Abstract
Heritable epigenetic signatures are proposed to serve as an important regulatory mechanism in lineage fate determination. To investigate this, we profiled chromatin modifications in murine hematopoietic stem cells, lineage-restricted progenitors, and CD4+ T cells using modified genome-scale mini-chromatin immunoprecipitation technology. We show that genes involved in mature hematopoietic cell function associate with distinct chromatin states in stem and progenitor cells, before their activation or silencing upon cellular maturation. Many lineage-restricted promoters are associated with bivalent histone methylation and highly combinatorial histone modification patterns, which may determine their selective priming of gene expression during lineage commitment. These bivalent chromatin states are conserved in mammalian evolution, with a particular overrepresentation of promoters encoding key regulators of hematopoiesis. After differentiation into progenitors and T cells, activating histone modifications persist at transcriptionally repressed promoters, suggesting that these transcriptional programs might be reactivated after lineage restriction. Collectively, our data reveal the epigenetic framework that underlies the cell fate options of hematopoietic stem cells.
Introduction
The murine hematopoietic system is a hierarchically organized process that arises from a small pool of self-renewing hematopoietic stem cells (HSCs). Upon induction of differentiation, HSCs lose self-renewal ability and develop through a series of specialized progenitor cell types that possess restricted differentiation potential.1 Although several cell-intrinsic and microenvironmental factors that can control these processes have been identified, the precise molecular circuitry controlling HSC self-renewal and lineage restriction has yet to be fully elucidated.
Recent observations suggest that epigenetic-based mechanisms play an important role in controlling HSC self-renewal or differentiation.2,3 Epigenetic regulation of gene expression is largely controlled by the posttranslational modification of histones and DNA methylation, resulting in the alteration of chromatin structure and function at genes throughout cellular differentiation.4 Core histones can be covalently modified, for example, by acetylation and methylation at multiple residues, offering combinatorial codes with diverse functional outcomes.5 We and others have hypothesized previously that HSCs possess unique epigenetic signatures, whose inheritance by progenitor subsets allows for differentiation into mature blood cell types via highly coordinated gene activation and silencing.4,6–9 These unique chromatin states may allow for the preassembling of critical transcription factors at lineage-specifying promoters in HSC and progenitor cells, before full gene expression in differentiated subsets.10–13 This process, known as multilineage gene priming, is supported by the low-level transcription of several lineage-affiliated genes of lymphoid, myeloid, and erythroid genetic programs which occurs in HSCs and early progenitor cells.8,14–16 Most recently, genome-wide profiling of human hematopoietic stem/progenitor cells and differentiated erythrocyte precursor cells has revealed epigenetic signatures that are proposed to be important for maintaining HSC multipotency.17 Despite the insights gained from such studies, most have been based on either selected loci or global analysis of cell populations with heterogeneous lineage potentials. As a result, the true epigenetic status of functionally homogeneous stem and progenitor cell compartments may have been underestimated.
We have undertaken a global analysis of highly purified and functionally validated murine HSCs, early hematopoietic progenitors, and mature CD4+ T cells to reveal the epigenetic features associated with their unique functional properties. We show that promoters of genes affiliated with regulation of hematopoietic cell maturation are occupied by bivalent histone modifications in HSCs and their immediate progeny. In addition, many lineage-specifying promoters in these primitive cells possess a diverse range of histone modification patterns, together suggesting that specific combinations prepare these genes for selective expression or silencing during lineage commitment. While differentiation into progenitors and T cells leads to the establishment of lineage-specific gene expression programs, their promoters remained associated with activating histone modifications, implying that residual epigenetic priming is retained during lineage restriction. In summary, the chromatin maps reveal a complex epigenetic framework that underscores the lineage relationships between HSCs, early progenitors, and mature hematopoietic cells.
Methods
Mice
Wild-type 2- to 3-month-old C57BL/6 mice were used throughout these studies. Mice were maintained at the Lund University animal facility, and procedures were performed with the approval of the Lund University ethics committee.
Hematopoietic cell purification
HSCs, multipotent progenitors (MPPs), and megakaryocyte/erythrocyte progenitors (PreMegEs) were isolated from murine bone marrow via c-kit enrichment by using c-kit–conjugated magnetic beads (Miltenyi Biotec) and subsequent staining with antibodies as previously described.18,19 CD4+ splenic T cells were isolated as previously described.20 Cells were sorted on a FACSAria cell sorter (BD Biosciences).
Transplantation experiments
Two hundred prospectively purified cells were transplanted into lethally irradiated recipients to evaluate their in vivo capacities for multilineage cell generation using the CD45.1/CD45.2 congenic strain system as previously described.19
In vitro culture assays
Freshly sorted hematopoietic stem and progenitor cells were cultured to evaluate single-cell clonogenic activity and lineage potentials of isolated cell populations as previously described.19
miniChIP-qPCR and promoter tiling array experiments using 10 000 cells
The 10 000-cell mini-chromatin immunoprecipitation–quantitative polymerase chain reaction (miniChIP-qPCR) and miniChIP-chip methods were established from previously described methods.6,21–23 Detailed protocols can be found in supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article). Raw and processed data for the miniChIP-chip experiments are deposited under accession number GSE18734 in the Gene Expression Omnibus (National Center for Biotechnology Information).
qRT-PCR
Affymetrix gene expression
RNA was extracted from purified hematopoietic cells using an RNeasy Micro Kit (QIAGEN). Subsequent handling was performed at the Faculty for Health Sciences, Linköping University, and as previously described.19 The Affymetrix gene expression datasets for CD4+ T cells were obtained from previous work.20 Raw and processed data for the Affymetrix microarray experiments are deposited under accession number GSE18669 in the Gene Expression Omnibus.
Results
Chromatin signatures of HSCs during lineage commitment using miniChIP-chip
To profile the genome-wide epigenetic changes associated with early hematopoietic stem and progenitor cell differentiation using cells that are rare in vivo, we established a simplified and reproducible miniChIP-chip array technology (supplemental Figures 1-2). This was a refinement of our previously described miniChIP-qPCR method, which allowed histone modifications to be identified at selected genes using 50 000 hematopoietic stem and progenitor cells.6 Thus, to investigate global chromatin modification patterns of HSCs, we made further modifications to enable rapid (< 3 days) genome-scale analysis using 10 000 cells.6,21 As part of the optimization of miniChIP-chip, we compared 2 NimbleGen (Roche Applied Science) promoter tiling array platforms. These experiments revealed that the performance of the high-density 2.1-million–feature 11-kb promoter tiling arrays (HD 2.1M) was superior to the lower-density 385 000 4-kb promoter 2-array set (385K) due to increased sensitivity and peak detection ability (supplemental Figures 3-8).
MiniChIP-chip provided a novel advance for the identification of chromatin states of rare murine hematopoietic stem and progenitor subsets. We chose to study HSCs and early progenitors (MPPs and PreMegEs) as defined by the recently revised developmental scheme for early hematopoiesis.19,24 Using a fluorescence-activated cell sorter (FACS), HSCs were isolated as lineage−, Flt3/Flk2−, Sca-1+, c-kit+, and CD150+ (LSKCD150+ cells), whereas MPP progenitors were identified as lineage−, Sca-1+, c-kit+, and CD150− (LSKCD150− cells).18,25,26 We isolated PreMegE progenitors as lineage−, Sca-1−, c-kit+ and CD150+, CD105−, CD41−, FcgRI/IIlow.19 The functional characteristics of the FACS-purified populations were confirmed by in vitro differentiation and in vivo transplantation assays (supplemental Figure 9). These findings recapitulated that the phenotypically defined HSC population was highly enriched for multipotent HSC activity, which can form non–self-renewing and lineage-committed MPPs and PreMegEs. As a differentiated hematopoietic cell population, we used CD4+ T cells from mouse spleen.20
Chromatin signatures for each cell type were determined using miniChIP-chip HD 2.1M arrays to detect activating (H3K4me3, H3K79me2, acetylated histone H3 [H3ac]) and silencing histone modifications (H3K9me3, H3K27me3) as well as RNA polymerase II (PolII) occupancy. To allow for a direct link to gene activity, we performed transcriptional profiling by Affymetrix mRNA microarray analysis. To verify the amplified miniChIP samples and the resultant genome scale maps, we conducted quality control analyses using the approaches performed for miniChIP-chip optimization (supplemental Figures 10-13). All subsequent miniChIP-chip HD2.1M array analyses were restricted to promoters comprising at least one peak with a false discovery rate (FDR) of 0.05 or lower in the promoter region closest to annotated transcriptional start sites (TSSs).
Chromatin signatures of HSCs, progenitor cells, and CD4+ T cells
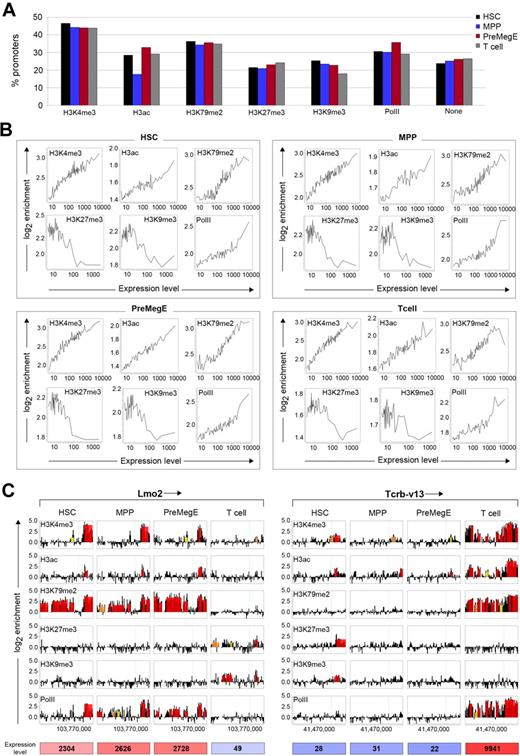
To investigate the chromatin signatures during the process of HSC differentiation into MPPs and PreMegEs, and farther downstream into T cells, we identified the association of the different histone modifications and PolII with 22 492 gene promoters. According to the analysis, H3K4me3, H3K79me2, H3ac, and PolII showed the highest association with promoters ranging from approximately 30% to 45% bound (Figure 1A). While we found that promoter occupancies of these modifications and PolII were highly similar across the 4 cell types, reduced H3ac was detected in MPPs with only 17% of promoters bound compared with approximately 30% in HSCs, PreMegEs, and T cells. The H3K27me3 and H3K9me3 modifications bound fewer promoters, ranging from approximately 18% to 25% across each cell type. This result may also reflect their broader distribution into intergenic regions and gene bodies, genomic features that are not currently resolved by the HD 2.1M arrays (supplemental Figure 12).27
Chromatin modification profiling of hematopoietic stem cell lineage commitment. (A) Genome-wide distribution of histone H3 modifications and PolII on promoters in HSCs, MPPs, PreMegEs, and T cells using HD 2.1M promoter arrays. (B) Correlation between histone modifications and gene expression in the different cell types. Genes were grouped into bins of 100 genes based on their expression levels. The average log2 enrichment of each histone modification was determined for each bin and plotted to assess correlation trends. (C) Association of histone modifications and PolII with the promoters of Lmo2 (chr2; left panel) and TcR Vβ13 (chr6; right panel) in HSCs, MPPs, PreMegEs, and T cells. Each box represents the tiled region (−8.2 to +3 kb from TSS), with the arrow near gene name indicating the direction of transcription and the TSS is shown as a green vertical line. Data are displayed as log2 ChIP/input probe signal ratios (black) with the overlaid red, orange, and yellow boxes representing peaks with FDR ≤ 0.05, ≤ 0.1, and ≤ 0.2, respectively. The x-axis shows the chromosomal coordinates, and the y-axis shows the log2 enrichment values of probes and peaks.
Chromatin modification profiling of hematopoietic stem cell lineage commitment. (A) Genome-wide distribution of histone H3 modifications and PolII on promoters in HSCs, MPPs, PreMegEs, and T cells using HD 2.1M promoter arrays. (B) Correlation between histone modifications and gene expression in the different cell types. Genes were grouped into bins of 100 genes based on their expression levels. The average log2 enrichment of each histone modification was determined for each bin and plotted to assess correlation trends. (C) Association of histone modifications and PolII with the promoters of Lmo2 (chr2; left panel) and TcR Vβ13 (chr6; right panel) in HSCs, MPPs, PreMegEs, and T cells. Each box represents the tiled region (−8.2 to +3 kb from TSS), with the arrow near gene name indicating the direction of transcription and the TSS is shown as a green vertical line. Data are displayed as log2 ChIP/input probe signal ratios (black) with the overlaid red, orange, and yellow boxes representing peaks with FDR ≤ 0.05, ≤ 0.1, and ≤ 0.2, respectively. The x-axis shows the chromosomal coordinates, and the y-axis shows the log2 enrichment values of probes and peaks.
To investigate the transcriptional states of genes associated with the various chromatin signatures, we directly correlated histone modification and gene expression profiles. This was performed by determining gene expression levels of promoters associated with each of the histone modifications or PolII and plotting this against log2 ChIP/input signal ratios. In each cell type, PolII and the histone modifications such as H3K4me3, H3ac, and H3K79me2 showed positive correlations with gene expression (Figure 1B). This result was consistent with previous studies linking these particular histone modifications to active gene transcription.4 In contrast, H3K27me3 and H3K9me3 were associated with promoters of genes expressed at low and intermediate levels and therefore showed negative correlations. These trends were highly discernable by examining the relationship between peak number and gene expression level at promoters (supplemental Figure 14).
To illuminate how these changes in histone modifications correlate with gene expression at specific loci, we analyzed 2 hematopoietic-related genes that are expressed differentially in HSCs, MPPs, and PreMegEs compared with T cells. As shown in Figure 1C, the Lmo2 gene is expressed in HSCs, MPPs, and PreMegEs and not in T cells. Consistent with expression, the Lmo2 gene promoter exhibited enrichment of H3K4me3, H3K79me2, H3ac, and PolII in HSCs, MPPs, and PreMegEs. In T cells, these peaks were replaced by the H3K27me3 and H3K9me3 modifications. Similar correlations between histone modification profiles and gene expression were seen for the TcR Vβ13 promoter. As expected, the TcR Vβ13 gene was silent in HSCs, MPPs, and PreMegEs and became induced in T cells. The entire TcR Vβ13 promoter was enriched in H3K4me3, H3K79me2, H3ac, and PolII in T cells. MPPs and PreMegEs showed some enrichment for H3K4me3 and H3ac, but the promoter region lacked H3K27me3 and H3K9me3. However, in HSCs the TcR Vβ13 promoter region was enriched for decreased levels of H3K4me3 and H3ac, together with H3K27me3 and H3K9me3. This combination suggests that the TcR Vβ13 promoter is marked with a bivalent histone methylation pattern in HSCs that comprises both H3K4me3 and H3K27me3.28,29 Analysis of additional hematopoietic cell regulators using both miniChIP-chip and miniChIP-qPCR further confirmed the association between gene expression and histone modifications (supplemental Figure 15). The data from these global based promoter analyses indicate that HSCs, early progenitors, and T cells exhibit dynamic epigenetic signatures that correlate well with gene expression during lineage commitment.
Bivalent chromatin states in HSCs and during lineage commitment
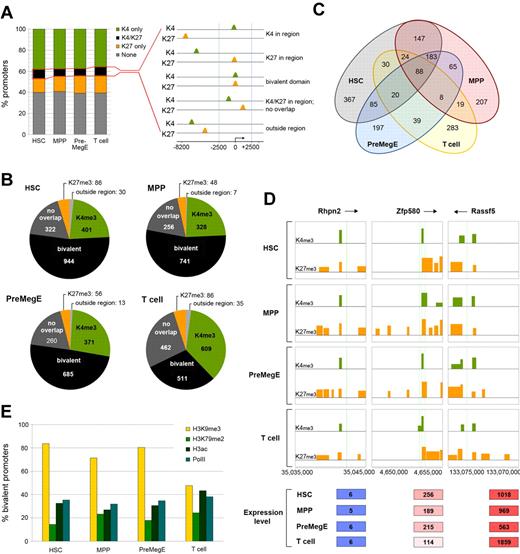
Bivalent chromatin signatures at promoters are proposed to poise key developmental genes for lineage-specific activation or repression during differentiation.28,30,31 To investigate this in murine HSCs and downstream progeny, a computational strategy was devised that enabled us to determine coincident H3K4me3 and H3K27me3 bivalent peaks across 22 492 gene promoters. We first determined the percentage of tiled promoter regions bound with (1) H3K4me3, (2) H3K27me3, (3) both H3K4me3 and H3K27me3, or (4) none of these marks in each of the cell types (Figure 2A). The percentages of the 4 categories were similar with 1783, 1380, 1385, and 1703 (7.9%, 6.13%, 6.15%, and 7.5%) promoter regions associated with H3K4me3 and H3K27me3 in HSCs, MPPs, PreMegEs, and T cells, respectively. Next, the promoters belonging to both H3K4me3 and H3K27me3 were analyzed in greater detail for bivalent overlapping peaks coincident at TSSs.31 We set the promoter region from −2.5 kb to +2.5 kb relative to TSSs to encompass the H3K4me3 genomic distribution profile (supplemental Figure 12). Bivalent modifications were found to be associated with 944, 741, 685, and 511 (4.2%, 3.3%, 3%, and 2.3%) bivalent promoters in HSCs, MPPs, PreMegEs, and T cells, respectively (Figure 2B). This analysis revealed that HSCs had the highest number of bivalent promoters compared with the other cell types. Interestingly, they were more similar to the bivalent promoters in MPPs and PreMegEs than the T-cell bivalent promoters, a result that may reflect their developmental position in the hematopoietic hierarchy (Figure 2C and supplemental Figure 16). In each cell type, promoters bound by the bivalent modification category associated with low to intermediate levels of gene expression (Figure 2D and supplemental Figure 17).30
H3K4me3 and H3K27me3 bivalent promoter methylation profiles in hematopoietic stem cells, early progenitors and T cells. (A) Total H3K4me3 (green) and H3K27me3 (orange) enriched promoters in HSCs, MPPs, PreMegEs, and T cells (left panel). Promoters associated with both H3K4me3 and H3K27me3 (black) were assessed for overlapping peaks (FDR ≤ 0.05) within a 5-kb region of TSS, and bivalent promoter methylation was defined as presence of concurrent H3K4me3 and H3K27me3 peaks within the −2.5- to +2.5-kb region surrounding TSS (right panel). (B) Comparison of the number of bivalent promoters in the different cell types according to the above criteria. Note that many promoters comprised both modifications but were not overlapping within the 5-kb region (dark gray). The H3K4me3 and H3K27me3 modifications present at tiled intervals outside the 5-kb region are also indicated (light gray). (C) Venn diagram showing the overlap of bivalent promoters in HSCs, MPPs, PreMegEs, and T cells. (D) The bivalent histone modification profiles at the Rhpn2 (chr7), Zfp580 (chr7), and Rassf6 (chr5) promoters in HSCs, MPPs, PreMegEs, and T cells. All figures showing the bivalent profile are labeled in the same way. Peaks of H3K4me3 (green) and H3K27me3 (orange) across tiled promoter regions (−8.2 to +3 kb) are indicated in each box diagram. TSS is shown as green vertical lines, and arrows near gene names show the direction of transcription. The x-axis shows the chromosomal coordinates, and the y-axis shows the log2 enrichment for peaks. The gene expression values are indicated below for each gene and the different cell types are given below. (E) Percentage of bivalent promoters associated with H3K9me3, H3K79me2, H3ac, and PolII in HSCs, MPPs, PreMegEs, or T cells.
H3K4me3 and H3K27me3 bivalent promoter methylation profiles in hematopoietic stem cells, early progenitors and T cells. (A) Total H3K4me3 (green) and H3K27me3 (orange) enriched promoters in HSCs, MPPs, PreMegEs, and T cells (left panel). Promoters associated with both H3K4me3 and H3K27me3 (black) were assessed for overlapping peaks (FDR ≤ 0.05) within a 5-kb region of TSS, and bivalent promoter methylation was defined as presence of concurrent H3K4me3 and H3K27me3 peaks within the −2.5- to +2.5-kb region surrounding TSS (right panel). (B) Comparison of the number of bivalent promoters in the different cell types according to the above criteria. Note that many promoters comprised both modifications but were not overlapping within the 5-kb region (dark gray). The H3K4me3 and H3K27me3 modifications present at tiled intervals outside the 5-kb region are also indicated (light gray). (C) Venn diagram showing the overlap of bivalent promoters in HSCs, MPPs, PreMegEs, and T cells. (D) The bivalent histone modification profiles at the Rhpn2 (chr7), Zfp580 (chr7), and Rassf6 (chr5) promoters in HSCs, MPPs, PreMegEs, and T cells. All figures showing the bivalent profile are labeled in the same way. Peaks of H3K4me3 (green) and H3K27me3 (orange) across tiled promoter regions (−8.2 to +3 kb) are indicated in each box diagram. TSS is shown as green vertical lines, and arrows near gene names show the direction of transcription. The x-axis shows the chromosomal coordinates, and the y-axis shows the log2 enrichment for peaks. The gene expression values are indicated below for each gene and the different cell types are given below. (E) Percentage of bivalent promoters associated with H3K9me3, H3K79me2, H3ac, and PolII in HSCs, MPPs, PreMegEs, or T cells.
Global studies of bivalent domains in many different cell types have indicated a high degree of association with additional histone modifications to form combinatorial marked chromatin domains.17,31–33 To investigate this further, we determined the presence of additional marks at bivalent promoters in each of the 4 cell types (Figure 2E). Our analysis revealed that bivalent promoters in HSCs were also associated with H3K79me2 (15.5%), H3ac (32.6%), PolII (35.6%), and H3K9me3 (83.5%). In MPPs and PreMegEs, similar percentages of association with the other marks were seen, but there was a large reduction in H3K9me3 in T cells compared with the progenitor cell types (47% compared with ∼70%-84%). This result suggested that different groups of bivalent gene promoters might exist in hematopoiesis, consistent with the shared bivalent promoters in HSCs, MPPs, and PreMegEs compared with T cells (Figure 2C and supplemental Figure 16). Thus, the H3K4me3 and H3K27me3 bivalent domain promoters may cooperate with combinatorial histone modifications to prime genes for expression or silencing during lineage commitment.
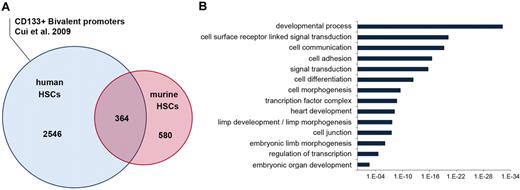
Parallel studies of mouse and human HSCs have indicated that conserved gene regulatory networks may be important for stem cell function and self-renewal across species.34 Thus, we hypothesized that similar gene promoters would be marked with bivalent histone methylation patterns in human and mouse HSCs. To perform this analysis, we compared the bivalent modifications present at orthologous gene promoters, using the recently described human CD133+ HSC/hematopoietic progenitor cell (HPC) ChIP-sequencing data.17 Roughly 40% of bivalent mouse promoters were occupied with the bivalent chromatin state in human HSCs/HPCs (Figure 3A), a finding consistent with previous comparisons of human and mouse embryonic stem (ES) cells.35 To gain insight into their possible biological roles, we determined the gene ontology (GO) terms enriched with the bivalent pattern. Bivalent promoters were overrepresented in genes encoding cell–cell signaling molecules, developmental regulators, cell adhesion molecules, and embryonic morphogenic proteins, consistent with previous studies30 (Figure 3B). Visual inspection of individual genes further revealed extensive representation of these pluripotent ES cell promoters in mouse and human HSCs while also highlighting the enrichment of hematopoietic regulators (supplemental Figure 18).
Bivalent promoter methylation states in hematopoietic stem cells are evolutionarily conserved. (A) Venn diagram showing the conservation between promoters occupied with bivalent states in mouse and human HSCs. The human bivalent dataset was taken from a recent study describing chromatin modification profiles of CD133+ HSCs/HPCs.17 (B) Distribution of shared mouse and human bivalent promoters in different gene ontology categories.
Bivalent promoter methylation states in hematopoietic stem cells are evolutionarily conserved. (A) Venn diagram showing the conservation between promoters occupied with bivalent states in mouse and human HSCs. The human bivalent dataset was taken from a recent study describing chromatin modification profiles of CD133+ HSCs/HPCs.17 (B) Distribution of shared mouse and human bivalent promoters in different gene ontology categories.
Dynamic resolution of bivalent domains after HSC differentiation
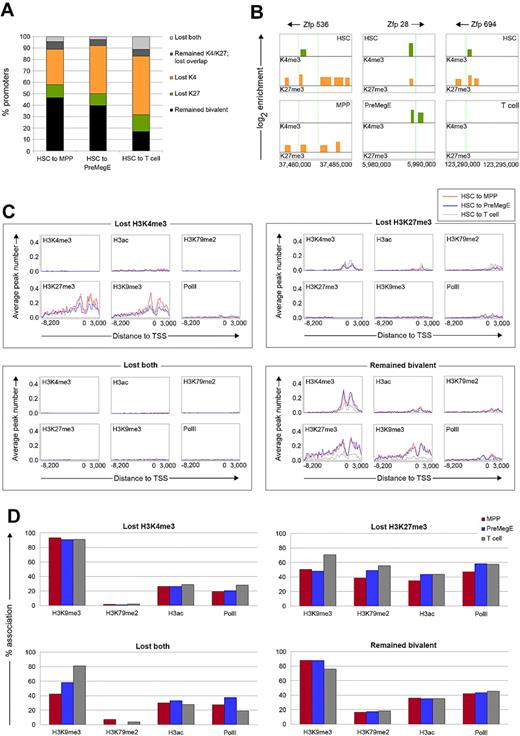
Differentiation into defined lineages can lead to the resolution of bivalent promoters according to their gene expression status, namely H3K4me3 at expressed genes and H3K27me3 at silenced genes.31 To trace the 944 bivalent promoters during HSC differentiation, we characterized their changing chromatin states into MPPs, PreMegEs, or T cells. A large majority (82%) of promoters lost their bivalent profile in the T cells, and roughly half resolved their bivalent profile in MPPs and PreMegEs (Figure 4A). In MPPs, 105 (11%) lost H3K27me3, 292 (31%) lost H3K4me3, and 40 (4.2%) lost both. Similar changes occurred during the transition into PreMegEs, with slightly more bivalent promoters losing H3K4me3 compared with MPP (42% vs 31%). In contrast, only 162 (17%) of promoters retained the bivalent profile in T cells, while 137 (14.5%) lost H3K27me3, 484 (51%) lost H3K4me3, and 104 (11%) lost both. This analysis provided a resource for the identification of new regulators of hematopoiesis. For example, we noted that 3 bivalent zinc finger proteins (Zfp536, Zfp28, Zfp694) in HSCs became resolved into monomethylation or none of the marks in the differentiating progeny (Figure 4B).
The resolution of bivalent domains during differentiation is coupled to their association with distinct epigenetic signatures in HSCs. (A) The percentage of promoters changing from a bivalent chromatin state in HSCs after differentiation into MPPs, PreMegEs, and T cells. Promoters either remained bivalent (black), lost H3K27me3 (green), lost H3K4me3 (orange), remained both H3K4me3/H3K27me3 without overlapping peaks (dark gray), or lost both marks (light gray). (B) Resolution of bivalent histone modification profiles at the Zfp536, Zfp28, and Zfp694 promoters located on chromsome 7 in HSCs after differentiation into MPPs, PreMegEs, and T cells, respectively. (C) Subsequent histone modification and PolII profiles for HSC bivalent promoters that lose H3K4me3, H3K27me3, both, or remain bivalent in MPPs (blue line), PreMegEs (red line), and T cells (gray line). For each plot, the x-axis shows the tiled promoter regions, and the y-axis is the average number of peaks at promoters. (D) Subsequent histone modification and PolII profiles for HSC bivalent promoters also occupied with H3K9me3, H3K79me2, H3ac, and PolII that lose H3K4me3, H3K27me3, both, or remained bivalent in MPPs (blue), PreMegEs (red), or T cells (gray).
The resolution of bivalent domains during differentiation is coupled to their association with distinct epigenetic signatures in HSCs. (A) The percentage of promoters changing from a bivalent chromatin state in HSCs after differentiation into MPPs, PreMegEs, and T cells. Promoters either remained bivalent (black), lost H3K27me3 (green), lost H3K4me3 (orange), remained both H3K4me3/H3K27me3 without overlapping peaks (dark gray), or lost both marks (light gray). (B) Resolution of bivalent histone modification profiles at the Zfp536, Zfp28, and Zfp694 promoters located on chromsome 7 in HSCs after differentiation into MPPs, PreMegEs, and T cells, respectively. (C) Subsequent histone modification and PolII profiles for HSC bivalent promoters that lose H3K4me3, H3K27me3, both, or remain bivalent in MPPs (blue line), PreMegEs (red line), and T cells (gray line). For each plot, the x-axis shows the tiled promoter regions, and the y-axis is the average number of peaks at promoters. (D) Subsequent histone modification and PolII profiles for HSC bivalent promoters also occupied with H3K9me3, H3K79me2, H3ac, and PolII that lose H3K4me3, H3K27me3, both, or remained bivalent in MPPs (blue), PreMegEs (red), or T cells (gray).
These differences in bivalent domain resolution that we found between early progenitors and mature T cells prompted us to examine the link with additional histone modifications. As shown in Figure 4C, the loss of H3K4me3 correlated closely with the acquisition and/or retention of additional silencing marks in the downstream cells, whereas the loss of H3K27me3 was associated with the presence of additional activating marks. A strong increase in H3K27me3 and H3K9me3 was observed at genes that lost H3K4me3, correlating with an overall reduction in gene expression (Figure 4C and supplemental Figure 19). A reciprocal trend was seen for genes that lost H3K27me3 and became transcriptionally active upon differentiation. For the genes that lost both H3K4me3 and H3K27me3, all other investigated histone modifications or PolII were no longer detected at gene promoters. Gene expression analysis revealed that these promoters became transcriptionally silent (supplemental Figure 19). In contrast, the promoters that remained bivalent in MPPs, PreMegEs, and T cells retained all of the other investigated marks and were expressed at low to intermediate levels.
Next, we sought to determine whether promoters that lost H3K4me3 or H3K27me3 in the downstream progeny showed differences in bivalent chromatin states in HSCs. Promoters that subsequently lost H3K4me3 were associated with less activating modifications (H3K79me2, H3ac, and PolII) compared with promoters that lost H3K27me3 (Figure 4D). Their association of H3K9me3 was high (∼ 90%), consistent with a role in gene silencing. Promoters in HSC that would become changed in T cells exhibited a different chromatin modification pattern compared with those that changed in MPPs and PreMegEs. Specifically, for promoters that lost H3K27me3 in T cells, a higher percentage was linked with H3K9me3 compared with the promoters that lost H3K27me3 in MPPs and PreMegEs (70% vs ∼ 48%; Figure 4D). The same was true for HSC bivalent promoters that lost both H3K4me3 and H3K27me3 in T cells. Taken together, our data indicate that HSC bivalent promoters associate with distinct chromatin states depending on their selective expression during differentiation.
Distinct chromatin signatures of hematopoiesis regulators during differentiation
To further our understanding of early hematopoietic cell lineage commitment, we explored the chromatin states of genes that were associated with development into MPPs and PreMegEs. First, we identified genes showing 3-fold or higher differential expression before and after differentiation into either progenitor. Gene ontology analysis revealed an expected association of gene categories for promoters specifically up-regulated in each progenitor (data not shown).19,36 Next we determined the changing chromatin states of these progenitor-specific genes by grouping these as activating (H3K4me3, H3K79me2, H3ac, and PolII), silencing (H3K27me3 and H3K9me3), or both activating and silencing (all investigated marks) categories. The largest fractions of promoters that displayed a change in chromatin state after differentiation into MPPs or PreMegEs were occupied initially by activating marks in HSCs (Figure 5A-B). Unsurprisingly, promoters of this kind acquired activating modifications at genes that were up-regulated in the progenitors, as these genes were linked with progenitor cell functions, including Klf1, Plxdc1, Tek, Itga4, and Gp5.19,36 Likewise, very few up-regulated genes in MPPs and PreMegEs that had activating marks in HSCs acquired silencing or act/sil modifications in the progenitors. Intriguingly, a high proportion of promoters also showed activating modifications in the down-regulated gene group in MPPs with an even higher fraction detected in PreMegEs, where most genes were no longer expressed. Furthermore, compared with MPPs, a much larger number of genes down-regulated in PreMegE changed from having activating marks in HSCs to having silencing or act/sil modifications. Many of these genes down-regulated in PreMegEs were involved in mature granulocyte and T- and B-cell functions, including Lsp1, Sell, CD34, Ly6a, Tgfb1, Mpa2, Hpse, and Gata3. These patterns were confirmed when the histone modifications and PolII profiles were examined directly at the differentially regulated promoters (supplemental Figure 20). Our analysis indicated that during rapid and selective transcriptional induction and repression of gene expression programs in the earliest stages of hematopoiesis, many lineage-affiliated promoters remained associated with activating histone modification profiles upon gene silencing.
Maintenance of transcriptionally poised chromatin modifications states after HSC differentiation. (A-C) Changing histone modification profiles at promoters of genes showing at least 3-fold up-regulation (red bars) and down-regulation (blue bars) in MPPs (A), PreMegEs (B), and T cells (C). Up-regulated and down-regulated promoters were grouped according to whether they acquired activating (active), silencing (silent), or both activating and silencing (act/sil) modifications at each developmental stage. (D) Histone methylation and PolII profiles of promoters showing at least 3-fold up- or down-regulation of gene expression in T cells. Left panels show gene expression in HSCs, MPPs, PreMegEs, and T cells as assessed by Affymetrix. Right panels indicate the histone methylation and PolII profiles at up- and down-regulated promoters in all 4 cell types, sorted according to increasing numbers of activating histone modifications in T cells relative to HSCs.
Maintenance of transcriptionally poised chromatin modifications states after HSC differentiation. (A-C) Changing histone modification profiles at promoters of genes showing at least 3-fold up-regulation (red bars) and down-regulation (blue bars) in MPPs (A), PreMegEs (B), and T cells (C). Up-regulated and down-regulated promoters were grouped according to whether they acquired activating (active), silencing (silent), or both activating and silencing (act/sil) modifications at each developmental stage. (D) Histone methylation and PolII profiles of promoters showing at least 3-fold up- or down-regulation of gene expression in T cells. Left panels show gene expression in HSCs, MPPs, PreMegEs, and T cells as assessed by Affymetrix. Right panels indicate the histone methylation and PolII profiles at up- and down-regulated promoters in all 4 cell types, sorted according to increasing numbers of activating histone modifications in T cells relative to HSCs.
To compare the histone modification patterns in a mature hematopoietic cell type, we performed a similar analysis for genes showing differential expression between HSCs and T cells (Figure 5C). Unlike for the progenitor cells, genes that were actively marked in HSCs and down-regulated in T cells showed enrichment of H3K27me3 and H3K9me3 (Figure 5C-D). However, the activating marks were still retained at the down-regulated promoters. Furthermore, a larger number of down-regulated genes changed from having activating marks in HSCs to the sil/act state in T cells compared with the 2 early progenitors. Most of these promoters, which became silenced, corresponded to key effectors of HSC and progenitor cell function that are repressed during differentiation. Some examples included Gata2, CD150, Mpo, and Pu.1. These data indicate that T-cell differentiation is linked to increased H3K27me3- and H3K9me3-mediated silencing of genes required for other hematopoietic cell lineages. However, as in progenitor cells, many down-regulated HSC genes retained activating modifications in T cells.
Epigenetic lineage priming of T-cell genes in HSCs
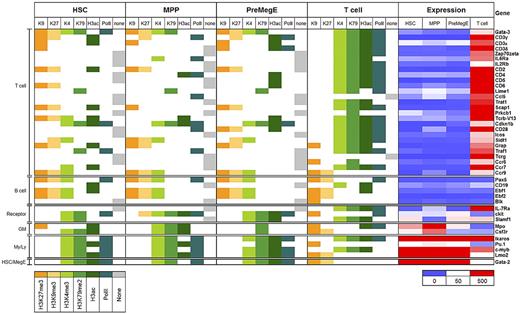
We and others have proposed that lineage priming is an epigenetic-based mechanism initiated in HSCs that can act as a memory system for hematopoietic cell lineage commitment.6–8,14,15 To gain further insight into this phenomenon, we focused our analysis on a panel of well-described T-cell genes described in the literature. Consistent with a previous study, our microarray analysis confirmed that most genes were expressed selectively in T cells and showed little or no expression in HSCs, MPPs, and PreMegEs (Figure 6).7 For comparison, we analyzed genes that were differentially expressed in one or more of the 3 primitive cells types and not mature T cells, including c-Kit, CD150, Ikaros, c-Myb, and Gata2 as well as B cell–associated genes Pax5, CD19, and Ebf-1.
Epigenetic priming of T cell–specific gene promoters in early hematopoiesis. T cell–specific genes were selected from the literature and their promoters analyzed for the presence of activating (green) or silencing (orange) histone modifications, PolII (teal), or no marks (gray) in HSCs, MPPs, PreMegEs. and T cells. Genes were grouped according to lineage-associated functions: T cells, B cells; cell surface signaling receptors, granulocyte/monocytes (GM); myeloid/lymphoid cells (My/Ly), hematopoietic stem cells/megakaryocytes and erythrocytes (HSC/MegE). Gene expression values are shown as a heat map using a cutoff value of 50 (white) to determine genes with no detectable expression (blue) and expressed genes (red).
Epigenetic priming of T cell–specific gene promoters in early hematopoiesis. T cell–specific genes were selected from the literature and their promoters analyzed for the presence of activating (green) or silencing (orange) histone modifications, PolII (teal), or no marks (gray) in HSCs, MPPs, PreMegEs. and T cells. Genes were grouped according to lineage-associated functions: T cells, B cells; cell surface signaling receptors, granulocyte/monocytes (GM); myeloid/lymphoid cells (My/Ly), hematopoietic stem cells/megakaryocytes and erythrocytes (HSC/MegE). Gene expression values are shown as a heat map using a cutoff value of 50 (white) to determine genes with no detectable expression (blue) and expressed genes (red).
MiniChIP-chip analysis showed that many T cell–specific promoters were associated with combinatorial activating and silencing histone modifications in the hematopoietic stem and progenitor cells (Figure 6). These combinatorial patterns frequently comprised H3K27me3 and H3K9me3 together with at least one of the activating modifications. Consistent with previous work, we found genes that were associated with activating marks comprising H3K4me3, H3K79me2, H3ac, and PolII while being maintained in a transcriptionally repressed state in HSCs, MPPs, and PreMegEs. Some examples included Lime1, Cdkn1b, CD28, Traf1, and Ccr7. In addition, many of the T cell–specific promoters were not found with any of the modifications that we analyzed in HSCs, MPPs, or PreMegEs, including Zap70, Prkcb, Icos, and IL7Rα, most of which had activating modifications in T cells consistent with expression. The Gata3, TcR Vβ13, Sidt1, and Ccr9 promoters exhibited bivalent marking in HSCs and progenitors as did the B cell–associated genes Pax5 and Ebf1 (Figure 6 and supplemental Figure 13).6 To avoid any bias that might have occurred by selecting these genes in the literature, we repeated and confirmed the analysis using a list of 170 genes expressed in T cells through comparisons of the Affymetrix expression data (supplemental Figure 21). MiniChIP-qPCR analysis validated the respective bivalent and activating patterns for 2 genes, c-Myb and Ccr9 (supplemental Figure 22). Thus, most T cell–specific promoters are enriched in combinatorial histone modifications, including bivalent marks in hematopoietic stem and early progenitor cells that together may constitute an epigenetic lineage-priming mechanism important for T-cell development.
Discussion
HSCs undergo dramatic changes in morphology, cell-cycle status, and gene expression as they differentiate into progenitor subsets with defined lineage potentials. Such alterations are proposed to result from chromatin reorganization of the genome during differentiation, allowing for the establishment and maintenance of lineage-specific transcriptional networks. Here we have documented the chromatin reconfiguration that occurs in some of the earliest stages of murine HSC lineage commitment during normal hematopoiesis. This analysis was made possible by developing a genome-scale 10 000 cell miniChIP array technology that allows for rare primary cell subsets to be investigated. Our study reveals key epigenetic features that document the dynamic relationship between histone modification patterns and the regulation of gene expression as early progenitor cells commit to hematopoietic lineages in vivo.
We found bivalent histone methylation at promoters in HSCs, early progenitor subsets, and T cells, which is consistent with previous work.17,37 Furthermore, many lineage-affiliated promoters occupied by a bivalent chromatin state in HSCs could be either lost or maintained during differentiation into the progenitor subsets or mature T cells. Therefore, our bivalent miniChIP-chip data provide a resource for the identification of novel regulators of hematopoiesis. Roughly half of the bivalent mouse promoters we identified showed a similar profile in human HSCs/HPCs reported by Cui et al, consistent with what has been found in comparable studies of human and murine ES cells.17 Our result was remarkable given the differences in ChIP technologies and in cell isolation techniques that were used to generate the respective human and mouse datasets. In particular, the murine HSCs represent functionally homogeneous populations in vivo, whereas the human CD133+ population was exposed to growth factors during in vitro culture, which could also account for these differences. Furthermore, we found that bivalent marks were highly overrepresented at promoters encoding for embryonic developmental regulators, in addition to key transcription factors and growth factors implicated in hematopoiesis.30 This result suggested that the establishment of hematopoietic bivalent domains might occur during embry-onic development and before the emergence of multipotent HSCs in adult bone marrow.
Studies of ES cells and human HSCs/HPCs have revealed that the fate of lineage-restricted promoters is controlled by epigenetic chromatin modifications encoded initially in SCs.17,35,38 In support of this, we found differences in the chromatin states of bivalent promoters in HSCs that we propose could influence their gene expression pattern during hematopoiesis. This was particularly evident for H3K9me3, which associated with a higher percentage of bivalent promoters in HSC that lost H3K27me3 in T cells compared with the more primitive progenitor cells. H3K9me3 at bivalent promoters in HSC predestined for activation in T cells suggests that it may be needed as an extra silencing cue to maintain a silenced but poised state as T-cell precursors differentiate in the thymus. It will be interesting to determine at what stage of differentiation, and by what mechanism, H3K9me3 and H3K27me3 are removed from the promoters needed for mature T-cell development.39 Together, we suggest that threshold levels of H3K9me3 methylation in the context of bivalent domains in HSCs may be a general mechanism for limiting the scope of transcriptional activation in developmentally distant lineages. Thus, our data are compatible with a model in which bivalent domain promoters cooperate functionally with combinatorial chromatin modifications to provide an epigenetic memory system that regulates tightly controlled gene programs critical for hematopoietic lineage commitment.
The loss of multipotency and acquisition of lineage restriction are determined by the induction and silencing of genes that enable oligopotent progenitors to generate mature blood cells. The molecular characterization of the gene expression programs underlying these hematopoietic cell transitions has demonstrated the complexity of the lineage commitment process.40 However, the epigenetic plasticity of this process remains unclear, and, in particular, it is unknown at what stage cell fate choices become irreversible. Previously, technical limitations have made it impossible to obtain a global view of the chromatin signatures that might be involved in these processes, and therefore our novel miniChIP array technology represents a major advance in tackling these questions.
Our data showed that T-cell maturation is linked to H3K27me3- and H3K9me3-mediated silencing of genes that are required for other hematopoietic cell lineages. However, the chromatin architecture in mature T cells also exhibited transcriptionally poised states, which could allow dynamic gene expression changes during differentiation. The presence of bivalent marks, as well as the retention of activating marks at repressed promoters in T cells, suggested that it might be possible to induce the transcription of these genes given the correct conditions. Recent work suggests that the ability of naive CD4+ T cells to differentiate into different helper and regulatory population is underpinned by an epigenetic framework controlled by bivalent chromatin states.37 It is therefore highly probably that the retention by mature T cells of bivalent histone modifications at promoters may poise them to differentiate into different effector T-cell populations. Interestingly, we found a reduced association of bivalent promoters with H3K9me3 in T cells compared with the primitive cell types. It is tempting to speculate that the absolute levels of H3K9me3 at bivalent gene promoters might be important for staging the correct gene expression patterns in downstream lineages.
Thus, the data reveal that the chromatin architecture is poised for dynamic multilineage expression in early progenitor stages, and that these flexible epigenetic signatures persist in T cells. Further studies are needed to determine how lineage-irrelevant genes are repressed in the context of activating histone modifications at their promoters. This could involve loss of activating marks at upstream enhancers and locus control regions, gain of DNA methylation at CpG-poor promoters, or active antagonism of transcription factors.41–44 Importantly, our finding that activating histone modification marks were maintained at transcriptionally repressed gene promoters indicates that histone modifications do not truly reflect transcriptional status; instead, they may be predictors of expression or reporters for recently transcribed genes. This hypothesis is gaining support as histone modifications are studied globally in the context of cellular development.31,33,45
The multilineage priming hypothesis is supported by the evidence that there is low-level transcription of lineage specifying genes in HSCs/HPCs, suggesting that genes destined for activation exist in a transcriptionally permissive chromatin configuration8,14–16,34. We show herein that T cell–affiliated genes in HSCs and progenitors have a poised chromatin structure before their transcriptional induction during differentiation. Despite this poised chromatin state, robust transcriptional activation of these genes may not occur in HSCs or early progenitor subsets due to an absence of transcriptional activators or DNA methylation. Nonetheless, propagation of genes with this chromatin architecture to downstream cells would provide a form of epigenetic memory during the differentiation process, allowing stage-specific transcription factors to activate genes as they become available. Consistent with this hypothesis, our data revealed that although activating histone modifications were present at promoters of a large number of T cell–specifying genes in hematopoietic stem and early progenitors, they showed little or no detectable expression. Similar observations have been recently described for human cord blood–derived hematopoietic precursors and T cell– and B cell–affiliated promoters.7 Therefore, this analysis extends the lineage priming hypothesis that lineage-affiliated gene expression programs are governed by a complex interplay of chromatin-modifying enzymes and transcription factors acting in a gene- and cell-specific context.
In summary, the genome-wide chromatin profiles we have generated will further our understanding of the epigenetic control of hematopoietic stem and progenitor cells. These data will provide an important framework for understanding the epigenetic mechanisms of lineage commitment throughout normal and aberrant hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank K. Zhao for sharing of the ChIP-seq bivalent dataset. We thank B. Ren and G. Hon for advice on ChIP-chip normalization, and A. Mowat for critical reading of the manuscript. J.L.A. is a recipient of a Vetenskapsrådet Medicine Visiting Scientist Position and is supported by the Hemato-Linne Program and the Faculty of Medicine (Lund University), as well as by CancerFonden.
Authorship
Contribution: H.W. and J.L.A. performed the research and analyzed the results; M.S. performed Affymetrix gene expression microarrays; and J.L.A. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joanne L. Attema, Institute for Experimental Medical Science, BMC D14, Immunology Unit, Lund University, 221 84 Lund, Sweden; e-mail: joanne.attema@med.lu.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal