 | TTTTCCAAGT-TTTAATATGG |
CFHR1 indicates complement factor H–related; and MLPA, multiplex ligation-dependent probe amplification.
CFHR4 copy number was measured by the use of a multiplex polymerase chain reaction (PCR) assay designed to amplify CFHR4 exon 2 and intron 1. CFHR1 copy number was measured by the use of the same multiplex assay designed to amplify CFHR1 introns 3 and 5. This acted as an internal control for the CFHR4 assay. PCRs were carried out in 25-μL volumes with the use of 150 ng of DNA and contained 0.5mM each dNTP, 6.7mM MgCl2, 12.5pM of each primer, 1 U of a hot-start Taq polymerase (Immolase, Bioline) in a buffer of 16mM (NH4)2SO4, 67mM Tris-HCI, and 0.01% Tween-20. PCR cycling conditions were such that amplification remained in the linear phase of the reaction (95°C for 10 minutes, with 20 cycles of 94°C for 45 seconds, 60°C for 45 seconds, 72°C for 1 minute, and a final extension of 10 minutes at 72°C). Primers that amplify KCNT2 exon 9 and KCNT2 exon 17 were included in the assay as controls for normal dosage. One of each pair of primers was fluorescently labeled (5′FAM), and all primers are shown in Table 2. After PCR, amplification products were analyzed by capillary electrophoresis (ABI, PerkinElmer). Peak areas were obtained for each sample and dosage quotients calculated.
Primer sequences for the CFHR1 and CFHR4 QF-PCR dosage analysis
| Exon . | Forward (5′-3′) . | Reverse (5′-3′) . | bp . |
|---|---|---|---|
| KCNT2, exon 17 | TGAATCGATGATGTATCTGCAA | AGTGGCCCTACCCTGTCTCT | 98 |
| CFHR1, intron 5 | CAAAATAATCACAGATTATTGAACAACC | CACACCCAGCCCTAAAGAGA | 106 |
| CFHR4, exon 2 | CGCGTAGACCATACTTTCCA | ACCCATCTTGTGTGCAGTGA | 117 |
| CFHR4, intron 1 | TCTAAACACTCAGCTTCCCTCT | TCTCACAAAATATGCTACTTCTGC | 126 |
| CFHR1, intron 3 | ACGGAATCAAAAATGTCGAAATAG | TCATCAGAGAGTTTCAGGTCCA | 135 |
| KCNT2, exon 9 | GTAGAACCACAAGAGCAACAC | ATAGGAAAGAAGCTGAATCTC | 145 |
| Exon . | Forward (5′-3′) . | Reverse (5′-3′) . | bp . |
|---|---|---|---|
| KCNT2, exon 17 | TGAATCGATGATGTATCTGCAA | AGTGGCCCTACCCTGTCTCT | 98 |
| CFHR1, intron 5 | CAAAATAATCACAGATTATTGAACAACC | CACACCCAGCCCTAAAGAGA | 106 |
| CFHR4, exon 2 | CGCGTAGACCATACTTTCCA | ACCCATCTTGTGTGCAGTGA | 117 |
| CFHR4, intron 1 | TCTAAACACTCAGCTTCCCTCT | TCTCACAAAATATGCTACTTCTGC | 126 |
| CFHR1, intron 3 | ACGGAATCAAAAATGTCGAAATAG | TCATCAGAGAGTTTCAGGTCCA | 135 |
| KCNT2, exon 9 | GTAGAACCACAAGAGCAACAC | ATAGGAAAGAAGCTGAATCTC | 145 |
bp indicates base pair; CFHR1, complement factor H–related; and QF-PCR, quantitative fluorescence polymerase chain reaction.
Complement assays
C3 and C4 levels were measured by rate nephelometry (Beckman Array 360). Factor H and factor I levels were measured by radioimmunodiffusion (Binding Site). The normal ranges were C3 (0.68-1.38 g/L), C4 (0.18-0.60 g/L), factor H (0.35-0.59 g/L), and factor I (38-58 mg/L).
Mutation screening and genotyping
In all patients found to have factor H autoantibodies mutation, screening of CFH, CD46, CFI, CFB, and C3 was or had previously been undertaken by the use of direct fluorescent sequencing as described.2-4,6,19 Genotyping of the following SNPs was undertaken by the use of direct sequencing: CD46 -652A>G (rs2796267), CD46 -366A>G (rs2796268), CD46 c.4070T>C (rs7144), CFH -331C>T (rs3753394), CFH c.2016A>G p.Gln672Gln (rs3753396), and CFH c.2808G>T p.Glu936Asp (rs1065489).
Results
Factor H autoantibodies
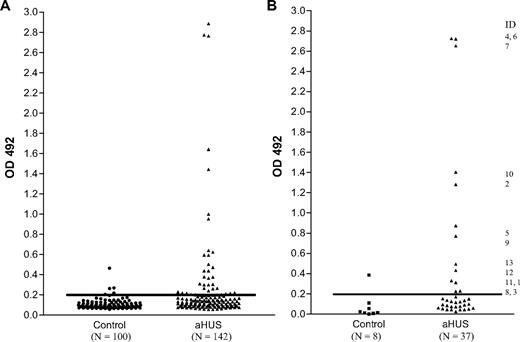
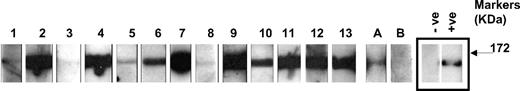
In enzyme-linked immunosorbent assays (ELISAs), the mean optical density plus 2SD in the control group before BSA subtraction was 0.215. Eight control patients and 37 aHUS patients had an OD reading greater than or close to this value (Figure 1A). BSA subtraction was undertaken in all these samples. Subsequently 13 patients and 1 control patient had an OD greater than 0.2 (Figure 1B). This cutoff was used to identify the presence of a factor H autoantibody. The specificity of the autoantibodies was confirmed in all 13 patients and the 1 control subject by Western blotting (Figure 2). By the use of a standard strip blot approach, patient sera was exposed to factor H immobilized on nitrocellulose and any factor H–specific human IgG detected. By the use of this approach, autoantibody binding to factor H was demonstrated for all subjects originally identified by ELISA. Signal intensity varied between patients, but in general strong responses via ELISA were mirrored in the western blot analysis.
Detection of factor H autoantibodies. Factor H autoantibodies in aHUS patients and control subjects were detected by the use of a sensitive ELISA as described in “Factor H autoantibody assay.” (A) OD492 for 100 control subjects and 142 aHUS patients. (B) OD492 for 8 control subjects and 37 aHUS patients with an uncorrected OD value greater than or close to 0.215 (the mean + 2SD for the 100 control subjects) after background reactivity to BSA has been subtracted to exclude false-positive interactions. The identity of the 13 factor H autoantibody–positive patients is shown.
Detection of factor H autoantibodies. Factor H autoantibodies in aHUS patients and control subjects were detected by the use of a sensitive ELISA as described in “Factor H autoantibody assay.” (A) OD492 for 100 control subjects and 142 aHUS patients. (B) OD492 for 8 control subjects and 37 aHUS patients with an uncorrected OD value greater than or close to 0.215 (the mean + 2SD for the 100 control subjects) after background reactivity to BSA has been subtracted to exclude false-positive interactions. The identity of the 13 factor H autoantibody–positive patients is shown.
Analysis of antifactor H binding by Western blotting. Purified factor H was run out on 10% SDS-PAGE and transferred to nitrocellulose. Strips of nitrocellulose were then incubated with sera collected from subjects and bound antibody detected as described in “Methods.” ECL Western blotting substrate was used to visualize bound antibody. The same secondary reagent and exposure times were used throughout. Sera from known factor H autoantibody–positive and –negative subjects were used to allow standardization across multiple experiments. The positive (+ve) and negative controls (−ve) are on adjacent strips on the same autorad film and are shown as representative control signals (black box). The factor H autoantibody patients (1-13) and 2 normal subjects, A and B, are shown. These data are from a collection of sequential experiments. Molecular weight markers are shown, and the data are representative of at least 3 independent experiments.
Analysis of antifactor H binding by Western blotting. Purified factor H was run out on 10% SDS-PAGE and transferred to nitrocellulose. Strips of nitrocellulose were then incubated with sera collected from subjects and bound antibody detected as described in “Methods.” ECL Western blotting substrate was used to visualize bound antibody. The same secondary reagent and exposure times were used throughout. Sera from known factor H autoantibody–positive and –negative subjects were used to allow standardization across multiple experiments. The positive (+ve) and negative controls (−ve) are on adjacent strips on the same autorad film and are shown as representative control signals (black box). The factor H autoantibody patients (1-13) and 2 normal subjects, A and B, are shown. These data are from a collection of sequential experiments. Molecular weight markers are shown, and the data are representative of at least 3 independent experiments.
Binding of autoantibodies to CFH and CFHR1 fragments
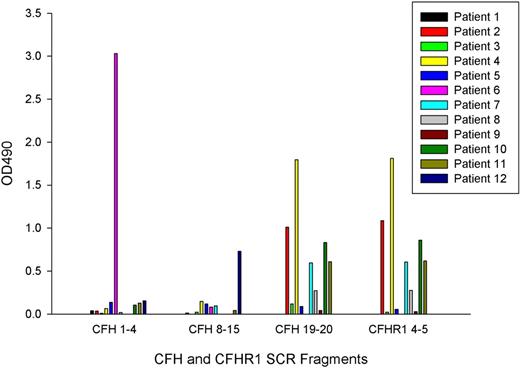
The site of binding of the autoantibodies was determined with the use of factor H fragments SCRs 1 to 4, 8 to 15, 19 to 20, and a factor H–related protein 1 fragment SCR 4 to 5 (Figure 3). Antibodies from 1 patient (number 6, CFHR1 copy number 2) bound only to factor H SCR 1 to 4, from 1 patient (number 12, CFHR1 copy number 0) they bound only to factor H SCR 8 to 15, and from 7 patients (numbers 2, 3, 4, 7, 8, 10, and 11) they bound to both factor H SCR 19 to 20 and factor H–related protein 1 SCR 4 to 5 in a similar pattern. Of these 7 patients, 6 had 0 copies of CFHR1, and 1 (number 7) had 2 copies of CFHR1. Three patients (numbers 1, 5, and 9) demonstrated binding to full-length factor H but not to any of these fragments. The control subject with a positive factor H autoantibody demonstrated binding to factor H SCR fragment 8 to 15 with an OD 490 of 1.18.
Autoantibody reactivity with short factor H fragments. Autoantibody binding to factor H fragments (corresponding to SCRs 1-4, 8-15, and 19-20) and a factor H–related protein 1 fragment (SCR 4-5) was assessed by the use of ELISA in a similar manner to the original autoantibody screen. Molar equivalent concentrations of the SCR fragments were coated onto separate ELISA plates, and a BSA subtraction was performed. Results are representative of 3 separate experiments.
Autoantibody reactivity with short factor H fragments. Autoantibody binding to factor H fragments (corresponding to SCRs 1-4, 8-15, and 19-20) and a factor H–related protein 1 fragment (SCR 4-5) was assessed by the use of ELISA in a similar manner to the original autoantibody screen. Molar equivalent concentrations of the SCR fragments were coated onto separate ELISA plates, and a BSA subtraction was performed. Results are representative of 3 separate experiments.
CFHR1, CFHR3, and CFHR4 copy number
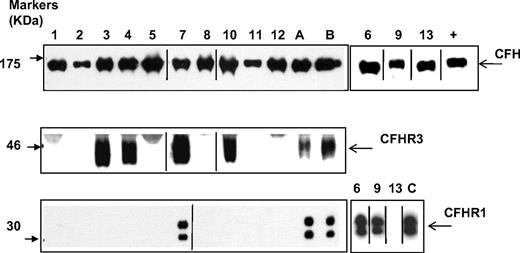
Homozygous deletion of CFHR1 was significantly more frequent in the aHUS patients than control subjects (aHUS 12.8% vs control 3.0%). Likewise the allele frequency of CFHR1 deletion was significantly greater in aHUS patients than in control subjects (aHUS 26.5% vs control 17.3%; Tables 3–4). The CFHR1 deletion frequency in the aHUS patients is not consistent with Hardy-Weinberg equilibrium (P = 2.8 × 10−4). This finding is the result of an excess of homozygous CFHR1 deletion in the aHUS patients; the frequency of heterozygous CFHR1 deletion was very similar in aHUS patients and control subjects (aHUS 27.6% vs control 28.7%). In the 13 factor H autoantibody–positive patients, 10 had 0 copies of CFHR1. However, 3 patients (6, 7, and 9) had 2 copies of CFHR1, as did the 1 control patient who had detectable factor H autoantibodies. This patient's status was confirmed at a protein level by Western blotting (Figure 4). In the 128 aHUS patients with paired serum and DNA samples, there were 6 patients who had no copies of either CFHR1 or CFHR3 whose serum samples did not show evidence of factor autoantibodies. In the 10 patients who were positive for factor H autoantibodies and had 0 copies of CFHR1, there were 3 (patients 3, 4, and 10) who had a single copy of CFHR3 rather than none. This finding was confirmed by Western blotting (Figure 4) with the use of factor H–related protein 3 antisera (kindly provided by Professor Peter Zipfel). In these 3 patients we demonstrated that this finding was caused by the presence of a novel deletion (∼ 125 kb) in the regulators of complement activation (RCA) cluster that incorporates CFHR1 and CFHR4, leaving CFHR3 intact (Figure 5).
CFHR1 copy number in aHUS patients and control subjects
| CFHR1 copy number . | aHUS (n = 196) . | Controls (n = 505) . |
|---|---|---|
| 0 | 25 | 15 |
| 1 | 54 | 145 |
| 2 | 115 | 342 |
| 3 | 2 | 3 |
| CFHR1 copy number . | aHUS (n = 196) . | Controls (n = 505) . |
|---|---|---|
| 0 | 25 | 15 |
| 1 | 54 | 145 |
| 2 | 115 | 342 |
| 3 | 2 | 3 |
aHUS indicates atypical hemolytic uremic syndrome; and CFHR1, complement factor H–related 1.
χ2 = 25.9; P = 1.0 × 10−5.
CFHR1 allele frequency in aHUS patients and control subjects
| CFHR1 allele frequency . | aHUS patients (n = 196) . | Controls (n = 505) . |
|---|---|---|
| Deleted | 104 | 175 |
| Present | 288 | 835 |
| CFHR1 allele frequency . | aHUS patients (n = 196) . | Controls (n = 505) . |
|---|---|---|
| Deleted | 104 | 175 |
| Present | 288 | 835 |
aHUS indicates atypical hemolytic uremic syndrome; CFHR1, complement factor H–related 1.
χ2 = 15.0; P = 1.1 × 10−4.
Analysis of factor H, factor H–related protein 3, and factor H–related protein 1 in patients with factor H autoantibodies. Sera from subjects were run out on 10% SDS-PAGE and transferred to nitrocellulose. Factor H, factor H–related protein 3, and factor H–related protein 1 were then detected by staining as described in the Methods section. ECL Western blotting substrate was used to visualize bound antibody. Sera was available from all 13 patients (samples 1-13) for analysis of factor H and factor H–related protein 1, but only from 9 patients for factor H–related protein 3. These 9 samples plus control samples were run on parallel gels. A, B, and C are normal controls known to have 2 copies of CFHR1 and CFHR3. Data from the remaining 3 samples are shown on the right. Purified factor H (equivalent to 0.5 mg/mL) was used as a positive control (+) in the smaller antifactor H blot. Black vertical lines indicate a repositioned gel lane, and black boxes illustrate the individual blots used. This figure is representative of several independent experiments.
Analysis of factor H, factor H–related protein 3, and factor H–related protein 1 in patients with factor H autoantibodies. Sera from subjects were run out on 10% SDS-PAGE and transferred to nitrocellulose. Factor H, factor H–related protein 3, and factor H–related protein 1 were then detected by staining as described in the Methods section. ECL Western blotting substrate was used to visualize bound antibody. Sera was available from all 13 patients (samples 1-13) for analysis of factor H and factor H–related protein 1, but only from 9 patients for factor H–related protein 3. These 9 samples plus control samples were run on parallel gels. A, B, and C are normal controls known to have 2 copies of CFHR1 and CFHR3. Data from the remaining 3 samples are shown on the right. Purified factor H (equivalent to 0.5 mg/mL) was used as a positive control (+) in the smaller antifactor H blot. Black vertical lines indicate a repositioned gel lane, and black boxes illustrate the individual blots used. This figure is representative of several independent experiments.
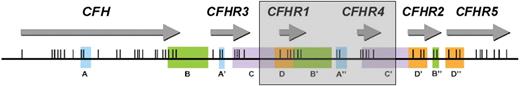
A novel deletion including CFHR1 and CFHR4. Position of the genes encoding factor H and the factor H–related proteins in a centromeric segment of the RCA cluster at 1q32. Regions of high sequence identity (originally determined by Male et al20 ) are indicated by the same letter and color. Exons are indicated as vertical lines. The shaded box shows the presence of a novel ∼ 125-kb deletion, which includes CFHR1 and CFHR4. This deletion occurs within the duplicons C/C′ as a result of nonallelic homologous recombination.
A novel deletion including CFHR1 and CFHR4. Position of the genes encoding factor H and the factor H–related proteins in a centromeric segment of the RCA cluster at 1q32. Regions of high sequence identity (originally determined by Male et al20 ) are indicated by the same letter and color. Exons are indicated as vertical lines. The shaded box shows the presence of a novel ∼ 125-kb deletion, which includes CFHR1 and CFHR4. This deletion occurs within the duplicons C/C′ as a result of nonallelic homologous recombination.
Using quantitative PCR we showed that these 3 patients (3, 4, and 10) had 1 copy of CFHR4, whereas the remaining 10 all had 2 copies of CFHR4. Thus, in these 3 patients there was on 1 allele a deletion of CFHR3/CFHR1 and on the other allele a CFHR1/CFHR4 deletion. In 143 aHUS patients, copy number of both CFHR3 and CFHR1 was measured. In 4 of these patients there was evidence of a heterozygous CFHR1/CFHR4 deletion, including the 3 aforementioned patients who were found to have factor H autoantibodies. A serum sample was not available for the other patient. In all 4 patients there was evidence of a CFHR3/CFHR1 deletion on the other allele. Therefore, the allele frequency of the CFHR1/CFHR4 deletion in aHUS patients is 1.4%. In 505 control subjects there were 9 individuals with evidence of a heterozygous CFHR1/CFHR4 deletion. In 2 of these there was evidence of a CFHR3/CFHR1 deletion on the other allele. Therefore, the allele frequency of the CFHR1/CFHR4 deletion is 0.9% in control subjects. There was no significant difference between aHUSpatients and control subjects in the allele frequency of the CFHR1/CFHR4 deletion (χ2 = 1.318, DF = 1, P = .251), but there was insufficient power for us to be certain. However, the combination of a CFHR3/CFHR1 deletion on one allele and a CFHR1/CFHR4 deletion on the other was significantly more frequent in the aHUS patients (χ2 = 7.004, DF = 1, P = .008).
Clinical details of the patients with factor H autoantibodies
The clinical details of the 13 patients with factor H autoantibodies are shown in Table 5. There were 6 male and 7 female patients, and the median age at presentation was 8 years (range, 1-11 years). The median length of follow-up was 6 years (range, 1-11 years). Seven patients have regained renal function, 2 of these are on prophylactic long-term plasma exchange, and 1 has had 3 relapses that were successfully treated on each occasion with plasma exchange. Six patients progressed to end-stage renal failure; of these, 3 have received a renal allograft without recurrence in a follow-up period ranging from 2 to 6 years. Levels of C3 were low in 3 patients (numbers 1, 2, and 12) at presentation. One patient (number 5) had a low factor H level, but mutation screening of CFH did not show any abnormality. Mutation screening of CFH, CD46, CFI, CFB, and C3 was undertaken in all patients with the use of direct fluorescent sequencing as described previously (Table 6).2-4,6,19 One patient had a heterozygous sequence variant in CFH (c.2850G>T, p.Gln950His), which has been described previously in aHUS but has also been reported by one group in healthy subjects.21 One patient who has been reported previously had a sequence variant in CD46 (c.718T>C, Ser240Pro), which has been shown to be functionally significant.4 A third patient had a previously unreported sequence variant in exon 11 of CFI (Exon 11, c.1216C>T, p.Arg406Cys), which is predicted to be functionally significant. Two novel sequence variants were identified in C3 in 2 patients. One was in the 5′UTR (c.-3_-2dup) and the other in exon 15 (c.1898A>G;p.Lys633Arg). The genotyping results for CD46 and CFH susceptibility factors are shown in Table 7. Ten of the 13 patients were heterozygous for the at-risk G allele of CD46 -652A>G (rs2796267), and 2 were homozygous. Seven of the 13 patients were heterozygous for the at-risk G allele of CD46 -366A>G (rs2796268), and 4 were homozygous. Six of the 13 patients were heterozygous for the at-risk C allele of CD46 c.4070T>C (rs7144), and 4 were homozygous. From these results we inferred that 8 patients (numbers 1, 3, 4, 5, 6, 7, 11, and 12) were heterozygous for the at-risk CD46 haplotype CD46GGAAC, and 2 (numbers 2 and 10) were homozygous. Six patients were heterozygous for the at risk T allele of CFH -331C>T (rs3753394), and 2 were homozygous. One patient was homozygous for the at-risk G allele of CFH c.2016A>G;p.Gln672Gln (rs3753396), and 1 patient was homozygous for the at-risk allele of CFH c.2808G > T;p.Glu936Asp (rs1065489). From these results we inferred that 1 patient (number 6) was homozygous for the at-risk CFHTGTGGT (H3) haplotype.
Clinical details of the aHUS patients with factor H autoantibodies
| Patient ID . | Age at presentation, y . | Sex . | Clinical outcome . | Length of follow-up, y . | Transplanted . | C3, g/L . | C4, g/L . | Factor H, g/L . | Factor I, mg/L . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | M | Recovered renal function. 2 relapses. On PE 2×/week | 2 | No | 0.61 | 0.1 | 0.37 | 39 |
| 2 | 11 | M | Recovered renal function | 5 | No | 0.57 | 0.15 | 0.44 | 70 |
| 3 | 8 | F | ESRF | 9 | No | 0.86 | 0.25 | 0.49 | 58 |
| 4 | 4 | F | ESRF | 1 | No | 0.89 | 0.3 | 0.52 | 41 |
| 5 | 10 | F | ESRF | 5 | Yes, 3 y no recurrence | 0.92 | 0.2 | 0.29 | n/a |
| 6 | 8 | M | ESRF | 6 | No | 1.08 | 0.36 | 0.5 | 74 |
| 7 | 6 | M | ESRF | 7 | Yes, 2 years no recurrence | 0.77 | 0.28 | 0.56 | 77 |
| 8 | 1 | M | Recovered renal function | 4 | No | 1.59 | 0.38 | 0.67 | 75 |
| 9 | 9 | F | Recovered renal function | 11 | No | n/a | n/a | 0.63 | n/a |
| 10 | 5 | M | Recovered renal function; multiple relapses; PE 1×/2 weeks | 3 | No | 1.06 | 0.13 | 0.45 | 68 |
| 11 | 4 | F | Recovered renal function; 3 relapses treated with PE | 6 | No | 0.85 | 0.24 | n/a | n/a |
| 12 | 10 | F | ESRF | 8 | Yes, 6 y no recurrence | 0.51 | 0.23 | 0.57 | 62 |
| 13 | 9 | F | Recovered renal function | 8 | No | n/a | n/a | n/a | n/a |
| Patient ID . | Age at presentation, y . | Sex . | Clinical outcome . | Length of follow-up, y . | Transplanted . | C3, g/L . | C4, g/L . | Factor H, g/L . | Factor I, mg/L . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | M | Recovered renal function. 2 relapses. On PE 2×/week | 2 | No | 0.61 | 0.1 | 0.37 | 39 |
| 2 | 11 | M | Recovered renal function | 5 | No | 0.57 | 0.15 | 0.44 | 70 |
| 3 | 8 | F | ESRF | 9 | No | 0.86 | 0.25 | 0.49 | 58 |
| 4 | 4 | F | ESRF | 1 | No | 0.89 | 0.3 | 0.52 | 41 |
| 5 | 10 | F | ESRF | 5 | Yes, 3 y no recurrence | 0.92 | 0.2 | 0.29 | n/a |
| 6 | 8 | M | ESRF | 6 | No | 1.08 | 0.36 | 0.5 | 74 |
| 7 | 6 | M | ESRF | 7 | Yes, 2 years no recurrence | 0.77 | 0.28 | 0.56 | 77 |
| 8 | 1 | M | Recovered renal function | 4 | No | 1.59 | 0.38 | 0.67 | 75 |
| 9 | 9 | F | Recovered renal function | 11 | No | n/a | n/a | 0.63 | n/a |
| 10 | 5 | M | Recovered renal function; multiple relapses; PE 1×/2 weeks | 3 | No | 1.06 | 0.13 | 0.45 | 68 |
| 11 | 4 | F | Recovered renal function; 3 relapses treated with PE | 6 | No | 0.85 | 0.24 | n/a | n/a |
| 12 | 10 | F | ESRF | 8 | Yes, 6 y no recurrence | 0.51 | 0.23 | 0.57 | 62 |
| 13 | 9 | F | Recovered renal function | 8 | No | n/a | n/a | n/a | n/a |
aHUS indicates atypical hemolytic uremic syndrome; ESRF, end-stage renal failure; F, female; M, male; n/a, not available; and PE, plasma exchange.
Mutation screening of CFH, CD46, CFI, CFB, C3, and measurement of CFHR1, CFHR3, and CFHR4 copy number
| Patient ID . | CFH . | CD46 . | CFI . | CFB . | C3 . | Copy number . | ||
|---|---|---|---|---|---|---|---|---|
| CFHR1 . | CFHR3 . | CFHR4 . | ||||||
| 1 | nmd | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
| 2 | nmd | nmd | nmd | nmd | c.-3_-2dup | 0 | 0 | 2 |
| 3 | nmd | nmd | nmd | nmd | nmd | 0 | 1 | 1 |
| 4 | nmd | nmd | Exon 11, c.1216C>T;p.Arg406Cys | nmd | nmd | 0 | 1 | 1 |
| 5 | nmd | nmd | nmd | nmd | c.1898A>G;p.Lys633Arg | 0 | 0 | 2 |
| 6 | nmd | nmd | nmd | nmd | nmd | 2 | 2 | 2 |
| 7 | nmd | nmd | nmd | nmd | nmd | 2 | 2 | 2 |
| 8 | nmd | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
| 9 | nmd | c.718T>C;p. Ser240Pro | nmd | nmd | nmd | 2 | 2 | 2 |
| 10 | nmd | nmd | nmd | nmd | nmd | 0 | 1 | 1 |
| 11 | nmd | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
| 12 | c.2850G>T; p.Gln950His | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
| 13 | nmd | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
| Patient ID . | CFH . | CD46 . | CFI . | CFB . | C3 . | Copy number . | ||
|---|---|---|---|---|---|---|---|---|
| CFHR1 . | CFHR3 . | CFHR4 . | ||||||
| 1 | nmd | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
| 2 | nmd | nmd | nmd | nmd | c.-3_-2dup | 0 | 0 | 2 |
| 3 | nmd | nmd | nmd | nmd | nmd | 0 | 1 | 1 |
| 4 | nmd | nmd | Exon 11, c.1216C>T;p.Arg406Cys | nmd | nmd | 0 | 1 | 1 |
| 5 | nmd | nmd | nmd | nmd | c.1898A>G;p.Lys633Arg | 0 | 0 | 2 |
| 6 | nmd | nmd | nmd | nmd | nmd | 2 | 2 | 2 |
| 7 | nmd | nmd | nmd | nmd | nmd | 2 | 2 | 2 |
| 8 | nmd | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
| 9 | nmd | c.718T>C;p. Ser240Pro | nmd | nmd | nmd | 2 | 2 | 2 |
| 10 | nmd | nmd | nmd | nmd | nmd | 0 | 1 | 1 |
| 11 | nmd | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
| 12 | c.2850G>T; p.Gln950His | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
| 13 | nmd | nmd | nmd | nmd | nmd | 0 | 0 | 2 |
CFHR indicates complement factor H–related; and nmd, no mutation detected.
CD46 and CFH susceptibility factors
| Patient ID . | CD46 −652A>G (rs2796267) . | CD46 −366A>G (rs2796268) . | CD46 c.4070T>C (rs7144) . | CFH −331C>T (rs3753394) . | CFH c.2016A>G Gln672Gln (rs3753396) . | CFH c.2808G>T Glu936Asp (rs1065489) . |
|---|---|---|---|---|---|---|
| 1 | AG | GG | CC | CT | AA | GG |
| 2 | GG | GG | CC | CC | AA | GG |
| 3 | AG | AG | TC | CC | AA | GG |
| 4 | AG | AG | TC | CC | AA | GG |
| 5 | AG | AG | TC | CC | AA | GG |
| 6 | AG | AG | TC | TT | GG | TT |
| 7 | AG | AG | TC | CT | AA | GG |
| 8 | AG | AA | TT | CC | AA | GG |
| 9 | AG | AG | TT | TT | AA | GG |
| 10 | GG | GG | CC | CT | AA | GG |
| 11 | AG | GG | CC | CT | AA | GG |
| 12 | AG | AG | TC | CT | AA | GG |
| 13 | AA | AA | TT | CT | AA | GG |
| Patient ID . | CD46 −652A>G (rs2796267) . | CD46 −366A>G (rs2796268) . | CD46 c.4070T>C (rs7144) . | CFH −331C>T (rs3753394) . | CFH c.2016A>G Gln672Gln (rs3753396) . | CFH c.2808G>T Glu936Asp (rs1065489) . |
|---|---|---|---|---|---|---|
| 1 | AG | GG | CC | CT | AA | GG |
| 2 | GG | GG | CC | CC | AA | GG |
| 3 | AG | AG | TC | CC | AA | GG |
| 4 | AG | AG | TC | CC | AA | GG |
| 5 | AG | AG | TC | CC | AA | GG |
| 6 | AG | AG | TC | TT | GG | TT |
| 7 | AG | AG | TC | CT | AA | GG |
| 8 | AG | AA | TT | CC | AA | GG |
| 9 | AG | AG | TT | TT | AA | GG |
| 10 | GG | GG | CC | CT | AA | GG |
| 11 | AG | GG | CC | CT | AA | GG |
| 12 | AG | AG | TC | CT | AA | GG |
| 13 | AA | AA | TT | CT | AA | GG |
Discussion
In this study we have found that 9.2% (13/142) of the Newcastle cohort of aHUS patients had factor H autoantibodies at the time of or shortly after presentation. This finding corresponds well with previous reports. Dragon-Durey et al7 screened serum samples from 48 children who presented with aHUS and found factor H autoantibodies in 3 (6.3%). Józsi et al8 screened plasma samples from 60 patients and found autoantibodies in 5 (8.3%). We have previously shown that deletion of the genes encoding factor H–related proteins 1 and 3 is associated with aHUS.11 In particular it has been shown that homozygous deletion of CFHR1 and CFHR3 is more frequent in aHUS than in control patients. It has recently been shown that the presence of factor H autoantibodies is strongly associated with complete deficiency of factor H–related proteins 1 and 3.9 Józsi et al9 showed that in an extended cohort of 147 aHUS patients, 16 (including those described in the previous study by Józsi et al8 ) had factor H autoantibodies. Of these 14 had complete absence of factor H–related proteins 1 and 3 on Western blotting, and 2 had low levels. In their cohort of 147 aHUS patients, there were 22 patients in total with complete absence of factor H–related proteins 1 and 3. Thus, 14 (64%) of 22 patients with complete deficiency of factor H–related proteins 1 had factor H autoantibodies.
This finding agrees well with our cohort, in which 10 (63%) of 16 patients with 0 copies of CFHR1 had factor H autoantibodies. In Józsi et al's cohort,8 14 (88%) of 16 patients with factor H autoantibodies had complete deficiency of both factor H–related proteins 1 and 3. Again this finding agrees well with our cohort, in which 10 (77%) of 13 patients with factor H autoantibodies had 0 copies of CFHR1. However, in contrast to Józsi et al's patients with factor H autoantibodies, we found that 3 patients had 2 copies of both CFHR1 and CFHR3 with no evidence of a deficiency of factor H–related protein 1 on Western blotting (Figure 4). Two of these patients had the highest autoantibody titers of the whole group. Thus, our data would suggest that a significant autoantibody response to factor H can be mounted in the presence of normal expression of factor H–related proteins 1 and 3.
A novel observation in our study is that in 3 patients the complete deficiency of factor H–related protein 1 is caused by a deletion encompassing CFHR3 and CFHR1 on one allele and CFHR1 and CFHR4 on the other. This novel deletion (CFHR1/CFHR4), like the previously described CFHR3/1 deletion, has probably occurred as a result of nonallelic homologous recombination through segmental duplications in this region of the RCA cluster (as shown in Figure 5). The allele frequency of this deletion in both our control patients and aHUS patients is low (0.9% vs 1.4%). With the power available to us, there was no significant difference between the 2 groups. However, the presence of this deletion on one allele and the CFHR3/1 deletion on the other resulting in complete deficiency of factor H–related protein 1 was significantly more common in the aHUS cohort. This finding suggests that it is probably complete deficiency of factor H–related protein 1 that is the significant factor associated with the production of factor H autoantibodies in aHUS. How complete deficiency of factor H–related protein 1 might predispose one to the development of factor H autoantibodies in aHUS is not known. It is possible that it results in a failure of central and/or peripheral tolerance.
Are factor H autoantibodies pathogenic in aHUS? It is well established that CFH mutations in aHUS cluster in the C-terminal SCRs and that these mutations are associated with impaired control of complement activation at cell surfaces.22 Moreover, a transgenic murine model in which the mouse factor H lacks the 5 C terminal SCRs (FHΔ16-20) spontaneously develops aHUS.23 Józsi et al8 used recombinant fragments of factor H to map the binding site of factor H autoantibodies in 5 patients. They showed in all 5 that the binding site was in the C-terminal SCRs. In our study we have used a similar approach. In agreement with Józsi et al,8 we found that autoantibodies from 7 patients showed binding to a fragment comprising SCRs 19 to 20. Thus, most factor H autoantibodies in aHUS bind to and impair the activity of the C-terminal SCRs of factor H. This is the same region in which functionally significant mutations cluster. Moreover, it has been shown that factor H autoantibodies impair the binding of factor H to C3b and are associated with enhanced hemolysis of sheep erythrocytes in patient plasma.8 In addition we have shown that the autoantibodies from the 7 patients with binding to the factor H fragment SCRs 19 to 20 cross-react with the homologous factor H–related 1 protein fragment SCRs 4 to 5.
Our study is the first to show that in addition to the presence of factor H autoantibodies some patients have mutations in complement genes. In 1 patient we found a CFH mutation, in another a CFI mutation, in another we had already found and reported a functionally significant CD46 mutation,4 and in 2 other patients we found novel C3 variants. Although the functional significance of some of these sequence variants remains to be established, this finding strengthens the suggestion that multiple concurrent susceptibility factors are necessary in some aHUS patients for the disease to become manifest.14
In patients with CFH, CFI, CD46, CFB, and C3 mutations it is now well recognized that additional variants (SNPs and haplotype blocks) in CFH and CD46 act as susceptibility factors for the development of the disease.24 One particular CFH haplotype, CFHTGTGGT (also known as the CFH-H3), is associated with an increased risk of aHUS. This haplotype is defined by the following SNPs -331C>T (rs3753394), c.184G>A Val62Ile (rs800292), c.1204T>C p.Tyr402His (rs1061170), c.2016A>G p.Gln672Gln (rs3753396), IVS15-543G>A intron 15 (rs1410996), and c.2808G>T p.Glu936Asp (rs1065489), where the at-risk alleles are in bold. The at-risk haplotype can be tagged by genotyping rs3753394, rs3753396, and rs1065489; we have done this in all 13 patients. This haplotype was present in only 1 patient (number 6) in homozygosity. Of interest, this patient had 2 copies of both CFHR3 and CFHR1. It has recently been established that the CFHR3/1 deletion is associated with particular CFH haplotypes in patients with age-related macular degeneration and control subjects.25-27 In the study undertaken by Spencer et al,27 all deletion homozygotes were homozygous for alleles GCGAAG at rs529825, rs2019724, rs1831281, rs6677604, rs3753396, and rs1065489. In our study all the factor H autoantibody–positive patients, who were either homozygous for the CFHR3/1 deletion or had the CFHR3/1 deletion on one allele and the CFHR1/4 deletion on the other allele, were homozygous for alleles “AG” at rs3753396 and rs1065489. Interestingly, 2 patients (numbers 7 and 9) who had 2 copies of CFHR1 and CFHR3 were also homozygous for alleles “AG” at rs3753396 and rs1065489. This finding raises the possibility that this particular CFH haplotype has a role in the pathogenesis of factor H autoantibodies.
As with CFH there is a particular CD46 haplotype, CD46GGAAC, associated with an increased risk of aHUS. This haplotype is defined by the following SNPs -652A>G (rs2796267), -366A>G (rs2796268), IVS9-78G>A (rs1962149), IVS12+638G>A (rs859705), and c.4070T>C (rs7144). The at-risk haplotype can be tagged by genotyping rs2796267, rs2796268, and rs7144; we have again undertaken this in all 13 factor H autoantibody–positive patients. We inferred that 8 (numbers 1, 3, 4, 5, 6, 7, 11, and 12) were heterozygous for the at-risk CD46 haplotype, CD46GGAAC, and 2 (numbers 2 and 10) were homozygous. This finding suggests that CD46 may be acting as an additional susceptibility factor for the development of aHUS in patients with factor H autoantibodies.
The clinical features of the 13 patients with factor H autoantibodies that we describe here are similar to those previously reported. All were children with an age at presentation ranging from 1 to 11 years. Of the 3 patients described by Dragon-Durey et al,7 only 1 developed end-stage renal failure. Of our 13 patients, 6 developed end-stage renal failure, and 7 have maintained renal function to date. Three of the patients who developed end-stage renal failure have undergone transplantation without recurrence of the disease, with a follow-up period ranging from 2 to 6 years. A recent report documented the pretransplant administration of the anti-CD20 monoclonal antibody rituximab followed by intensive plasma exchange postoperatively in a 10-year-old girl with factor H autoantibodies.28 Before the administration of rituximab, the patient had been treated with prednisolone, azathioprine, and plasma exchange in an attempt to decrease the autoantibody titer. However, this was not achieved until after the administration of rituximab. Subsequently there was no recurrence of the disease after transplant, with a follow-up period of 2 years. We do not have serum samples available from the immediate pretransplant period from patients 5, 7, and 12 to determine autoantibody titer at that time, but we have analyzed a recent convalescent sample from patient 12. This sample was negative for factor H autoantibodies. This finding suggests that the titer of factor H autoantibodies may spontaneously decline with time. A pragmatic approach to the transplant management of patients with factor H autoantibodies would be to regularly monitor antibody levels and administer rituximab to those with persistently elevated titers.
In summary we have confirmed that factor H autoantibodies are found in approximately 9% of patients with aHUS. While most have complete deficiency of factor H–related protein 1, we have shown that this is not a prerequisite. Moreover, we have shown that deficiency of factor H–related protein 1 can arise from a novel CFHR1/4 deletion and that mutations of CFH, CD46, CFI, and C3 are present in some patients. The latter observation provides further evidence that multiple “hits” are necessary in some patients before aHUS presents clinically.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the following physicians for sending samples and providing clinical information: Dr Jean-Claude Davin, Emma Children's Hospital, Academic Medical Center; Dr Nick Webb, Royal Manchester's Children's Hospital; Dr Milos Ognjanovic, Royal Victoria Infirmary; Dr Lesley Rees, Great Ormond Street Hospital for Children; Dr Kay Tyerman, St James's University Hospital; Dr Nesrin Besbas, Hacettepe University School of Medicine; Dr Eiske Dorresteijn, Erasmus Medical Center; Dr Carol Inward, Bristol Royal Hospital for Children; Dr Mary Waldron, Our Lady's Children's Hospital; Dr Salih Kavucku, Medical Faculty, Dokuz Eylül University; Dr Gaurav Kapur, Children's Hospital of Michigan; and Professor Bente Jespersen, Århus Universitetshospital.
This work was supported by the Medical Research Council (G0701325) and the Northern Counties Kidney Research Fund.
Authorship
Contribution: T.H.J.G. and K.J.M. designed the research, analyzed the data, and wrote the paper; and I.M., L.S., I.P., D.K., P.N.B., A.P.H., C.Q.S., S.J.S., L.V.H., R.W., L.M., and K.J.M. performed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Professor Tim Goodship, Institute of Human Genetics, Central Parkway, Newcastle upon Tyne, NE1 3BZ United Kingdom; e-mail: t.h.j.goodship@ncl.ac.uk.