Abstract
During inflammation, elevated granulocyte macrophage–colony-stimulating factor (GM-CSF) directs the development of new dendritic cells (DCs). This pathway is influenced by environmental factors, and we previously showed that physiologic levels of estradiol, acting through estrogen receptor alpha (ERα), promote the GM-CSF–mediated differentiation of a CD11b+ DC subset from myeloid progenitors (MPs). We now have identified interferon regulatory factor 4 (IRF4), a transcription factor induced by GM-CSF and critical for CD11b+ DC development in vivo, as a target of ERα signaling during this process. In MPs, ERα potentiates and sustains GM-CSF induction of IRF4. Furthermore, retroviral delivery of the Irf4 cDNA to undifferentiated ERα−/− bone marrow cells restored the development of the estradiol/ERα-dependent DC population, indicating that an elevated amount of IRF4 protein substitutes for ERα signaling. Thus at an early stage in the MP response to GM-CSF, ERα signaling induces an elevated amount of IRF4, which leads to a developmental program underlying CD11b+ DC differentiation.
Introduction
Dendritic cell (DC) subsets in vivo are distinguished based on location, surface markers, function, and migratory capacity, and whether they are present in steady-state conditions or develop as a consequence of inflammation.1 In non–lymphoid tissues such as dermis and lung, 2 major populations of CD11c+ MHCII+ DCs have been identified as CD11bhi CD103− and CD11blo CD103+ Langerin+ or −.2,3 These interstitial DCs derive from monocytes or blood-borne precursors that seed tissue, while a distinct population of epidermal Langerin+ DCs (LCs) are replenished slowly from local self-renewing precursors. The cytokines that mediate the development of interstitial DCs during homeostasis are not defined, and may include Flt3 ligand or micro amounts of granulocyte macrophage–colony-stimulating factor (GM-CSF) or M-CSF present in non–lymphoid tissue.3 During inflammation, DC differentiation from Gr-1hi monocytes is largely mediated by elevated amounts of GM-CSF.4 This GM-CSF–driven pathway is elevated in infection and autoimmune disease and leads to the appearance of de novo DC populations in lymphoid organs and tissues.1,5 In culture models, GM-CSF mediates CD11b+ DC differentiation from monocytes and myeloid progenitors (MPs), and a subset of the CD11bint DCs express Langerin.6,7 However, it remains unclear how these DCs relate to Langerin+ epidermal LCs or populations of CD11b+ or Langerin+ interstitial DCs.
The lineage fate of hematopoietic progenitor cells is determined by ordered expression of transcription factors and receptor tyrosine kinases such as Flt3, yet the molecular steps involved in the development of specific DC subtypes from common precursors, including myeloid and lymphoid progenitors, pro-DCs, and monocytes, are not completely defined. Genetic analyses have identified several transcription factors, including Irf4, to be crucial for development of specific DC subsets found in lymphoid organs.3 For example, mice deficient in relB, PU.1, or interferon regulatory factor 4 (IRF4) lack splenic CD11bhi CD8α− CD4+ DCs, whereas mice deficient in IRF8 lack splenic CD8α+ CD4− CD11b− DCs and plasmacytoid DCs. IRF4 also is necessary for the development of MHC class II+ DCs during GM-CSF–mediated differentiation from murine bone marrow (BM) or human monocytes in vitro.8-10 Less is known about transcription factor requirements for tissue DCs, and factors directing the development of recently defined interstitial CD11b+ or CD103+ Langerin+ DCs have not been reported. Analyses of IRF4−/− mice did not include assessments of tissue DCs.8,9
Transcription factor dosage is often important in cell fate specification. Relevant to our study, quantitatively different levels of IRF4 protein alter B-cell differentiation pathways.11 IRF4 acts alone or dimerizes with PU.1 or Spi-B to mediate transcription.12 Quantitative differences in PU.1 in hematopoietic progenitors also lead to alternate myeloid or lymphoid fates.13,14 Thus, IRF4 and PU.1 act as dosage-sensitive regulators to instruct distinct cell differentiation programs, and therefore, factors (eg, hormones or other transcription factors15,16 ) that regulate IRF4 expression are likely to alter IRF4-mediated developmental pathways.
Environmental stimuli can influence DC differentiation, and we previously have defined a role for estrogen receptor (ER) signaling in DC development.17 ERs are ligand-dependent transcription factors that regulate gene expression by forming complexes with chromatin-modifying coregulators.18 Immune cells and their progenitors express ERs, suggesting that ER-mediated responses to circulating 17-β-estradiol (E2) regulate cellular differentiation and function. Indeed, elevated E2 levels decrease lymphopoiesis in vivo by depleting primitive hematopoietic progenitors.19 E2 and ERα promote murine and human DC differentiation mediated by GM-CSF.20-22 We demonstrated that E2 specifically promotes the GM-CSF–mediated differentiation of murine CD11c+ CD11bint Ly6C− Langerin+ or − DCs, a population that may represent in vivo populations of tissue or inflammatory DCs.7 Estradiol acting via ERα, but not ERβ, was required for the development of this CD11bint Ly6C− DC population from highly purified MPs (Lin− c-kithi sca-1− IL-7Rα− flt3+),17 but the molecular mechanism of ERα action was unknown.
We now report that E2/ERα signaling increases Irf4 mRNA and protein expression in differentiating MPs and CD11c− precursors, and promotes the development of the CD11cint Ly6C− DC subset expressing the highest levels of IRF4. Enforced expression of Irf4 cDNA in differentiating ERα−/− BM cells restored the development of the E2/ERα-dependent CD11bint Ly6C− DC population, indicating that sufficiently high levels of IRF4 alleviate the requirement for ERα signaling during GM-CSF–mediated DC differentiation. Thus, an elevated amount of cellular IRF4 expression, which can be induced by E2/ERα signaling, is required for CD11b+ and Langerin+ DC differentiation mediated by GM-CSF.
Methods
Mice
Female C57BL/6 mice (CD45.2+) and congenic CD45.1+ C57BL/6 mice (8 to 12 weeks old) were purchased from the NCI Animal Production Program. ERα-deficient (B6.129-Esr1tm1KskN10) mice from JAX were bred at Oklahoma Medical Research Foundation. The Oklahoma Medical Research Foundation Institutional Animal Care and Use Committee approved the studies.
GM-CSF–mediated DC differentiation
The GM-CSF–driven culture model for DC differentiation, in which E2 levels or ER signaling are manipulated, was used as described.17 Cultures were initiated from total BM cells, Lin− BM cells, or purified MPs. Standard (hormone-replete) culture medium was RPMI, 10% fetal bovine serum (FBS). Steroid hormone-deficient culture medium was phenol red free RPMI, 10% charcoal dextran stripped FBS (Omega). E2 (Sigma-Aldrich) was dissolved in EtOH (vehicle), or was in water-soluble form as cyclodextrin-encapsulated E2 with cyclodextrin as vehicle, and added to a final concentration of 0.1nM to 1nM. ICI 182,780 was dissolved in DMSO (vehicle) and added to a final concentration of 100nM.
Flow cytometry and myeloid progenitor isolation
Cells were prepared for flow cytometry as described.17 The goat anti-IRF4 antiserum M-17 (Santa Cruz Biotechnology) was detected with donkey anti–goat Ig-Cy3, and isotype control goat IgG, or anti-IRF4 Ab preblocked with cognate peptide, controlled for the specificity of binding.8 The anti-Langerin mAb L31 was biotinylated and detected with streptavidin-allophycocyanin.23 Samples were run on an LSRII instrument (Becton Dickinson Biosciences) and analyzed with FlowJo software Version 8.8.5 (TreeStar). Lin− cells and MPs were isolated as described.17
BM chimeric mice
Mixed ERα+/ERα−/− BM chimeric mice were generated by transferring equal numbers of ERα+CD45.1+ and ERα−/−CD45.2+ BM cells into lethally irradiated [CD45.1xCD45.2]F1 recipients as described.24 On days 21 to 24 after reconstitution, BM was isolated and placed into GM-CSF–driven DC differentiation cultures, or DC populations in BM were analyzed directly ex vivo by flow cytometry.
Microarrays
MPs were incubated with 1nM E2 or vehicle for variable time periods between 0 and 72 hours. Biotinylated amplified RNA was produced from 175 ng of total RNA per sample using an Illumina TotalPrep RNA Amplification Kit (Ambion). Amplified RNA was hybridized overnight at 58°C to Sentrix Mouse-6 Expression BeadChips Version 1.1 (Illumina). These arrays contain 50-mer oligonucleotides coupled to beads that are mounted on glass slides. Each bead has an approximately 20- to 30-fold redundancy. Microarrays were washed under high stringency, labeled with streptavidin-Cy3, and scanned using an Illumina BeadStation 500 scanner. To gain statistical power, gene expression response over time was parameterized through a “mixed model”25 according to the following equation: Normalized Fluorescent Units = α*Time + β1*Trx + β2*Trx*Time, where α, β1, β2 are beta coefficients, Trx is Estrogen and No Estrogen, and Time is in hours. The Trx*Time interaction P values were adjusted with a False Discovery Rate (FDR).26 An FDR of 10% was considered statistically significant. A first order autoregressive covariance matrix was used to account for the correlation structure inherent in the longitudinal measurements of gene expression. All mixed model analysis was performed in the SAS system Version 9.1.3.
qPCR
RNA and cDNA were generated and quantitative real-time reverse transcriptase–polymerase chain reaction (qPCR) was done as described.17 Relative expression of genes was determined using the ΔΔCt method with normalization to Gapdh expression, which does not change with E2 stimulation. Specific primer sequences were: sense Gapdh 5′ AGGTCGGTGTGAACGGATTTG 3′, antisense Gapdh 5′ TGTAGACCATGTAGTTGAGGTCA 3′; sense Irf4 5′ GCCCAACAAGCTAGAAAG 3′, antisense Irf4 5′ TCTCTGAGGGTCTGGAAACT 3′; sense Irf8 5′ GTTTACCGAATTGTCCCCGAG 3′, antisense Irf8 5′ CTCCTCTTGGTCATACCCATGTA 3′; Sense Pu.1 5′ ATGTTACAGGCGTGCAAAATGG 3′, antisense Pu.1 5′ TGATCGCTATGGCTTTCTCCA 3′. Id2 primers (sequences proprietary) were obtained from QIAGEN.
Retrovirus-mediated gene delivery
The Irf4 cDNA was cloned from murine DCs and inserted in front of the Egfp gene in the retroviral vector MiG.27 To generate virus, MiG was transfected with pCL/Eco plasmid into 293T cells. Lin− BM cells from ERα−/− mice were isolated and cultured for 16 hours in stem cell factor (50 ng/mL), IL-3 (20 ng/mL), and IL-6 (50 ng/mL). For viral transduction, 1.5 × 105 Lin− BM cells were mixed with 2 mL of retroviral supernatant plus 10 μg/mL polybrene in 12-well plates and centrifuged for 1 hour. Transduced Lin− cells were cultured in GM-CSF in E2-deficient medium for 5 days.
Statistical analyses
Statistical significance (P < .05) of differences in IRF4 mean fluorescence intensity (MFI) values was determined using unpaired t tests in Prism software. Significant differences between ERα+ and ERα−/− DCs in mixed BM chimeras were determined using paired t tests.
Results
An early and brief exposure to estradiol significantly increases DC differentiation
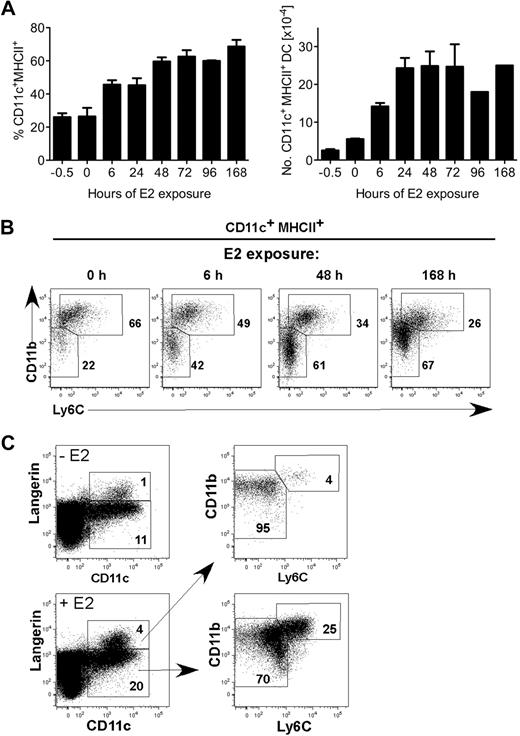
Two major subsets of CD11c+ MHCII+ DCs (CD11bhi Ly6C+ and CD11bint Ly6C− Langerin+ or −) differentiate from BM precursors in GM-CSF–driven cultures.7 We previously showed that ERα and physiologic amounts of E2 (0.1nM-1nM) were required for the GM-CSF–mediated differentiation of the CD11c+ CD11bint Ly6C− subset from MPs.17 To determine the temporal window of E2 exposure needed to augment DC differentiation, we incubated MPs (Lin− c-kithi sca-1− IL-7Rα− flt3+) in standard (hormone-replete) medium supplemented with E2 (0.1nM) for various periods of time (6 hours to 96 hours) before blockade of ER signaling by addition of ICI 182,780, an ER antagonist that induces ER degradation.28 Cells were cultured for 7 days, and the percentage and numbers of CD11c+ MHCII+ DCs were quantified by flow cytometry (Figure 1A). A 6-hour E2 exposure was sufficient to augment CD11c+ DC development approximately 2-fold with a marked increase in the CD11bint Ly6C− population (Figure 1B). Within 48 hours of E2 exposure, the percentages of total differentiated CD11c+ MHCII+ DCs and the CD11bint Ly6C− subset were comparable with that of a 7-day (168-hour) E2 exposure. Within the CD11bint Ly6C− population, E2 exposure also increases the percentage of Langerin+ DCs (Figure 1C). Thus, within 6 hours, E2/ERα signaling in MPs biases the developmental program initiated by GM-CSF toward development of the CD11c+ CD11bint Ly6C− DC subset.
A 6-hour E2 exposure to myeloid progenitors yields a significant increase in DCs after 7 days. (A) MPs were stimulated with GM-CSF and 0.1nM E2 in standard medium at time 0. At the indicated times (0-96 hours), ER signaling was blocked by ICI 182,780 (100nM), such that E2 responsiveness was limited to the hour shown on the x axis. All cells were analyzed by flow cytometry on day 7. Shown are the average and range of percentages (left panel) and numbers (right panel) of CD11c+ MHCII+ DCs in duplicate cultures. (B) Shown are the relative percentages of 2 DC subsets (CD11bhi Ly6C+ and CD11bint Ly6C−) present in cultures harvested on day 7 after E2 exposure for 0, 6, 48, or 168 hours. (C) Shown is the staining of anti-Langerin mAb L31 in DCs in the absence or presence of E2. Langerin+ DCs are within the CD11bint Ly6C− population. Data are representative of 2 independent experiments.
A 6-hour E2 exposure to myeloid progenitors yields a significant increase in DCs after 7 days. (A) MPs were stimulated with GM-CSF and 0.1nM E2 in standard medium at time 0. At the indicated times (0-96 hours), ER signaling was blocked by ICI 182,780 (100nM), such that E2 responsiveness was limited to the hour shown on the x axis. All cells were analyzed by flow cytometry on day 7. Shown are the average and range of percentages (left panel) and numbers (right panel) of CD11c+ MHCII+ DCs in duplicate cultures. (B) Shown are the relative percentages of 2 DC subsets (CD11bhi Ly6C+ and CD11bint Ly6C−) present in cultures harvested on day 7 after E2 exposure for 0, 6, 48, or 168 hours. (C) Shown is the staining of anti-Langerin mAb L31 in DCs in the absence or presence of E2. Langerin+ DCs are within the CD11bint Ly6C− population. Data are representative of 2 independent experiments.
Estradiol increases Irf4 mRNA levels in myeloid progenitors
To identify genes induced by E2/ERα signaling, we performed a microarray experiment in which MPs were incubated with vehicle or E2 (1nM) for variable times between 6 hours and 72 hours. This experiment identified Irf4 as an E2-induced gene. We focused on IRF4 because it is critical for development of CD11c+ CD11b+ splenic DCs in vivo and in GM-CSF–driven cultures.
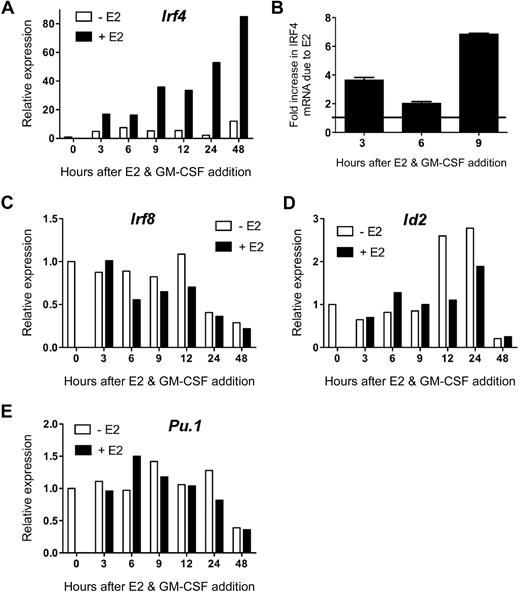
To confirm E2 induction of Irf4 mRNA, independently isolated MPs were incubated with GM-CSF and 1nM E2 (or vehicle) in hormone-deficient medium for 3 to 48 hours before analysis of Irf4 expression by qPCR. E2 induced a marked (∼ 3- to 4-fold over the vehicle control) increase in Irf4 mRNA by 3 hours, which continued to increase throughout the 48-hour time point (to 6- to 7-fold over the vehicle control; Figure 2A). In contrast, in the absence of E2, the GM-CSF–induced increase in Irf4 mRNA was significantly less and peaked at 6 hours. Due to these distinct kinetics, the difference in Irf4 mRNA levels in the absence or presence of E2 at the early time points was 2-fold at 6 hours, with greater differences at 3 hours and 9 to 48 hours (Figure 2B). These data show that upon stimulation with GM-CSF, ERα signaling induces greater and more sustained amounts of Irf4 mRNA in MPs within 3 hours.
E2 exposure markedly increases the amount of Irf4 mRNA present in GM-CSF–stimulated myeloid progenitors. MPs were stimulated with GM-CSF and 1nM E2 or vehicle in hormone-deficient medium. At the indicated time points, the amount of gene-specific mRNA was quantified using qPCR of the sample in triplicate. The relative expression of each mRNA was calculated using the ΔΔCt method. (A) Irf4 mRNA was not present at time 0 and was increased significantly in the presence of E2. (B) The fold increase in Irf4 mRNA due to the presence of E2 at early time points was reproducible. Error bars represent the range in 2 independent experiments. (C) Irf8, (D) Id2, and (E) Pu.1 mRNA were present at time 0, as determined by the Ct values, and the relative amounts did not differ in the presence or absence of E2. Data are representative of 2 to 4 independent experiments.
E2 exposure markedly increases the amount of Irf4 mRNA present in GM-CSF–stimulated myeloid progenitors. MPs were stimulated with GM-CSF and 1nM E2 or vehicle in hormone-deficient medium. At the indicated time points, the amount of gene-specific mRNA was quantified using qPCR of the sample in triplicate. The relative expression of each mRNA was calculated using the ΔΔCt method. (A) Irf4 mRNA was not present at time 0 and was increased significantly in the presence of E2. (B) The fold increase in Irf4 mRNA due to the presence of E2 at early time points was reproducible. Error bars represent the range in 2 independent experiments. (C) Irf8, (D) Id2, and (E) Pu.1 mRNA were present at time 0, as determined by the Ct values, and the relative amounts did not differ in the presence or absence of E2. Data are representative of 2 to 4 independent experiments.
We investigated whether E2 exposure altered MP expression of other transcription factors involved in DC differentiation. Unlike Irf4, unstimulated MPs contained detectable levels of Irf8, Pu.1, and Id2 mRNA. The mRNA levels of these factors did not change significantly (< 2-fold) within 24 hours of GM-CSF stimulation, nor in the presence of E2 (Figure 2C-E). Amounts of these 3 mRNA decreased at 48 hours, but this did not depend on the presence of E2. Therefore, ERα signaling rapidly and specifically increases Irf4 mRNA upon GM-CSF stimulation.
Estradiol increases IRF4 protein levels in differentiating myeloid progenitors
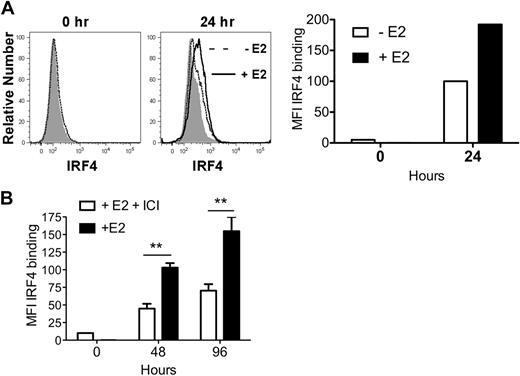
Within 24 hours, GM-CSF–stimulated MPs exposed to E2 showed a 2-fold increase in IRF4 protein, relative to vehicle-treated MPs (Figure 3A). An E2-induced increase in IRF4 protein was also detected in undifferentiated CD11c− precursors at 48 and 96 hours (Figure 3B).
E2 exposure increases the amount of IRF4 protein in differentiating MPs 2-fold by 24 hours. MPs were stimulated with GM-CSF in hormone-deficient medium plus 1nM E2 or vehicle. (A) Shown is the binding of anti-IRF4 Ab to differentiating MPs in the absence (dotted line) or presence (solid line) of E2 at 24 hours. The shaded histogram indicates the binding of anti-IRF4 Ab preincubated with blocking peptide. The mean fluorescence intensity (MFI; with value for anti-IRF4 Ab bound to blocking peptide subtracted) of anti-IRF4 binding at 0 and 24 hours is plotted. Data are representative of 2 experiments. (B) MPs were stimulated with GM-CSF in the presence of E2 or E2 plus ICI 182,780 (100nM). The amount of intracellular IRF4 protein at 48 and 96 hours was assessed using flow cytometry. Shown are the MFI values (mean ± SD) of anti-IRF4 binding (with value for anti-IRF4 Ab bound to blocking peptide subtracted) of cells in triplicate cultures. **P < .01, n = 3.
E2 exposure increases the amount of IRF4 protein in differentiating MPs 2-fold by 24 hours. MPs were stimulated with GM-CSF in hormone-deficient medium plus 1nM E2 or vehicle. (A) Shown is the binding of anti-IRF4 Ab to differentiating MPs in the absence (dotted line) or presence (solid line) of E2 at 24 hours. The shaded histogram indicates the binding of anti-IRF4 Ab preincubated with blocking peptide. The mean fluorescence intensity (MFI; with value for anti-IRF4 Ab bound to blocking peptide subtracted) of anti-IRF4 binding at 0 and 24 hours is plotted. Data are representative of 2 experiments. (B) MPs were stimulated with GM-CSF in the presence of E2 or E2 plus ICI 182,780 (100nM). The amount of intracellular IRF4 protein at 48 and 96 hours was assessed using flow cytometry. Shown are the MFI values (mean ± SD) of anti-IRF4 binding (with value for anti-IRF4 Ab bound to blocking peptide subtracted) of cells in triplicate cultures. **P < .01, n = 3.
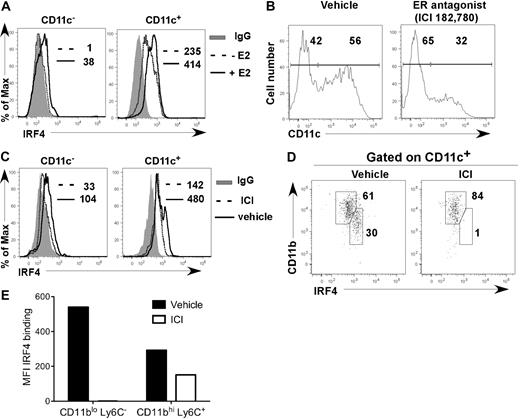
After 7 days of E2 exposure in cultures initiated from lineage-negative (Lin−) BM cells in hormone-deficient medium, CD11c− precursors and fully differentiated CD11c+ cells contained 2-fold more IRF4 protein than vehicle-treated cells (Figure 4A). ICI 182,780 inhibited the increase in IRF4 expression in CD11c− precursors that occurred in hormone-replete medium (Figure 4B-E). Within the CD11c+ cell fraction, the CD11bint Ly6C− DC subset, whose appearance is inhibited by ICI 182,780, expressed 2-fold more IRF4 than the CD11bhi Ly6C+ DC subset (Figure 4D-E). ER signaling also increased expression of IRF4 in the CD11bhi Ly6C+ DC subset (Figure 4E). These data show that when fully differentiated, the E2/ERα-dependent CD11bint Ly6C− DC subset normally expresses 2-fold higher levels of IRF4 than the CD11bhi Ly6C+ DC subset, suggesting that higher levels of IRF4 protein are required for their development and maintenance.
E2/ERα signaling promotes development of DCs that harbor increased amounts of IRF4 protein. (A) Lin− BM cells were incubated with 1nM E2 or vehicle in hormone-deficient medium. On day 7, IRF4 protein expression in CD11c− and CD11c+ cells was determined. Shown are: binding of anti-IRF4 Ab to cells from vehicle-treated cultures (dotted line); binding of anti-IRF4 Ab to cells from E2-treated cultures (solid line); binding of isotype control IgG on cells from the E2-treated cultures (shaded gray). MFI values for anti-IRF4 binding with or without E2 are indicated (with value for isotype control IgG subtracted). Data are representative of 3 experiments. (B-E) MPs were incubated in standard medium with ICI 182,780 (100nM) or vehicle. On day 7, the amount of IRF4 protein expression in CD11c− and CD11c+ cells was determined. (B) CD11c expression on all cells; the percentages of CD11c− and CD11c+ cells are indicated. (C) Anti-IRF4 binding on CD11c− and CD11c+ cells in the presence of ICI 182,780 (dotted line) or vehicle (solid line). The binding of isotype control IgG on cells in vehicle-treated cultures is indicated (shaded gray). MFI values for anti-IRF4 binding with or without ICI 182,780 are indicated (with value for isotype control IgG subtracted). (D) Within CD11c+ cells, CD11bint DCs express higher levels of IRF4 than CD11bhi DCs. The percentage of DCs within each gate is indicated. (E) MFI values for anti-IRF4 binding with or without ICI 182,780 are indicated for each DC subset. Data are representative of 2 experiments.
E2/ERα signaling promotes development of DCs that harbor increased amounts of IRF4 protein. (A) Lin− BM cells were incubated with 1nM E2 or vehicle in hormone-deficient medium. On day 7, IRF4 protein expression in CD11c− and CD11c+ cells was determined. Shown are: binding of anti-IRF4 Ab to cells from vehicle-treated cultures (dotted line); binding of anti-IRF4 Ab to cells from E2-treated cultures (solid line); binding of isotype control IgG on cells from the E2-treated cultures (shaded gray). MFI values for anti-IRF4 binding with or without E2 are indicated (with value for isotype control IgG subtracted). Data are representative of 3 experiments. (B-E) MPs were incubated in standard medium with ICI 182,780 (100nM) or vehicle. On day 7, the amount of IRF4 protein expression in CD11c− and CD11c+ cells was determined. (B) CD11c expression on all cells; the percentages of CD11c− and CD11c+ cells are indicated. (C) Anti-IRF4 binding on CD11c− and CD11c+ cells in the presence of ICI 182,780 (dotted line) or vehicle (solid line). The binding of isotype control IgG on cells in vehicle-treated cultures is indicated (shaded gray). MFI values for anti-IRF4 binding with or without ICI 182,780 are indicated (with value for isotype control IgG subtracted). (D) Within CD11c+ cells, CD11bint DCs express higher levels of IRF4 than CD11bhi DCs. The percentage of DCs within each gate is indicated. (E) MFI values for anti-IRF4 binding with or without ICI 182,780 are indicated for each DC subset. Data are representative of 2 experiments.
ERα signaling in hematopoietic cells increases IRF4 expression in a cell-autonomous manner
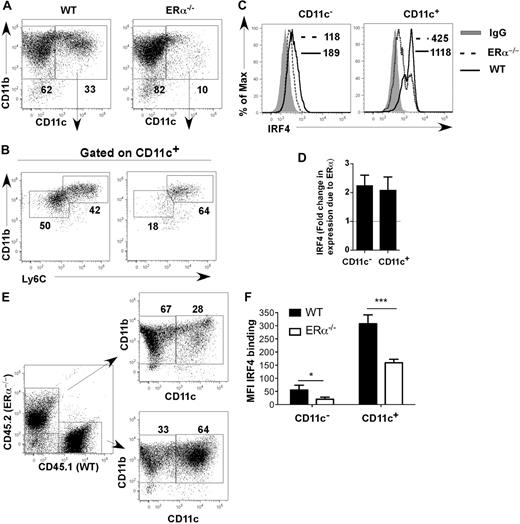
Comparison of differentiating ERα+ and ERα−/− BM cells showed that on day 7, ERα+ DCs (composed of CD11bint Ly6C− and CD11bhi Ly6C+ DCs) expressed 2-fold more IRF4 protein than ERα−/− DCs (composed primarily of CD11bhi Ly6C+ DCs; Figure 5A-D). One possibility was that DC precursors in ERα−/− mice are defective due to the absence of factors supplied by estrogen-responsive BM stromal cells in vivo.29 To determine whether the effect of ERα signaling on IRF4 expression was intrinsic to hematopoietic DC precursors, we generated ERα+/ERα−/− mixed BM chimeric mice, and BM cells from these chimeric mice were placed into GM-CSF–driven cultures. After 7 days, ERα+ cells expressed higher amounts of IRF4 and had developed into both CD11bhi and CD11bint DC subsets, whereas ERα−/− cells expressed lower amounts of IRF4 and had developed primarily into CD11bhi DCs (Figure 5E,F). These data show that ERα signaling acts in a cell-autonomous manner in DC precursors to increase expression of IRF4. Thus, E2/ERα signaling in MPs or CD11c− DC precursors leads to a reproducible 2-fold increase in IRF4 protein by 24 hours, which is maintained throughout the 7-day culture period and in fully differentiated CD11c+ CD11bint Ly6C− cells.
E2/ERα signaling acts in a cell-autonomous manner to increase expression of IRF4. (A-D) WT or ERα−/− BM cells were incubated with GM-CSF in standard medium for 7 days. (A) The percentage of CD11c+ CD11b+ DCs was determined. (B) WT CD11c+ cells harbor both CD11bhi Ly6C+ and CD11bint Ly6C− subsets, whereas ERα−/− CD11c+ cells primarily harbor the CD11bhi Ly6C+ subset. (C) Binding of anti-IRF4 Ab to WT (solid line) or ERα−/− (dotted line) cells. The binding of isotype control IgG on WT cells is indicated (shaded gray). MFI values for anti-IRF4 binding to cells are indicated (with value for isotype control IgG subtracted). (D) The average fold change in IRF4 expression (anti-IRF4 Ab MFI) due to ERα in CD11c− and CD11c+ is plotted; error bars represent the standard error in 5 independent experiments. (E-F) BM from ERα+/ERα−/− BM chimeric mice was incubated with GM-CSF in standard medium for 7 days. (E) The percentage of CD11c+ DCs in the WT/CD45.1 and ERα−/−/CD45.2 cell fractions was determined. (F) The bar graph shows MFI values of anti-IRF4 Ab binding to CD11c− and CD11c+ cells, averaged (mean ± SEM) from 4 BM chimeric mice generated from the same BM transfer. Identical results were obtained upon analyses of 4 mice from a second BM chimera experiment. ***P < .001; *P < .05, n = 4.
E2/ERα signaling acts in a cell-autonomous manner to increase expression of IRF4. (A-D) WT or ERα−/− BM cells were incubated with GM-CSF in standard medium for 7 days. (A) The percentage of CD11c+ CD11b+ DCs was determined. (B) WT CD11c+ cells harbor both CD11bhi Ly6C+ and CD11bint Ly6C− subsets, whereas ERα−/− CD11c+ cells primarily harbor the CD11bhi Ly6C+ subset. (C) Binding of anti-IRF4 Ab to WT (solid line) or ERα−/− (dotted line) cells. The binding of isotype control IgG on WT cells is indicated (shaded gray). MFI values for anti-IRF4 binding to cells are indicated (with value for isotype control IgG subtracted). (D) The average fold change in IRF4 expression (anti-IRF4 Ab MFI) due to ERα in CD11c− and CD11c+ is plotted; error bars represent the standard error in 5 independent experiments. (E-F) BM from ERα+/ERα−/− BM chimeric mice was incubated with GM-CSF in standard medium for 7 days. (E) The percentage of CD11c+ DCs in the WT/CD45.1 and ERα−/−/CD45.2 cell fractions was determined. (F) The bar graph shows MFI values of anti-IRF4 Ab binding to CD11c− and CD11c+ cells, averaged (mean ± SEM) from 4 BM chimeric mice generated from the same BM transfer. Identical results were obtained upon analyses of 4 mice from a second BM chimera experiment. ***P < .001; *P < .05, n = 4.
Enforced expression of IRF4 rescues the development of the ERα-dependent DC population from ERα−/− BM precursors
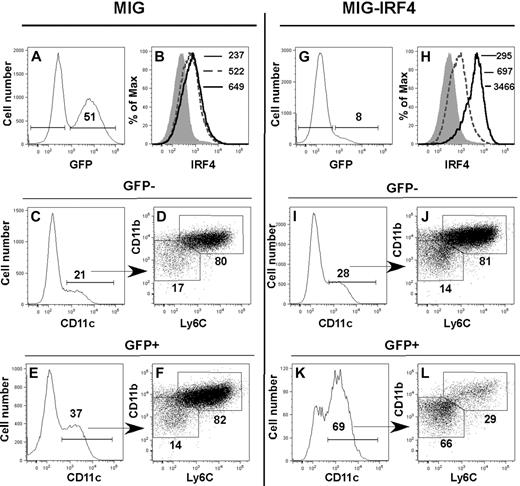
To determine whether elevated amounts of IRF4 protein were sufficient to induce differentiation of the CD11bint Ly6C− DC subset in the absence of ERα signaling, we expressed the Irf4 cDNA in Lin− ERα−/− BM cells (Figure 6). Lin− ERα−/− BM cells were transduced with MiG-GFP or MiG-IRF4-GFP retroviruses and subsequently stimulated with GM-CSF. After 5 days, the phenotypes of cells in GFP+ and GFP− fractions were determined by flow cytometry. Enforced expression of the Irf4 cDNA led to a 5-fold increase in the amount of intracellular IRF4 protein in GFP+ cells, relative to GFP− cells in MIG-IRF4-GFP–transduced cultures (Figure 6H) or to cells in the MIG-GFP–transduced cultures (Figure 6B). This was associated with a marked increase in the percentage of CD11c+ cells (Figure 6I-K). The difference in percentage of CD11c+ cells in GFP− and GFP+ MiG-GFP–transduced cultures (Figure 6C,E) was not reproducible. While the majority of DCs was CD11bhi Ly6C+ in the MiG-GFP–transduced cultures (Figure 6C-F) and in the GFP− fraction of the MiG-IRF4-GFP–transduced cultures (Figure 6I-J), the majority of DCs in the GFP+ fraction of the MiG-IRF4-GFP cultures was CD11bint Ly6C− (Figure 6K-L). The CD11bint Ly6C− population contained both Langerin+ and Langerin− DCs (not shown). Thus, increased expression of the Irf4 cDNA in differentiating ERα−/− Lin− cells restores the development of the E2/ERα-dependent CD11bint Ly6C− DC population, indicating that sufficiently high levels of IRF4 substitute for the requirement for E2/ERα signaling during GM-CSF–mediated DC differentiation.
Enforced IRF4 expression directs the development of the ERα/E2-dependent CD11bint Ly6C− DC subset in the absence of ERα signaling. Lin− ERα−/− BM cells were transduced with MiG-GFP (A-F) or MiG-IRF4-GFP (G-L). Transduced cells were incubated in GM-CSF for 5 days before assessment of DC subsets by flow cytometry. IRF4 expression was assessed in GFP− (dotted line) and GFP+ (solid line) fractions in (A-B) MiG-GFP– or (G-H) MiG-IRF4-GFP– transduced cells. The MFI of anti-IRF4 binding to GFP+ and GFP− cells is indicated in panels B and H. The shaded histogram indicates the binding of anti-IRF4 Ab precincubated with blocking peptide. In MiG-GFP–transduced cell cultures, CD11c+ DCs in the GFP− (C) and GFP+ (E) cell fractions are primarily CD11bhi Ly6C+ (panels D and F gated on CD11c+ cells shown in panels C and E). The percentage of cells with the bar or box gates is indicated. In MiG-IRF4-GFP–transduced cultures, CD11c+ DCs in the GFP− cell fraction (I) are also primarily CD11bhi Ly6C+ (J, gated on CD11c+ cells shown in panel I). In contrast, the percentage of CD11c+ DCs in the GFP+ fraction (K) is significantly increased, and the majority of DCs is CD11bint Ly6C− (L, gated on CD11c+ cells shown in panel K). Data are representative of 4 independent experiments.
Enforced IRF4 expression directs the development of the ERα/E2-dependent CD11bint Ly6C− DC subset in the absence of ERα signaling. Lin− ERα−/− BM cells were transduced with MiG-GFP (A-F) or MiG-IRF4-GFP (G-L). Transduced cells were incubated in GM-CSF for 5 days before assessment of DC subsets by flow cytometry. IRF4 expression was assessed in GFP− (dotted line) and GFP+ (solid line) fractions in (A-B) MiG-GFP– or (G-H) MiG-IRF4-GFP– transduced cells. The MFI of anti-IRF4 binding to GFP+ and GFP− cells is indicated in panels B and H. The shaded histogram indicates the binding of anti-IRF4 Ab precincubated with blocking peptide. In MiG-GFP–transduced cell cultures, CD11c+ DCs in the GFP− (C) and GFP+ (E) cell fractions are primarily CD11bhi Ly6C+ (panels D and F gated on CD11c+ cells shown in panels C and E). The percentage of cells with the bar or box gates is indicated. In MiG-IRF4-GFP–transduced cultures, CD11c+ DCs in the GFP− cell fraction (I) are also primarily CD11bhi Ly6C+ (J, gated on CD11c+ cells shown in panel I). In contrast, the percentage of CD11c+ DCs in the GFP+ fraction (K) is significantly increased, and the majority of DCs is CD11bint Ly6C− (L, gated on CD11c+ cells shown in panel K). Data are representative of 4 independent experiments.
ERα expression promotes CD11b+ DC development during inflammation in vivo
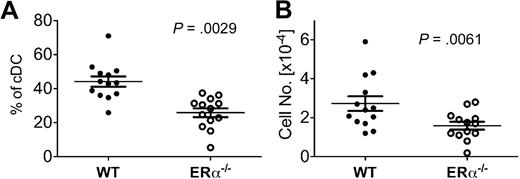
Our results in the GM-CSF–driven culture model suggest that ERα signaling promotes new DC differentiation during inflammation in vivo. To determine the relative efficiency of DC differentiation from ERα+ and ERα−/− BM precursors in vivo, we analyzed the CD11c+ CD11b+ DC subset present in BM of mixed ERα+/ERα−/− BM chimeric mice at an early time point (∼ day 23) after reconstitution when myeloid cells, but few lymphocytes, have developed. In this model, DC development occurs in the postradiation inflammatory environment, which is characterized by elevated serum IL-6 and high levels of costimulatory molecules on newly differentiated DCs, and normal levels of serum E2 (E.C., S. Bajaña, H. Agrawal, S.K., manuscript in preparation). In the BM of these chimeric mice, newly differentiated CD11b+ DCs were derived preferentially from ERα+ donor BM cells (∼ 2:1 ratio of wild type [WT] to ERα−/− DCs; Figure 7). Similar results were obtained upon analyses of splenic and lymph node DC populations in these chimeric mice (E.C., S. Bajaña, H. Agrawal, S.K., manuscript in preparation). Thus, during radiation-induced inflammation in vivo, ERα signaling in response to physiologic levels of endogenous estrogens promotes CD11b+ DC development from short-term repopulating BM progenitors.
ERα expression promotes DC development during inflammation in vivo. Using flow cytometry, conventional CD11c+ CD11b+ DCs (cDCs) were identified in BM of ERα+/ERα−/− BM chimeric mice on days 21 to 24 after reconstitution, a postradiation inflammatory environment. After gating on this DC population, the percentage of cells bearing CD45.1 (WT ERα+) or CD45.2 (ERα−/−) was determined. Shown are the (A) relative percentage and (B) number of cDCs derived from WT and ERα−/− donor BM in individual mice (n = 13), with the mean and SEM values indicated by the bars. Significant differences in the percentage and numbers of WT and ERα−/− DCs were calculated using paired t tests. The total number of BM cells in individual mice varied between 4 × 106 and 1 × 107.
ERα expression promotes DC development during inflammation in vivo. Using flow cytometry, conventional CD11c+ CD11b+ DCs (cDCs) were identified in BM of ERα+/ERα−/− BM chimeric mice on days 21 to 24 after reconstitution, a postradiation inflammatory environment. After gating on this DC population, the percentage of cells bearing CD45.1 (WT ERα+) or CD45.2 (ERα−/−) was determined. Shown are the (A) relative percentage and (B) number of cDCs derived from WT and ERα−/− donor BM in individual mice (n = 13), with the mean and SEM values indicated by the bars. Significant differences in the percentage and numbers of WT and ERα−/− DCs were calculated using paired t tests. The total number of BM cells in individual mice varied between 4 × 106 and 1 × 107.
Discussion
We have shown that an elevated amount of IRF4 protein, induced by ERα signaling, promotes the GM-CSF–mediated differentiation of CD11bint Ly6C− DCs from MPs. There is a precedent for high and low amounts of IRF4 protein directing distinct transcriptional programs in differentiating cells. Differentiating B cells expressed graded amounts of IRF4 protein, and high or low amounts of IRF4 specified plasma cell or germinal center gene expression programs, respectively.11 PU.1 pairs with IRF4 to regulate genes and, like IRF4, is required for the development of splenic CD11b+ DCs.3,12 Quantitatively different amounts (∼ 4-fold) of PU.1 in hematopoietic progenitors also specify distinct myeloid and lymphoid cell fates, in some cases because the ratio of PU.1 to other transcription factors is altered.14,30 This supports the significance of a 4- to 7-fold difference in IRF4 levels in MPs within 3 to 9 hours of E2 exposure, and suggests that the ERα/E2-induced changes in IRF4 expression could lead to altered interactions with other transcription factors and/or to distinct transcriptional programs.
IRF4 acts alone by binding ISRE or is recruited to composite IRF4/PU.1 binding sites by phosphorylated PU.1.12 Our data show that in MPs, Pu.1 mRNA changed little in response to GM-CSF or E2 stimulation over the 24-hour time period in which Irf4 mRNA is significantly increased. Therefore, it is possible that increased amounts of IRF4 protein alter the recruitment of IRF4/PU.1 complexes to target genes, as demonstrated for IRF8/PU.1 complexes.31 Autoregulation of the Irf4 gene also could be amplified by ERα signaling because IRF4/PU.1 complexes bind to the Irf4 promoter.10 Alternately, elevated levels of IRF4 might allow IRF4 to act independently of PU.1 to regulate genes that promote CD11b+ DC differentiation.
ERα binds directly to DNA at consensus estrogen response elements (EREs) proximal to genes.32 Algorithms within the ECR Browser (www.dcode.org) at the National Center for Biotechnology Information (NCBI) identified 4 conserved EREs proximal to the murine Irf4 gene. ERα also may complex with other factors such as Sp-1, NF-Y, FoxA1/HNF3α, or AP-1,18 and binding sites for these factors are also present proximal to the Irf4 gene. Unbiased genome-wide chromatin binding (ChIP) and sequencing approaches to identify ERα binding sites in E2-stimulated cells revealed that many ERα binding sites and EREs are more than 5 kb away from target genes.33 Intriguingly, ERα bound to a region that is approximately 12 kb proximal to the start site of the human IRF4 gene.34 This region contains a consensus ERE site, which is conserved in the same location 5′ to the murine Irf4 gene. This information is consistent with the hypothesis that E2-bound ERα participates in a chromatin-modifying complex that directly regulates the Irf4 gene.
Alternately, ERα signaling may modulate expression or activity of an intermediate molecule such as nuclear factor–κB (NF-κB). Indeed, GM-CSF stimulated IRF4 expression in differentiating human monocytes by activating NF-κB,10 and the NF-κB subunit Rel induced IRF4 expression in lymphocytes.35 RelB is also important for the development of IRF4-dependent CD11b+ splenic DCs.36,37 ER interactions with NF-κB in other cell types have been reported.38
In the GM-CSF–driven cultures, E2/ERα signaling promotes the development of CD11bint DCs, some of which are Langerin+. These could be the correlates of either epidermal Langerin+ DCs or populations of CD11b+ and CD103+ interstitial DCs found in the dermis and lung. CD103 is expressed on a significant portion of both the CD11bhi Ly6C+ and CD11bint Ly6C− DC subsets in our cultures (data not shown), so we could not use differential CD103 expression to correlate them with interstitial DC populations. A role for IRF4 in the development of these tissue DC populations was not studied in published reports of DC development in IRF4−/− mice.8,9 Our data now link IRF4 to the development of one or more of these tissue DC subsets in vivo.
The higher amount of IRF4 protein in E2-dependent CD11bint Ly6C− DCs may have functional implications. Fully developed human DCs, CD11b+ murine DCs, and macrophages retain expression of IRF4, and IRF4 negatively regulates Toll-like receptor signaling.39,40 Thus, IRF4hi DCs that develop in the context of E2/ERα signaling may have a greater capacity to regulate the magnitude of immune responses or promote tolerance.
During inflammation and autoimmune disease, elevated GM-CSF directs the development of new DCs from monocytes or other proliferating precursors that infiltrate tissues and secondary lymphoid organs.5 Increased estradiol synthesis also has been linked to inflammation.41,42 Our data show that one physiologic factor that regulates de novo inflammatory DC differentiation is E2/ERα signaling, which leads to increased expression of the IRF4 transcription factor that is critical to CD11b+ DC development.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank J. Bass and D. Hamilton in the Oklahoma Medical Research Foundation flow cytometry core facility, Drs H. Agrawal and S. Bajaña for assistance with BM chimeras, and our Oklahoma Medical Research Foundation colleagues for helpful discussions.
This work was supported by the Oklahoma Center for the Advancement of Science & Technology grants HR06-157 (S.K.) and AR081-006 (M.C.); and National Institutes of Health grants AI079616 (S.K.), RR020143 (E.C., M.C., S.K.), RR015577 (S.K., M.C.), and RR16478 (M.B.F.).
National Institutes of Health
Authorship
Contribution: E.C. designed and performed research, analyzed and interpreted data, and wrote the manuscript; S.T. performed research and analyzed and interpreted data; M.B.F. designed research and analyzed and interpreted data; N.K. performed statistical analysis; J.O. and A.S. performed research; M.C. analyzed and interpreted data; C.G.P. contributed vital new reagent; J.A.-I. designed research and analyzed and interpreted data; and S.K. designed research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Susan Kovats, Arthritis & Immunology Program, Oklahoma Medical Research Foundation, 825 NE 13th St, Oklahoma City, OK 73104; e-mail: susan-kovats@omrf.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal